Abstract
Aerobic heterotrophs were isolated from subsurface soil samples obtained from the U.S. Department of Energy's (DOE) Field Research Center (FRC) located at Oak Ridge, Tenn. The FRC represents a unique, extreme environment consisting of highly acidic soils with cooccurring heavy metals, radionuclides, and high nitrate concentrations. Four hundred isolates obtained from contaminated soil were assayed for heavy metal resistance, and a smaller subset was assayed for tolerance to uranium. The vast majority of the isolates were gram-positive bacteria and belonged to the high-G+C- and low-G+C-content genera Arthrobacter and Bacillus, respectively. Genomic DNA from a randomly chosen subset of 50 Pb-resistant (Pbr) isolates was amplified with PCR primers specific for PIB-type ATPases (i.e., pbrA/cadA/zntA). A total of 10 pbrA/cadA/zntA loci exhibited evidence of acquisition by horizontal gene transfer. A remarkable dissemination of the horizontally acquired PIB-type ATPases was supported by unusual DNA base compositions and phylogenetic incongruence. Numerous Pbr PIB-type ATPase-positive FRC isolates belonging to the genus Arthrobacter tolerated toxic concentrations of soluble U(VI) (UO22+) at pH 4. These unrelated, yet synergistic, physiological traits observed in Arthrobacter isolates residing in the contaminated FRC subsurface may contribute to the survival of the organisms in such an extreme environment. This study is, to the best of our knowledge, the first study to report broad horizontal transfer of PIB-type ATPases in contaminated subsurface soils and is among the first studies to report uranium tolerance of aerobic heterotrophs obtained from the acidic subsurface at the DOE FRC.
The remediation of hazardous mixed-waste sites, particularly those cocontaminated with heavy metals and radionuclides, is one of the most costly environmental challenges currently faced by the United States and other countries. Interactions between microorganisms, radionuclides, and metals that promote their precipitation and immobilization in situ are promising strategies for treatment and cleanup of the contaminated subsurface (1, 15). At mixed-waste sites where the concentrations of metal contaminants can reach toxic levels, the metal resistance of indigenous microbial populations could be critical for the success of in situ biostimulation efforts. For example, while a number of microbes can carry out reductive precipitation of radionuclides (e.g., Desulfovibrio sp., Geobacter sp., and Shewanella sp.) (28, 44, 63), the sensitivity of these organisms to heavy metals could possibly limit their in situ activities. Thus, the metal sensitivity of some radionuclide-reducing microbes suggests that the acquisition of metal resistance traits (e.g., PIB-type ATPases that regulate the transport of heavy metals) might be conducive to facilitating and/or enhancing microbial metabolism during subsequent biostimulation activities in metal- and radionuclide-contaminated subsurface environments.
The P-type ATPases represent a chromosomally encoded superfamily of ion-translocating proteins present in all three domains of life (2). The prokaryotic heavy metal-translocating PIB-type ATPases detoxify the cell cytoplasm by effluxing the divalent ions of cadmium, cobalt, lead, nickel, and zinc (3, 39, 50). The PIB-type ATPases represent one of three mechanisms for promoting microbial heavy metal resistance or tolerance, including (i) metal reduction (28), (ii) metal complexation (30), and (iii) ATP-dependent metal efflux (40). In previous studies workers have also determined the presence of PIB-type ATPase genes on mobile genetic elements (i.e., plasmids and transposons) in both gram-positive bacteria (24, 42, 43) and gram-negative bacteria (22, 33).
Analysis of completed microbial genomes has indicated that horizontal gene transfer (HGT) continues to be an important factor contributing to the innovation of microbial genomes (5, 17, 36). HGT driven by mobile genetic elements, such as plasmids (16), insertion sequences (31), integrons (37), transposons (45), and phages (9), has been shown to provide microbes with a wide variety of adaptive traits for microbial survival and proliferation (e.g., antibiotic and heavy metal resistance and diverse metabolic capabilities, including xenobiotic compound degradation and virulence). While point mutations contribute to microbial adaptation, horizontal dissemination of genes has proven to be more critical in promoting rapid genomic flexibility and microbial evolution (57). However, HGT among some subsurface microbial populations, particularly those present in the deep subsurface, has been postulated to be unlikely to occur owing to the low cell densities and the low permeability of the soil strata (61). Recently, detectable HGT of genes encoding bacterial PIB-type ATPases in bacterial isolates from a deep subsurface environment free of heavy metal contamination has been reported (11). Although HGT of genes encoding PIB-type ATPases was detected in only a few isolates, the extent of HGT may have been underestimated due to the close relatedness of the bacterial lineages studied (11).
