Abstract
Single-nucleotide polymorphisms (SNPs) are targets to discriminate intraspecies diversity of bacteria and to correlate a genotype with a potential pathotype. Quantification of polygenotypic populations supports this task for in vitro and in vivo applications. We present a novel assay capable of quantifying mixtures of two genotypes differing by only one SNP.
Intracellular bacteria and bacterial endosymbionts are characterized by a reduced horizontal gene transfer and, thus, are often clonal. Point mutations are a major source of genetic variability, and single-nucleotide polymorphisms (SNPs), fixed in the population and accumulating with time, can define genotypes and potentially account for differing pathogenicity. The obligate intracellular bacterium Chlamydia pneumoniae shows a genetic homology of >99% within its species (5, 11, 13). Different genotypes are supposed to be related to disease manifestations and tropism, e.g., vascular C. pneumoniae are suggested to be a genetic variant of a polyclonal respiratory population with a propensity for persistence (3). To further analyze the relevance of SNPs for disease manifestation, a quantitative, SNP-specific detection within a polyclonal population can elucidate the pathogenic background. However, specificity of most amplification techniques is not sufficient to discriminate between SNPs.
“Locked” nucleic acid (LNA) is an RNA homologue with a methylene bridge between the ribose 2′-oxygen and 4′-carbon atoms. LNA increases the melting temperature of LNA-DNA heteroduplexes by up to 3°C per LNA base and, thus, enhances the discriminatory power between match and mismatch under stringent annealing conditions (1, 10). SNP genotyping with LNA chemistry has been used in hybrid probe capture assays in microwell plates (4, 9), with LNA-modified oligomers in a clamped-probe assay (12), and with modified primers and thermal melt analysis in a real-time PCR (7). To our knowledge, LNA technology has not yet been used to quantify allelic variants. In this study, using LNA-modified primers, we developed an assay of two PCRs, each of them specific for one allele of the SNP. This novel approach is a promising tool to identify C. pneumoniae genotypes and to quantify different isolates within a mixed population.
C. pneumoniae isolates AR-39 (ATCC 53592) and CWL-029 (ATCC VR-1310) were cultivated in HeLa cells and purified by centrifugation over 30% diatrizoate (Urografin; Schering, Berlin, Germany) as described previously (2). DNA was extracted by the Nucleo Spin Tissue kit (MacheryNagel, Dueren, Germany). A quantitative PCR, targeting the gene for 16S rRNA (forward primer, CAT CGT TTA CGG CAA GGA CTA; reverse primer, AGG CCT TAG GGT TGT AAA GCA), was used to quantify Chlamydia DNA, and samples of both isolates were adjusted to the same DNA concentration. The genome sequence of CWL-029 (GenBank accession number AE001363) and AR-39 (GenBank accession number AE002161) was partially aligned by ClustalW (http://www.ebi.ac.uk/clustalw/) in order to identify a SNP differentiating between the two isolates. A polymorphism at position 89897 of the CWL-029 genome (G in CWL-029 is C in AR-39) was chosen for primer design (forward primer specific for CWL-029, AAC TAT AGT CAC GGA AAG AG+G), forward primer specific for AR-39, ACT ATA GTC ACG GAA AGA C+G; reverse primer, GGC AGA AAG AAT GGT TGG AG). The polymorphic base was located at the 3′-1 position of the forward primers, and a single-LNA base was incorporated at this position (+) (TIB MOLBIOL, Berlin, Germany). The polymorphism is located in the tufA gene that codes for the elongation factor Tu promoting the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis. Quantitative PCR was run on a LightCyler (Roche Diagnostics, Mannheim, Germany) using the SYBR green kit (Roche). Cycling conditions were as follows: 10 min at 95°C, 40 cycles at 95°C for 5 s, 67°C for 5 s, and 72°C for 16 s. Each sample was run in triplicates, crossing points of replicates differed not more than ±0.1 cycles from the median.
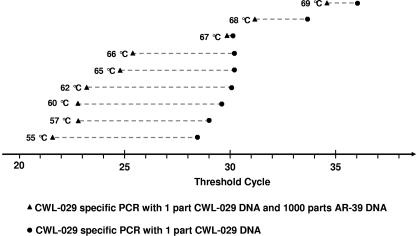
To optimize the specificity of the CWL-029-specific PCR, we quantified a sample of one part CWL-029 DNA and a second sample of a mixture of one part CWL-029 DNA plus a background of 1,000 parts AR-39 DNA under different annealing temperatures. The first sample (pure CWL-029) served as a reference of what could be expected from the second sample (mixture) under conditions of absolute specificity. Thus, differences of the threshold cycles between the two samples were a marker for unspecificity, and differences decreased with rising temperature up to an optimum at 67°C (Fig. 1). The sensitivity of the PCR decreased slightly with increasing temperature. When an equivalent PCR primer without LNA modification was tested, differences of the threshold cycles between the two samples were ≥5 under all conditions. The specificity for the AR-39-specific PCR was optimized in the same way (data not shown). All following experiments were run with an annealing temperature of 67°C.
