Abstract
A screening study of the presence of metallo-β-lactamases (IMP and VIM types and SPM-1) in isolates from different nonhospital sources was conducted, and it revealed the presence of blaVIM-2, associated with the In58 class 1 integron, in two unrelated Pseudomonas aeruginosa strains from aquatic habitats. The results suggest that the hospital setting was the possible origin of these blaVIM-2-carrying strains.
Metallo-β-lactamases (MBLs) of the IMP and VIM types are emerging worldwide as a source of acquired carbapenem resistance in gram-negative bacteria, and they are compromising the efficacy of several β-lactam antibiotics. Recently, the new MBLs SPM-1 and GIM-1 have also emerged, which are restricted to Brazil and Germany, respectively (17). During the last few years, several MBL-producing gram-negative bacteria have been reported in Portugal (2-4, 12). These bacteria were classified either as organisms that produce IMP-5, which has been observed only in Acinetobacter baumannii (4) and Pseudomonas aeruginosa (A. Duarte, A. Brizio, F. Grosso, T. Conceição, and G. Da Silva, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-2024, 2003), or as organisms that produce VIM-2, the most commonly observed MBL, which is present in several isolates of P. aeruginosa (2, 12; L. Peixe, unpublished observation) and has recently been found in Klebsiella oxytoca (3). Interestingly, these isolates were found almost exclusively in a hospital setting. Recently, a VIM-2-producing Pseudomonas pseudoalcaligenes strain was isolated outside hospital boundaries in a hospital sewage sample (11), which indicated that MBLs could spread into new, previously unreported niches. The notion of the apparent confinement of these clinically relevant enzymes to bacteria present in a hospital setting may be the result of a strongly biased sampling effort. The role of nonclinical niches as a reservoir for bacteria that carry acquired MBL genes is still poorly established, and clearly there is a need for further evaluation.
Acquired MBL genes usually are present as gene cassettes that are inserted into integrons found in the chromosome or in plasmids (9). Diverse blaVIM-2-carrying integron structures are commonly observed in different countries. In Portugal, although several distinct integrons have been associated with the blaVIM-2 gene, In58 seems to be the most widespread integron (Peixe, unpublished observation). The recently discovered association of integrons carrying these genes with transposon-like structures (15) may account for their variable locations and spread.
In this study, the presence of acquired MBLs (IMP and VIM types and SPM-1) in gram-negative bacilli obtained from nonhospital sources in northern Portugal between 2001 and 2005 was evaluated in order to obtain epidemiological data on the dissemination of these enzymes.
Twenty-four samples from healthy human volunteer feces, 83 samples from ambulatory patients, 30 samples from poultry skin (from the main brands sold in Portugal), 4 samples from swine feces, 13 samples from rivers, and 29 samples from urban sewage that received hospital wastewater (14 samples from upstream of the hospital wastewater discharge site and 15 samples from downstream of the hospital wastewater discharge site) were collected and used to screen for MBL production. Sample aliquots were plated on MacConkey agar and/or Pyocyanosel agar (Oxoid, Basingstoke, United Kingdom) supplemented with imipenem (2 μg/ml; Merck, Sharpe & Dohme, Portugal), and colonies having different morphologies were collected. Species identification was performed with API 32GN (bioMérieux, Marcy l'Étoile, France). Imipenem susceptibility was evaluated by the disk diffusion method (7). The MICs of β-lactam antibiotics for VIM-2 producers were determined by the Etest method (AB Biodisk, Solna, Sweden), and the susceptibilities to aminoglycosides and ciprofloxacin were determined by the disk diffusion method for some strains (7).
Isolates that were grown on MacConkey agar and/or Pyocyanosel agar supplemented with imipenem were evaluated for MBL production by a disk diffusion test (18). An increase (>2 mm) in the inhibition zone around an imipenem disk with EDTA was used as a preliminary criterion for detection of metallo-β-lactamases. Dot blot hybridization was also performed with all of the isolates in order to detect the presence of MBL genes. Chromosomal DNA was transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech, Orsay, France) and hybridized to blaVIM-, blaIMP-, and blaSPM-1-specific probes generated by PCR. Labeling and detection were performed with the ECL Random Prime labeling and detection system (Amersham Life Science) by following the manufacturer's instructions. A multiplex PCR assay with blaVIM- and blaIMP-specific primers was also used (2, 13). The MBL genetic context was characterized by PCR using class 1 integron-specific primers (5) and sequencing. Clonal relatedness among isolates was assessed by pulsed-field gel electrophoresis (PFGE) using SpeI (New England Biolabs, Izasa, Portugal). Additional comparisons were performed by using a Portuguese VIM-2-producing P. aeruginosa hospital strain collection. Total DNA digestion with the I-CeuI enzyme (New England Biolabs, Izasa, Portugal), separation by PFGE, and hybridization with blaVIM-2 and rRNA probes, as described previously (12), were performed in order to assess the genetic location of blaVIM-2.