In the present study, we examined the extent of horizontally transferred PIB-type ATPases in bacterial isolates cultured from subsurface soils with a history of radionuclide and heavy metal contamination. The isolates were obtained from soil samples from the Department of Energy (DOE) Field Research Center (FRC) located in the Oak Ridge National Laboratory Reservation (Oak Ridge, TN). The FRC subsurface is an extreme geochemical environment that places a number of stresses on the extant microbial community, including low pH (e.g., pH <4), nitrate concentrations that can exceed 100 mM, and the cooccurrence of heavy metals and radionuclides (U and other actinides) (http://www.esd.ornl.gov/nabirfrc) (47). Our main objective was to examine the role of HGT in the evolution of metal homeostasis by performing phylogenetic analyses of sequences of zntA/cadA/copA-like genes amplified from the genomes of 50 lead-resistant (Pbr) subsurface bacteria. The PIB-type ATPases were amplified from genomic DNA using a previously described set of PCR primers specific for this subfamily of ATPases (11). Analyses of 28 amplified zntA/cadA/pbrA-like loci revealed evidence of horizontal transfer among 10 Pbr Arthrobacter spp. and Bacillus spp. strains. Our results indicate that dissemination of PIB-type ATPases by HGT has occurred recently among isolates representing the Actinobacteria, Firmicutes, and Proteobacteria phyla present in metal- and radionuclide-contaminated soils of the FRC.
MATERIALS AND METHODS
Sampling site.
Contaminated subsurface soils were collected from the DOE Natural and Accelerated Bioremediation Field Research Center located in the Oak Ridge National Laboratory Reservation at Oak Ridge, Tenn. The contaminated soils are adjacent to an asphalt parking area that covers three former waste ponds (S-3 ponds) used during weapons production activities. The waste ponds and surrounding soils contain uranium (U) and other radionuclides, nitric acid, organic solvents, and heavy metals (http://www.lbl.gov/NABIR/). Soil cores (diameter, 3.75 cm; length, approximately 180 cm) were collected on 18 to 20 February 2003 as described by Petrie et al. (47). In this study, contaminated subsurface soil samples were obtained from the saturated zone, where elevated U and nitrate concentrations have been reported (http://www.lbl.gov/NABIR/). Soil core samples were obtained from boreholes FB058 and FB059 (area 1; maximum depth, 19 ft); FB051, FB053, and FB054 (area 2; maximum depth, 26 ft); and FB055 and FB057 (area 3; maximum depth, 19 ft). These samples were handled aseptically and were preserved under an argon atmosphere. Cores were shipped chilled overnight to the Georgia Institute of Technology and were processed for chemical and microbiological analyses immediately. The dry/wet ratio values ranged from 0.8 to 0.88. The soil pH, as determined by McLean (32), ranged from 4.0 (area 3) to 7.5 (area 2). Detailed geology, chemistry, and site descriptions are available on the DOE Natural and Accelerated Bioremediation website (http://www.lbl.gov/NABIR/).
Strain isolation, determination of plasmids, and metal resistance.
Aerobic chemoheterotrophs were isolated by homogenizing triplicate 3-g soil samples (e.g., samples obtained along the length of the intact subsectioned core; n = 3) in sterile saline and plating serial dilutions onto a variety of media, including full-strength (100%) PTYG, which contained (per liter) 5 g peptone, 5 g tryptone, 5 g yeast extract, 10 g glucose, 0.6 g MgSO4 · 7H2O, and 0.06 g CaCl2, 1% PTYG, 1% tryptone, which contained 10 g tryptone per liter, and R2A, which contained (per liter) 0.5 g yeast extract, 0.5 g proteose peptone, 0.5 g Casamino Acids, 0.5 g glucose, 0.5 g soluble starch, 0.3 g sodium pyruvate, 0.3 g K2HPO4, 0.05 g MgSO4 · 7H2O (Becton Dickinson), and 15 g agar (Sigma). Areas 1 and 2 yielded the highest number of isolates, and area 3, whose soils contained the highest concentrations of nitrate and uranium (http://www.esd.ornl.gov/nabirfrc/), yielded the lowest number of isolates. Due to the heterogeneous nature of the soil samples and low numbers of CFU, triplicate samples were used to obtain as many culturable isolates as possible. Plates were incubated for 1 to 5 days at 25°C and 30°C. Strains were purified by repeated streaking onto the same agar that was used for initial isolation. The Gram reaction of each of the 400 isolates was determined as described by Powers (48). PCR amplification of 16S rRNA genes from each isolate was performed as previously described (35). Isolates were grouped on the basis of the restriction fragment length polymorphism band patterns of their 16S rRNA amplicons after digestion with MspI and HhaI as described by Mills et al. (35). Multiple representatives of each restriction fragment length polymorphism group were subjected to 16S rRNA sequence analysis. Long-term storage of the FRC isolates was in dimethyl sulfoxide/glycerol at −80°C. Nutrient broth (NB) (3 g beef extract per liter, 5 g peptone per liter) was used for maintenance of the purified FRC strains. Isolates were screened for the presence of plasmids as previously described by Reyes et al. (51). For determination of resistance to the metal salts cadmium chloride, potassium chromate, lead citrate, and mercuric chloride, 400 FRC isolates were assayed as previously described (6, 51). The following concentrations of metals were tested: 500 nmol (C6H5O7)2Pb3 · 3H2O, 50 nmol Hg2Cl2, 2 μmol K2CrO4, and 500 nmol CdCl2 · 5/2H2O. Metal-resistant and -sensitive control strains were used as described by Benyehuda et al. (6) to confirm metal resistance phenotypes.
Uranium tolerance assays.