FIG. 1.
Optimization of annealing temperature with CWL-029-specific primers. Specificity increases with decreasing differences between both PCRs. Annealing temperatures of 55°C to 66°C show cross-reactivity with AR-39 DNA as background. Optimal annealing temperature was 67°C with almost no cross-reactivity. Sensitivity and specificity decrease at higher temperatures.
The performance of each PCR was tested with 10-fold dilutions of the homologous DNA (Table 1) and calculated according the formula (e × 2)n × c0 = cn (e, effectivity [ranging from 0 to 1]; n, number of cycles; c0, copy number at the beginning; cn, copy number at n cycles). The median effectivity was 0.915. To calculate the ratio of the mixed genotypes from both quantitative PCRs, the difference in the cycle numbers at which each PCR became positive was determined. Using this value for n and the median effectivity of 0.915 for e, the ratios of both allelic variants were calculated with the formula mentioned above [cn/c0 = (e × 2)n].
TABLE 1.
Performance characteristics of the assay: ratio of AR-39 to CWL-029 calculated from the difference between the threshold cycles of the two PCRs
| AR-39/CWL-029 input ratio | Threshold cyclea
|
ΔTcb | AR-39/CWL-029 calculated ratioc | |
|---|---|---|---|---|
| AR-39 specific | CWL-029 specific | |||
| 1:1 | 30.9 | 31.0 | 0.1 | 1.1:1 |
| 1:10 | 31.0 | 26.7 | −4.3 | 1:13.4 |
| 1:100 | 30.8 | 23.0 | −7.8 | 1:115 |
| 1:1,000 | 30.7 | 19.4 | −11.3 | 1:924 |
| 1:5,000 | 29.3 | 16.9 | −12.4 | 1:1,796 |
| 1:10,000 | 28.7 | 15.8 | −12.9 | 1:2,430 |
| 1:20,000 | 28.0 | 14.9 | −13.1 | 1:2,742 |
| 1:100,000 | 27.6 | 12.6 | −15.0 | 1:8,645 |
| 10,000:0 | 15.4 | ND | 13.0d | |
| 0:10,000 | 28.4 | ND | ||
| 10:1 | 27.1 | 30.8 | 3.7 | 9.4:1 |
| 100:1 | 23.0 | 30.5 | 7.5 | 93:1 |
| 1,000:1 | 19.5 | 30.4 | 10.9 | 725:0 |
| 5,000:1 | 17.5 | 29.0 | 11.5 | 1,043:1 |
| 10,000:1 | 15.4 | 28.4 | 13.0 | 2,581:1 |
| 20,000:1 | 14.5 | 27.7 | 13.2 | 2,913:1 |
| 100,000:1 | 11.5 | 26.0 | 14.5 | 6,391:1 |
| 0:10,000 | ND | 15.8 | 12.6d | |
| 10,000:0 | ND | 28.4 | ||
ND, not done.
ΔTc, threshold cycle of the CWL-029-specific PCR minus threshold cycle of the AR-39-specific PCR.
The ratio was determined as (0.915 × 2)ΔTc; 0.915 is the median effectivity, calculated from the ΔTc of 10-fold dilutions of AR-39 and CWL-029.
ΔTc of PCR with mismatch DNA and PCR with match DNA (e.g., AR-39-specific PCR with 10,000 copies of CWL-029 minus AR-39-specific PCR with 10,000 copies of AR-39).
The PCR assay was able to quantify each genotype within mixtures of >1:1,000 with high precision, showing less than 35% divergence between predicted and calculated ratios (Table 1). Genotype mixtures of <1:1,000 could be analyzed only in a semiquantitative way. The limit of the method became obvious when a PCR assay was run in the presence of only the mismatch DNA. PCR showed a positive reaction that was delayed for 12.6 or 13.0 cycles, respectively, in comparison to the match DNA (ratios of 10,000:0 and 0:10,000) (Table 1). Thus, genotype mixtures of 1:10,000 and lower could not be differentiated from pure samples. An approach for genotyping a sample known to be pure could be achieved by analyzing only the PCR with the lowest crossing point of both allele-specific PCRs.