A total of 273 gram-negative bacilli (21 lactose fermenters and 252 strains that did not ferment lactose) were isolated from the different sources used in the multidimensional screening for MBLs. All poultry isolates (n = 56) showed imipenem susceptibility. Decreased susceptibility to imipenem (isolates exhibiting resistance and intermediate susceptibility) was observed for 74 isolates from sewage (8 isolates from urban sewage and 17 isolates from sewage receiving hospital wastewater), rivers (17 isolates), swine feces (12 isolates), ambulatory patients (10 isolates), and healthy volunteer feces (10 isolates). Fifty-six of these isolates were putative MBL producers as determined by the disk diffusion test (24 isolates from sewage, 17 isolates from rivers, 7 isolates from swine feces, 7 isolates from ambulatory patients, and 1 isolate from a healthy human). The blaIMP and blaSPM-1 genes were not detected in any of these isolates. However, two P. aeruginosa isolates (strain R2, obtained from a river, and strain E58, obtained from sewage downstream from the Hospital Porto 1 discharge site) showed a hybridization signal with the blaVIM probe. It is important to stress that these two isolates were collected from geographically distant locations and that there was no physical link between the two aquatic ecosystems. The R2 and E58 strains exhibited resistance to imipenem (>32 μg/ml and 16 μg/ml, respectively) and cefepime (>32 μg/ml), intermediate susceptibility to ceftazidime (16 μg/ml), and variable susceptibility to meropenem (8 μg/ml and 1 μg/ml, respectively), but they remained susceptible to piperacillin (16 μg/ml), piperacillin-tazobactam (16 μg/ml), and aztreonam (4 μg/ml). The R2 isolate also exhibited resistance to ciprofloxacin. The two isolates were also resistant to tobramycin, gentamicin, amikacin, and netilmicin, and they had the corresponding aminoglycoside-modifying enzyme genes located in an integron which contained the blaVIM-2 gene. Sequencing resulted in identification of this integron as In58. This class 1 integron contains four gene cassettes: aacA7 in the first position, followed by the blaVIM-2 gene cassette, an aacC1 gene cassette, and an aacA4 gene cassette (10). The I-CeuI technique results revealed a chromosomal location for the blaVIM-2 gene (data not shown). PFGE of the two environmental P. aeruginosa VIM-2 producers resulted in distinct genomic fingerprints (Fig. 1). Nevertheless, there was a difference in five bands between the R2 strain and a VIM-2-producing P. aeruginosa clinical isolate (IPO-3) from a hospital (Hospital Porto 2) (Fig. 1), suggesting possible genetic relatedness (14), even though there was no physical connection between the river and hospital from which the strains were isolated.
FIG. 1.
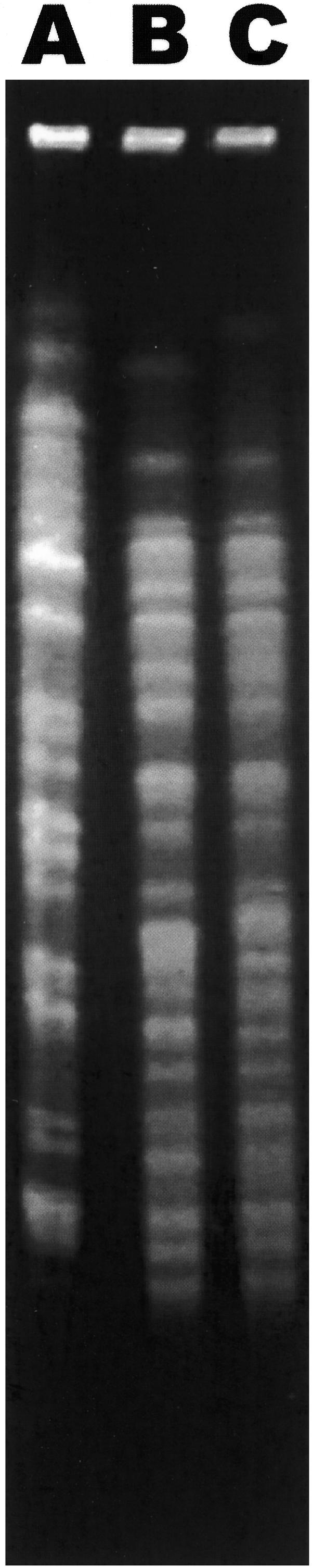
PFGE patterns of SpeI-digested genomic DNA of blaVIM-2-positive P. aeruginosa isolates. Lane A, isolate E58; lane B, hospital isolate IPO-3; lane C, isolate R2.