Tolerance to uranium (U) at pH 4 was determined as previously described (55), using 200 μM uranyl acetate [50 ppm U(VI)]. Five-milliliter overnight broth cultures of FRC isolates and control strains (Arthrobacter histidinolovorans ATCC 11442, Bacillus cereus ATCC 14579, and Escherichia coli JM109) were diluted 1:100 and grown to the mid-log phase either at 25°C in nutrient broth (FRC isolates and control strains) or at 37°C in Luria-Bertani (LB) broth (E. coli). Three 1-ml aliquots of mid-log-phase cells were centrifuged (10 min, 10,000 × g), the cell pellets were washed twice with 0.1 M NaCl (pH 4), and each aliquot was assayed as follows: (i) diluted in sterile saline and immediately plated on nutrient agar; (ii) incubated for 1 h in 0.1 M NaCl (pH 4); and (iii) incubated for 1 h in 0.1 M NaCl (pH 4) containing 200 μM U(VI). Following incubation, cells were serially diluted in sterile saline, plated on either LB or NB agar, incubated for 24 to 48 h, and enumerated. Triplicate assays were conducted, and all data were analyzed for statistical significance.
PCR amplification of 16S rRNA and PIB-ATPases.
Genomic DNA was isolated from FRC isolates by a rapid boiling method (19). Briefly, FRC isolates were incubated in NB for 6 h at 30°C. A 100-μl cell suspension was centrifuged, and the pellet was resuspended in 20 μl sterile distilled water and heated (100°C for 10 min). Lysates were centrifuged, and the supernatants were decanted and transferred to sterile tubes for storage at −20°C prior to use. PCR amplification of 16S rRNA genes was performed as previously described (35), and PCR amplification of the PIB-type ATPase genes was also performed as previously described (11). Specifically, the PCR primers used in reaction 1 targeted conserved sequences found in all P-type ATPases for the target genera indicated in Table 1. The PCR primers used in reaction 2 targeted domain sequences that are found only in heavy metal-transporting (PIB-type) ATPases (Table 1) (11).
TABLE 1.
Oligonucleotide primers used during nested PCRs to amplify PIB-type ATPases and numbers of isolates which yielded amplicons
| PCR | Primerc | Primer sequence | Target genus/genera | Annealing temp (°C) | No. of Pbr isolates that yielded a PCR product |
|---|---|---|---|---|---|
| 1a | 79JC | 5′ TGACTGGCGAATCGGTBCCBG 3′ | Bacillus, Acinetobacter, Pseudomonas | 59 | 8 |
| 84JC | 5′ GGAGCATCGTTAATDCCRTCDCC 3′ | ||||
| 132JC | 5′ CTAACTGGCGAATCAGTCCC 3′ | Arthrobacter | 55 | 25 | |
| 84JC | 5′ GGAGCATCGTTAATDCCRTCDCC 3′ | ||||
| 2b | 81JC | 5′ GGATGTCCTTGTGCTYTART 3′ | Bacillus, Acinetobacter, Pseudomonas | 49 | 5 |
| 84JC | 5′ GGAGCATCGTTAATDCCRTCDCC 3′ | ||||
| 133JC | 5′ CCCTCACCTTGTGCYCTGG 3′ | Arthrobacter | 49 | 23 | |
| 84JC | 5′ GGAGCATCGTTAATDCCRTCDCC 3′ |
Expected product size, approximately 1.2 kb.
Expected product size, approximately 0.75 kb.
See reference 11 for specific thermocycling parameters.
Sequencing and analyses.
Sequencing of the zntA/cadA/pbrA-like and 16S rRNA amplicons was performed at the School of Biology Genome Center (Georgia Institute of Technology) using a BigDye Terminator v3.1 cycle sequencing kit and an automated capillary sequencer (model 3100 gene analyzer; Applied Biosystems). The primers used to sequence the PIB-type ATPase loci are listed in Table 1, and primers 27f and 1522r (35) were used for 16S rRNA gene PCR products. Multiple sequences of PCR products were initially aligned using the program BLAST 2 Sequences (56) available through the National Center for Biotechnology Information and were assembled with the program BioEdit v5.0.9 (18). Sequences from this study and reference sequences, as determined by BLAST analysis, were aligned using ClustalX v1.81 (58). Neighbor-joining trees were created from these alignments. On average, 780 and 560 nucleotides were included in the phylogenetic analyses of the 16S rRNA gene and PIB-ATPase sequences, respectively. The tree topologies of 16S rRNA gene phylogenies were identical when they were analyzed using maximum likelihood and maximum parsimony. Similar analyses of PIB-ATPase amino acid sequences resulted in some tree topology differences but did not alter the outcome of the data generated by neighbor joining. The bootstrap data indicate percentages for 1,000 samplings. The final trees were viewed using NJPlot (46) and TreeView v1.6.6 available at http://taxonomy.zoology.gla.ac.uk/rod/treeview.html.
Nucleotide sequence accession numbers.
The 17 16S rRNA nucleotide sequences have been deposited in the GenBank database under accession numbers DQ224387 to DQ224403, and the 19 PIB-ATPase nucleotide sequences have been deposited in the GenBank database under accession numbers DQ234600 to DQ234618.
RESULTS AND DISCUSSION
Viable bacterial populations from contaminated soils and metal resistance phenotypes.