In bacteria with reduced lateral gene transfer, single-nucleotide polymorphisms can reveal the phylogeny of the genus and define pathotypes. In the case of C. pneumoniae, genotypes present in the vasculature are suggested to be a genetic variant of a polyclonal respiratory population with a propensity for persistence (3). To further analyze the relevance of SNP for disease manifestation, a SNP-specific detection within a polyclonal population is mandatory. In the present study, we developed a PCR assay specific for a single base and capable of quantifying mixtures of two genotypes differing by only one SNP. The performance of the assay can be described by three characteristics: (i) two genotypes present in ratios of 1:1 to 1:1,000 can be quantified with satisfactory precision; (ii) qualitative information on the presence of a minority genotype can be given in mixtures up to 1:10,000; and (iii) the assay can be used to easily genotype pure samples. A prerequisite for all applications is that the sequences of the allelic variants are known and specific primers are designed. To our knowledge, this is the first quantitative SNP-specific assay using LNA chemistry offering a broad range of applications. Simultaneous infections with different bacterial genotypes can be reliably diagnosed and quantified. Shifts within the ratios of two competing genotypes can be monitored under well-defined conditions in vitro. This allows to characterize the dominance of one genotype over the other and to investigate the possible relevance of a point mutation for pathogenesis. Alternatively, the assay allows the early detection of mutants in populations under selection pressure, e.g., under antimicrobial therapy. The quantification can give an estimate for the mutation frequency. Some of these applications can also be achieved by the clamped probe assay, in which the dominant genotype can be suppressed in a PCR without knowledge of the sequence of the mutated, minority genotype (6, 7, 8). Nevertheless, the clamped probe assay is a qualitative approach; its sensitivity was described for chloroquine-resistant Plasmodium falciparum to be 1:1,600 (12) and, thus, that qualitative assay is only equally as sensitive as our quantitative approach. In summary, LNA modification of primers allows the design of a quantitative SNP-specific PCR assay. This will facilitate the elucidation of the relevance of SNP and point mutations for bacterial pathogenesis in the future.
Acknowledgments
J.G. was supported by grants from the Deutsche Forschungsgemeinschaft (GI 344/2-1) and the University of Lübeck (N23-2003).
The excellent technical assistance of K. Rossdeutscher and S. Paetzmann is gratefully acknowledged.
REFERENCES
- 1.Braasch, D. A., and D. R. Corey. 2001. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 8:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Gieffers, J., H. Fullgraf, J. Jahn, M. Klinger, K. Dalhoff, H. A. Katus, W. Solbach, and M. Maass. 2001. Chlamydia pneumoniae infection in circulating human monocytes is refractory to antibiotic treatment. Circulation 103:351-356. [DOI] [PubMed] [Google Scholar]
- 3.Gieffers, J., L. Durling, S. P. Ouellette, J. Rupp, M. Maass, G. I. Byrne, H. D. Caldwell, and R. J. Belland. 2003. Genotypic differences in the Chlamydia pneumoniae tyrP locus related to vascular tropism and pathogenicity. J. Infect. Dis. 188:1085-1093. [DOI] [PubMed] [Google Scholar]
- 4.Jacobsen, N., M. Fenger, J. Bentzen, S. L. Rasmussen, M. H. Jakobsen, J. Fenstholt, and J. Skouv. 2002. Genotyping of the apolipoprotein B R3500Q mutation using immobilized locked nucleic acid capture probes. Clin. Chem. 48:657-660. [PubMed] [Google Scholar]
- 5.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 6.Kreuzer, K. A., P. Le Coutre, O. Landt, I. K. Na, M. Schwarz, K. Schultheis, A. Hochhaus, and B. Dorken. 2003. Preexistence and evolution of imatinib mesylate-resistant clones in chronic myelogenous leukemia detected by a PNA-based PCR clamping technique. Ann. Hematol. 82:284-289. [DOI] [PubMed] [Google Scholar]
- 7.Latorra, D., K. Campbell, A. Wolter, and J. M. Hurley. 2003. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum. Mutat. 22:79-85. [DOI] [PubMed] [Google Scholar]
- 8.Murdock, D. G., and D. C. Wallace. 2002. PNA-mediated PCR clamping. Applications and methods. Methods Mol. Biol. 208:145-164. [DOI] [PubMed] [Google Scholar]
- 9.Orum, H., M. H. Jakobsen, T. Koch, J. Vuust, and M. B. Borre. 1999. Detection of the factor V Leiden mutation by direct allele-specific hybridization of PCR amplicons to photoimmobilized locked nucleic acids. Clin. Chem. 45:1898-1905. [PubMed] [Google Scholar]
- 10.Petersen, M., and J. Wengel. 2003. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 21:74-81. [DOI] [PubMed] [Google Scholar]
- 11.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senescau, A., A. Berry, F. Benoit-Vical, O. Landt, R. Fabre, J. Lelievre, S. Cassaing, and J. F. Magnaval. 2005. Use of a locked-nucleic-acid oligomer in the clamped-probe assay for detection of a minority Pfcrt K76T mutant population of Plasmodium falciparum. J. Clin. Microbiol. 43:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of. Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]