It seems clear that MBL resistance genes have already crossed hospital boundaries, reinforcing the possible role of the aquatic environment as a reservoir for carbapenemase genes (1, 11). The “sewage habitat” has to be considered a gathering place for many bacterial species with distinct resistance profiles (6), and it has great potential for bacterial dissemination downstream into rivers (8) and coastal waters. Since sewage treatment plants are currently inefficient for selective elimination of bacteria (16) and since Pseudomonas spp. are ubiquitous and extremely adaptable, there seem to be few obstacles to the persistence and transfer of resistance genes, such as blaVIM-2.
Acknowledgments
We thank Maria Isaura Sousa and Helena Ferreira for kindly providing ambulatory strains and some of the aquatic samples, respectively, and Cristina Réu for technical assistance. We are also grateful to Nuno Monteiro for helpful discussions and a critical review of the manuscript. Sandra Quinteira is a Ph.D. student in Programa PRODEP.
REFERENCES
- 1.Aubron, C., L. Poirel, R. Ash, and P. Nordmann. 2005. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 11:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardoso, O., R. Leitão, A. Figueiredo, J. C. Sousa, A. Duarte, and L. Peixe. 2002. Metallo-β-lactamase VIM-2 in clinical isolates of Pseudomonas aeruginosa from Portugal. Microb. Drug Resist. 8:93-97. [DOI] [PubMed] [Google Scholar]
- 3.Conceição, T., A. Brízio, A. Duarte, and R. Barros. 2005. First isolation of blaVIM-2 in Klebsiella oxytoca clinical isolates from Portugal. Antimicrob. Agents Chemother. 49:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Da Silva, G., M. Correia, C. Vital, G. Ribeiro, J. C. Sousa, R. Leitão, L. Peixe, and A. Duarte. 2002. Molecular characterization of blaIMP-5, a new integron-borne metallo-β-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 215:33-39. [DOI] [PubMed] [Google Scholar]
- 5.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muniesa, M., A. García, E. Miró, B. Mirelis, G. Prats, J. Jofre, and F. Navarro. 2004. Bacteriophages and diffusion of β-lactamase genes. Emerg. Infect. Dis. 10:1134-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing, 11th informational supplement, M100-S12. NCCLS, Wayne, Pa.
- 8.Novais, C., T. M. Coque, H. Ferreira, J. C. Sousa, and L. Peixe. 2005. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl. Environ. Microbiol. 71:3364-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pallecchi, L., M. Riccio, J. Docquier, R. Fontana, and G. M. Rossolini. 2001. Molecular heterogeneity of bla(VIM-2)-containing integrons from Pseudomonas aeruginosa plasmids encoding the VIM-2 metallo-beta-lactamase. FEMS Microbiol. Lett. 195:145-150. [DOI] [PubMed] [Google Scholar]
- 10.Poirel, L., T. Lambert, S. Türkoglü, E. Ronco, J.-L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinteira, S., H. Ferreira, and L. Peixe. 2005. First isolation of blaVIM-2 in an environmental isolate of Pseudomonas pseudoalcaligenes. Antimicrob. Agents Chemother. 49:2140-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinteira, S., J. C. Sousa, and L. Peixe. 2005. Characterization of In100, a new integron carrying a metallo-β-lactamase and a carbenicillinase, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:451-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senda, K., Y. Harakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toleman, M. A., D. Biedenbach, D. Bennett, R. N. Jones, and T. R. Walsh. 2003. Genetic characterization of a novel metallo-β-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase genes in Europe: report from the SENTRY worldwide antimicrobial surveillance programme. J. Antimicrob. Chemother. 52:583-590. [DOI] [PubMed] [Google Scholar]
- 16.Vilanova, X., A. Manero, M. Cerdà-Cuéllar, and A. R. Blanch. 2004. The composition and persistence of faecal coliforms and enterococcal populations in sewage treatment plants. J. Appl. Microbiol. 96:279-288. [DOI] [PubMed] [Google Scholar]
- 17.Walsh, T., M. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yong, D., K. Lee, J. Yum, H. Shin, G. M. Rossolini, and Y. Chong. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 40:3798-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]