Cultivation-based methods were used to isolate aerobic heterotrophs present in radionuclide- and heavy metal-contaminated subsurface soils at the U.S. DOE FRC. In the current study, the numbers of heterotrophic bacteria recovered from the contaminated FRC soils were low, ranging from fewer than 10 CFU g−1 to 104 CFU g−1 (data not shown). All four media yielded similar CFU counts. Comparable low population densities for aerobic heterotrophs, isolated on the same media used in this study, have been reported previously for contaminated soils from a high-level waste plume at the Hanford Site (14). While enrichment-based studies targeting Fe(III)-reducing populations in FRC soils have also reported similar low population densities (47), fewer studies have reported data for aerobic populations from the FRC. Such information is particularly relevant as areas within contaminated FRC sites are already oxygenated and/or can become reoxygenated during rain-driven recharge events. Moreover, microorganisms that maintain metabolic activity in the presence of oxygen may play key roles in sequestration and immobilization of toxic radionuclides, such as U(VI), via bioprecipitation processes (4, 26). These biologically mediated processes could represent alternative remediation strategies as recent studies have reported that enzymatic and/or abiotic reoxidation of bioreduced U occurred (12, 13). Additionally, previous kinetic studies have also shown that, in the presence of electron shuttles, Fe(III) and Mn(IV) oxides promote the abiotic oxidation of U(IV) to U(VI)] (25, 38).
The majority (392 of 400) of the FRC isolates recovered from contaminated soils were gram positive and belonged to the high-G+C- and low-G+C-content genera Arthrobacter and Bacillus, respectively. As many as 40% of all cultured isolates recovered from FRC soil were related to Arthrobacter based on 16S rRNA gene analysis (data not shown). Gram-negative isolates recovered from the FRC sediments were most closely related to Rahnella (Fig. 1). As the acidic FRC subsurface soils are cocontaminated with heavy metals (8), it was of interest to determine the metal resistance phenotypes of the isolates, particularly as it has been postulated that Arthrobacter sp. (55). and Bacillus sp. (34) could be important in promoting the remediation of uranium through either intracellular sequestration or bioadsorption mechanisms. The percentages of FRC isolates (n = 400) that were resistant to the heavy metals cadmium, chromium, lead, and mercury were 10%, 11%, 44%, and 49%, respectively. The majority of the FRC isolates also exhibited resistance to two or more metals (data not shown). A comparison of the metal resistance characteristics of gram-positive FRC isolates and gram-positive strains previously isolated from saturated deep subsurface sediments at DOE's Hanford and Savannah River (SRS) sites revealed marked differences in the resistance phenotypes (6). Higher percentages of isolates from Hanford and SRS than of isolates from the FRC (11%) were shown to be resistant to chromium (38% and 37%, respectively [6]). The percentages of Hanford and SRS gram-positive strains shown to be resistant to lead were 48% and 20%, respectively (6). While 49% of the FRC gram-positive isolates were found to be resistant to mercury, only 8% of the SRS isolates were found to be mercury resistant (6). Cadmium resistance was not determined for the Hanford or SRS isolates in the previous study (6). It is important to note that the isolates from Hanford and SRS were cultured from uncontaminated soils from deeper saturated strata, whereas the FRC isolates were recovered from the contaminated subsurface. Such site and depth variation could have contributed to the observed differences in metal resistance phenotypes among subsurface bacteria isolated from the SRS, Hanford, and FRC sites.
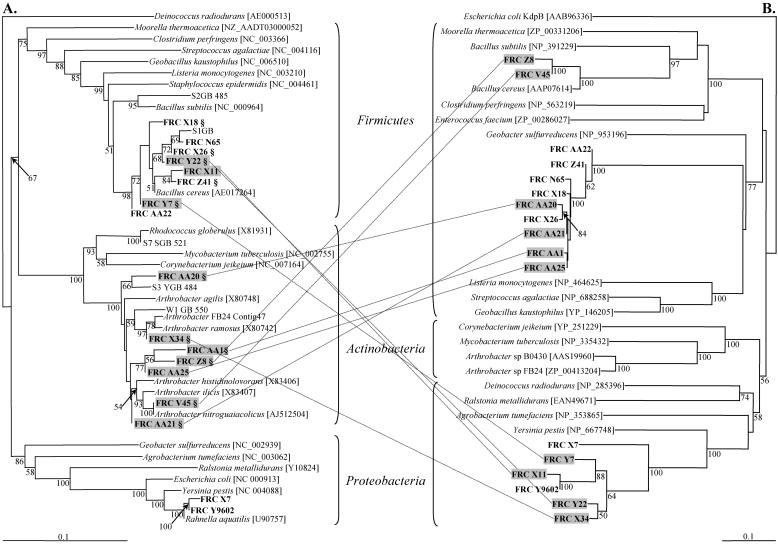
FIG. 1.
Neighbor-joining analysis of 16S rRNA gene (A) and zntA/cadA/prbA-like (B) sequences from either subsurface FRC isolates or completed genomes. Accession numbers are indicated in brackets. A section sign indicates an FRC strain containing one or more plasmids. Subsurface isolates in shaded boxes and connected by dotted lines are positive for horizontally acquired PIB-type ATPases related to zntA/cadA/prbA loci. Bootstrap values greater than 50% are indicated at the nodes. The scale bars for the 16S rRNA gene and zntA/cadA/prbA phylogenies indicate 0.1 change per nucleotide position and 0.1 change per amino acid position, respectively.
Horizontal transfer of PIB-type ATPases.
A cellular enzymatic detoxification mechanism to remove toxic metals is efflux pumping of mono- and divalent cations via chromosomally encoded metal homeostasis proteins (e.g., P-type ATPases [21]). Mobile genetic elements, including broad-host-range conjugative plasmids, have also been shown to encode resistance determinants, thus promoting their horizontal transfer to unrelated microorganisms (33). Recently, Coombs and Barkay (11) investigated the role of HGT in the evolution of lead resistance in 105 deep subsurface strains from uncontaminated Hanford and SRS saturated soils. Using nested PCR primers designed to specifically amplify PIB-type ATPases (e.g., zntA/cadA/pbrA-like), this study produced evidence for HGT of zntA/cadA/pbrA-related loci among gram-negative Proteobacteria. A total of 48 amplicons were obtained from 105 Pbr isolates, although only 4 of these amplicons, belonging to the genera Acinetobacter, Comamonas, and Ralstonia, exhibited criteria (phylogenetic incongruence, G+C content [17]) that were suggestive of acquisition by HGT.
To trace the possible evolutionary path(s) of the metal homeostasis genes in FRC bacteria from the contaminated subsurface, ATPase genes derived from the genomic DNA of 50 randomly chosen Pbr isolates were amplified using a nested PCR approach with the same primer sets used by Coombs and Barkay (11) (Table 1); 33 and 28 FRC isolates yielded PCR products in reactions 1 and 2, respectively (Table 1). The 28 amplicons obtained in reaction 2 (i.e., zntA/cadA/pbrA loci) represented a frequency similar to that reported by Coombs and Barkay (11). Sequences of 26 of the 28 amplicons were subsequently obtained; two of the 26 strains yielded products with two different primer sets (Table 1). Sequence analyses of these 26 amplicons revealed the presence of numerous signature regions of PIB-type ATPases, including phosphorylation and ATP-binding domains (29), thus demonstrating the utility of the primer sets. Seventeen of the 26 amplicons were selected for further sequence analyses as all of them contained complete regions of the signature domains, and only these isolates were subjected to 16S rRNA gene sequencing. The remaining nine amplicons were not long enough for phylogenetic analyses due to truncations in one or more of the expected domain sequences (possibly due to insertions, deletions, or other recombination events). The only gram-negative strains among the 50 randomly chosen Pbr strains that had amplifiable PIB-type ATPases were isolates FRC-X7 and FRC-Y9602. These isolates had PCR-amplifiable copA-related PIB-type ATPases (data not shown) along with the expected zntA/cadA/prbA loci (Fig. 1). This result is not surprising given the limited number of control strains tested in the initial primer sets for the PIB-type zntA/cadA/prbA-specific ATPases (11). Overall, the neighbor-joining and maximum likelihood tree analyses of the zntA/cadA/pbrA-like deduced amino acid sequences (e.g., 500 to 600 bp) resulted in a tree with remarkable incongruence between the 16S rRNA and ATPase gene phylogenies (Fig. 1). Seven of the 17 amplicons exhibited no unusual or unexpected incongruence which would have been suggestive of HGT, such as an atypical G+C content or phylogenetic incongruence (Fig. 1 and Table 2) (23). However, all seven of the zntA/cadA/pbrA-like sequences derived from Arthrobacter isolates exhibited evidence of recent acquisition by HGT. Specifically, the zntA/cadA/pbrA-like gene sequences amplified from isolates FRC-V45 and FRC-Z8, belonging to the phylum Actinobacteria as determined by 16S rRNA phylogeny (Fig. 1), grouped in one of the two bifurcated nodes within the Firmicutes (Fig. 1). In addition, the FRC-V45 and FRC-Z8 isolates contained zntA/cadA/pbrA-like genes with G+C contents much lower than those expected for other Arthrobacter spp. (35 to 36 mol% instead of 59 to 70 mol% [20]) (Table 2). A second set of PIB-type ATPases in isolates FRC-AA1, FRC-AA20, FRC-AA21, and FRC-AA25, also belonging to the phylum Actinobacteria (Fig. 1), clustered with ZntA/CadA/PbrA-like sequences most closely related to the second bifurcated node within the Firmicutes (Fig. 1). These four isolates also had unusual G+C contents (38 mol%) (Table 2). The ATPase-related sequences amplified from isolates FRC-X11, FRC-Y7, and FRC-Y22, belonging to the phylum Firmicutes, clustered in one distinct γ-Proteobacteria ZntA/CadA/PbrA-like clade (Fig. 1). Horizontal acquisition of the ATPase genes is also supported by unusual DNA base compositions, as indicated by the G+C contents (Table 2). Strains FRC-X11, FRC-Y7, and FRC-Y22 contained zntA/cadA/pbrA-like genes with G+C contents of 58 to 59 mol% (Table 2). These contents differed considerably from the content of the most closely related culturable isolate, Bacillus cereus, which has a G+C content of 32 mol% (10). Together, such phylogenetic incongruence and unusual G+C contents provide strong evidence for horizontal acquisition of the PIB-type ATPases genes. As determined by 16S rRNA gene analysis, Bacillus spp. isolates FRC-X11, FRC-Y7, and FRC-Y22 exhibited more than 96% identity, and the ATPase sequences amplified from these isolates exhibited more than 97% amino acid identity (data not shown). Interestingly, the γ-Proteobacteria-related ATPase amino acid sequences from isolates FRC-X34 and FRC-Y22 (Arthrobacter sp. and Bacillus sp., respectively) (Fig. 1) exhibited 98% identity but represented two distinct phyla based on the 16S rRNA gene sequences (Actinobacteria and Firmicutes) (Fig. 1). Although the G+C content of the zntA/cadA/pbrA-like sequence (59 mol%) amplified from isolate FRC-X34 is similar to the previously reported G+C contents of other Arthrobacter spp., many γ-Proteobacteria have comparable G+C contents (range, 38 mol% to 63 mol%) (20a). Thus, horizontal transfer of the zntA/cadA/pbrA-like gene to isolate FRC-X34 may still be supported by our results; however, we denoted it a “maybe” for the purposes of this study (Table 2).
TABLE 2.
Support for acquisition of PIB-type ATPases by HGT in subsurface isolates from radionuclide- and metal-contaminated soils
| Genus | Strain | Phylogenetic incongruence | G+C content (%) | Support for HGTa | Most closely related PIB-type ATPase |
|---|---|---|---|---|---|
| Arthrobacter | FRC-AA1 | + | 38 | + | Firmicutes |
| FRC-AA20 | + | 38 | + | Firmicutes | |
| FRC-AA21 | + | 38 | + | Firmicutes | |
| FRC-AA25 | + | 38 | + | Firmicutes | |
| FRC-V45 | + | 36 | + | Firmicutes | |
| FRC-X34 | + | 59 | Maybe | γ-Proteobacteria | |
| FRC-Z8 | + | 35 | + | Firmicutes | |
| Bacillus | FRC-AA22 | − | 38 | − | Firmicutes |
| FRC-N65 | − | 38 | − | Firmicutes | |
| FRC-X11 | + | 59 | + | γ-Proteobacteria | |
| FRC-X18 | − | 38 | − | Firmicutes | |
| FRC-X26 | − | 38 | − | Firmicutes | |
| FRC-Y7 | + | 58 | + | γ-Proteobacteria | |
| FRC-Y22 | + | 58 | + | γ-Proteobacteria | |
| FRC-Z41 | − | 38 | − | Firmicutes | |
| Rahnella | FRC-X7 | − | 59 | − | γ-Proteobacteria |
| FRC-X7 (copA) | − | 59 | − | γ-Proteobacteria | |
| FRC-Y9602 | − | 59 | − | γ-Proteobacteria | |
| FRC-Y9602 (copA) | − | 59 | − | γ-Proteobacteria |
The evidence for HGT was supported by analysis of both phylogenetic incongruence and the G+C content.
The FRC isolates were also screened for the presence of plasmids (Fig. 1). Of the 10 strains that fulfilled the criterion that the PIB-type ATPases may have been acquired by HGT, 8 contained plasmids that were large enough to encode such genes and to be mobilizable and/or self-transmissible (data not shown). Studies are being conducted to determine whether plasmid-encoded ATPase genes are indeed present in these isolates. Taken together, all of these findings are highly suggestive of considerable broad and remarkable dissemination of horizontally acquired PIB-type ATPase genes from both the Firmicutes and Proteobacteria phyla to bacteria isolated from contaminated subsurfaces (Fig. 1 and Table 2). However, it is important to note that we cannot conclude, based on the current study, whether such HGT events occurred prior to or following FRC soil contamination. Indeed, a host of other biological and/or environmental factors likely contribute to such horizontal gene exchanges (54, 57).
Tolerance to uranium.
In addition to heavy metals, microorganisms in the FRC subsurface are subjected to other contaminants, including radionuclides and organic solvents (http://www.esd.ornl.gov/nabirfrc). Thus, to determine whether the heterotrophic FRC strains isolated in this study were capable of tolerating toxic concentrations of U, tolerance assays were conducted under acidic conditions with numerous isolates representing the most commonly isolated genera (Arthrobacter, Bacillus, and Rahnella).
The isolates that were the least tolerant of both the acidic pH and U toxicity assay conditions were the FRC Bacillus spp. isolates (Table 3). A similar result was reported for Bacillus spp. previously isolated from an acidic inactive open-pit U mine (55). The exception was Bacillus sp. strain FRC-X18, which remained viable at pH 4 (Table 3) and exhibited the smallest reduction in cell number in the presence of U (Table 3). Strains FRC-N65, FRC-X18, and FRC-Y9-2 exhibited greater tolerance to the acidic pH conditions than the Bacillus control strain (Table 3). The gram-negative organism Rahnella sp. strain FRC-Y9602 exhibited only a slight decrease in cell viability (Table 3) due to the pH conditions but a >100-fold loss of viability upon exposure to U(VI). The Arthrobacter strains exhibited the greatest tolerance to the low-pH and U conditions (Table 3). Of the six Arthrobacter strains tested, the A. histidinolovarans control strain exhibited the greatest decrease in cell viability during incubation at pH 4 with or without U (Table 3). In contrast, the FRC strains exhibited little or no decrease in cell viability (Table 3). A similar tolerance to acid has also been reported for two other Arthrobacter spp. from an open-pit U mine (55). All five Arthrobacter FRC strains remained viable following exposure to U. Similarly, two of the three high-G+C-content microorganisms previously isolated from the open-pit U mine site exhibited comparable tolerance to U (55).
TABLE 3.
Viable cell counts determined after washing and after 1 h of incubation at pH 4 either with or without 200 μM uranyl acetatea
| Genus | Strain | CFU (washed)b | CFU (without U)c | CFU (with U) |
|---|---|---|---|---|
| Arthrobacter | FRC-AA1 | (1.79 ± 0.48) × 108d | (1.59 ± 0.49) × 108 | (1.45 ± 0.49) × 108 |
| FRC-AA21 | (1.53 ± 0.37) × 108 | (7.78 ± 0.20) × 107 | (6.45 ± 0.38) × 107 | |
| FRC-AA25 | 2.31 × 108 | 1.39 × 108 | (9.20 ± 0.49) × 107 | |
| FRC-V45 | (1.67 ± 0.40) × 108 | (9.13 ± 0.15) × 107 | (6.85 ± 0.18) × 107 | |
| FRC-X34 | 1.83 × 108 | 1.71 × 108 | 1.07 × 108 | |
| A. histidinolovorans ATCC 11442 | (1.57 ± 0.13) × 108 | (6.10 ± 0.49) × 107 | (4.35 ± 1.04) × 107 | |
| Bacillus | FRC-N65 | 2.99 × 108 | (1.12 ± 0.38) × 108 | <1 × 104 |
| FRC-X18 | (1.87 ± 0.50) × 108 | (3.51 ± 1.14) × 107 | 4.03 × 105 | |
| FRC-Y9-2 | (2.55 ± 0.64) × 107 | (3.12 ± 1.05) × 106 | <1 × 104 | |
| B. cereus ATCC 14579 | (9.70 ± 0.52) × 107 | <1 × 104 | <1 × 104 | |
| Escherichia | E. coli JM109 | (8.00 ± 0.37) × 107 | (4.65 ± 0.30) × 107 | <1 × 104 |
| Rahnella | FRC-Y9602 | 1.10 × 108 | (5.40 ± 0.40) × 107 | (1.54 ± 0.92) × 106 |
At pH 4 U(VI) occurs primarily as the uranyl ion (UO22+).
All strains were plated immediately following two washes with 0.1 M NaCl at pH 4.
Strains were incubated for 1 h at pH 4.
Standard deviations greater than 10% are shown.
The tolerance to U exhibited by gram-positive and gram-negative microorganisms may be explained by several different cellular mechanisms, as previously reported by other investigators (30, 49, 55). One mechanism, bioadsorption, has recently been hypothesized to occur in a well-characterized Arthrobacter type strain, the A. nicotianae type strain (59). In this study as much as 80% of the uranyl ions were removed from an aqueous solution at pH 4 (59); however, cellular U localization was not determined. A second mechanism, U sequestration, which was shown to occur in Arthrobacter isolate S3Y (55), resulted in intracellular accumulation of uranium precipitates, perhaps as a means to limit U toxicity. In this case, the authors used transmission electron microscopy and energy-dispersive X-ray analysis to identify coprecipitation of U with phosphorus- and calcium-rich granules (55). Interestingly, strain S3Y exhibited no evidence of bioadsorption (55), suggesting that Arthrobacter species such as A. nicotianae may also be capable of intracellular U accumulation. Studies are currently under way to determine whether intracellular U sequestration promotes the tolerance to U that has been observed in Arthrobacter strains isolated from heavy metal- and radionuclide-contaminated subsurface soils.
Among some of the most promising strategies for remediation of contaminated subsurfaces are bioimmobilization of metals and radionuclides by microbial processes and their metabolic products (4). Numerous microbes carry out reductive processes that result in decreased solubility, and thus bioavailability and toxicity, of metals and radionuclides (27). Recent efforts to stimulate microbial communities to remove U from contaminated aquifers and groundwater by promoting the in situ activity of dissimilatory metal-reducing organisms highlight the important role of microbial processes in the subsurface (1, 41). In these studies, biostimulation strategies for subsurface remediation resulted in significant increases in Geobacter spp. and sulfate-reducing bacterial populations. As these populations are expected to function in environments that are affected by mixed wastes, the presence of toxic heavy metals and nitrates that cooccur at sites such as the FRC (13, 47) could potentially limit their activities. For example, Desulfovibrio desulfuricans G20, a model organism for immobilization of metals as metal sulfides, has been shown to be susceptible to micromolar concentrations of heavy metals, including Cu(II), Zn(II), and Pb(II) (52), while mixed cultures of sulfate reducers were inhibited by Cr(VI) (53), Cu(II), and Zn(II) (60). Likewise, some heavy metals, including Cr(VI), have also been shown to negatively affect the growth of Shewanella spp., which have been studied for their role in immobilizing metals and radionuclides by reduction to insoluble forms (62). Thus, the (heavy) metal sensitivity of some radionuclide-reducing microorganisms indicates that acquiring metal resistance could be highly conducive to facilitating and/or enhancing microbial metabolism in metal-contaminated subsurface environments. Such enhancement could be achieved by stimulating the transfer of broad-host-range metal resistance plasmids to metal- and radionuclide-reducing microbes in treated subsurfaces and/or promoting the efficient transformation and incorporation of key genes for adaptation via transformation- or transduction-mediated processes (16). To the best of our knowledge, this study is among the first to report the heavy metal resistance phenotypes and U tolerance of Arthrobacter spp. isolated from an acidic contaminated subsurface environment. These unrelated, yet synergistic, physiological traits observed in Arthrobacter isolates residing in contaminated FRC soils may contribute to the survival of these organisms in such an extreme environment. Thus, Arthrobacter species, particularly those residing in contaminated subsurface environments, such as the FRC, may represent a largely untapped group of microorganisms with considerable bioremediation potential.
Acknowledgments
We thank Cassie Black, Mike Humphrys, Kerri Lafferty, and Kristin Tuttle for providing excellent technical assistance and Dave Watson for providing FRC soil cores.
This research was supported by the Natural and Accelerated Bioremediation Research (NABIR) program, Office of Science (BER), U.S. Department of Energy (grants DE-FG02-99ER62864 and DE-FG02-04ER63906).
REFERENCES
- 1.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelsen, K. B., and M. G. Palmgren. 1998. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46:84-101. [DOI] [PubMed] [Google Scholar]
- 3.Axelsen, K. B., and M. G. Palmgren. 2001. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 126:696-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkay, T., and J. Schaefer. 2001. Metal and radionuclide bioremediation: issues, considerations and potentials. Curr. Opin. Microbiol. 4:318-323. [DOI] [PubMed] [Google Scholar]
- 5.Beiko, R. G., T. J. Harlow, and M. A. Ragan. 2005. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 102:14332-14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benyehuda, G., J. Coombs, P. L. Ward, D. Balkwill, and T. Barkay. 2003. Metal resistance among aerobic chemoheterotrophic bacteria from the deep terrestrial subsurface. Can. J. Microbiol. 49:151-156. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Brooks, S. 2001. Waste characteristics of the former S-3 ponds and outline of uranium chemistry relevant to NABIR Field Research Center studies. NABIR Field Research Center, Oak Ridge, Tenn.
- 9.Canchaya, C., G. Fournous, S. Chibani-Chennoufi, M. L. Dillmann, and H. Brussow. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417-424. [DOI] [PubMed] [Google Scholar]
- 10.Claus, D., and R. C. W. Berkeley. 1986. Genus Bacillus Cohn, 174AL*, p. 1105-1139. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 11.Coombs, J. M., and T. Barkay. 2004. Molecular evidence for the evolution of metal homeostasis genes by lateral gene transfer in bacteria from the deep terrestrial subsurface. Appl. Environ. Microbiol. 70:1698-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias, D. A., L. R. Krumholz, D. Wong, P. E. Long, and J. M. Suflita. 2003. Characterization of microbial activities and U reduction in a shallow aquifer contaminated by uranium mill tailings. Microb. Ecol. 46:83-91. [DOI] [PubMed] [Google Scholar]
- 13.Finneran, K. T., M. E. Housewright, and D. R. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 14.Fredrickson, J. K., J. M. Zachara, D. L. Balkwill, D. Kennedy, S. M. W. Li, H. M. Kostandarithes, M. J. Daly, M. F. Romine, and F. J. Brockman. 2004. Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford Site, Washington state. Appl. Environ. Microbiol. 70:4230-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredrickson, J. K., J. M. Zachara, D. W. Kennedy, M. C. Duff, Y. A. Gorby, S. M. W. Li, and K. M. Krupka. 2000. Reduction of U(VI) in goethite (alpha-FeOOH) suspensions by a dissimilatory metal-reducing bacterium. Geochim. Cosmochim. Acta 64:3085-3098. [Google Scholar]
- 16.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 17.Gogarten, J. P., and J. P. Townsend. 2005. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3:679-687. [DOI] [PubMed] [Google Scholar]
- 18.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 19.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 20.Keddie, R. M., M. D., Collins, and D. Jones. 1986. Genus Arthrobacter Conn and Dimmick 1947, 300AL, p. 1288-1301. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 20a.Krieg, N. R., and J. G. Holt (ed.). 1984. Bergey’s manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 21.Kuhlbrandt, W. 2004. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5:282-295. [DOI] [PubMed] [Google Scholar]
- 22.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383-397. [DOI] [PubMed] [Google Scholar]
- 24.LeBrun, M., A. Audurier, and P. Cossart. 1994. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J. Bacteriol. 176:3040-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, C. X., J. M. Zachara, J. K. Fredrickson, D. W. Kennedy, and A. Dohnalkova. 2002. Modeling the inhibition of the bacterial reduction of U(VI) by beta-MnO2(s). Environ. Sci. Technol. 36:1452-1459. [DOI] [PubMed] [Google Scholar]
- 26.Lovley, D. R., and J. D. Coates. 1997. Bioremediation of metal contamination. Curr. Opin. Biotechnol. 8:285-289. [DOI] [PubMed] [Google Scholar]
- 27.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 28.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 29.Lutsenko, S., and J. H. Kaplan. 1995. Organization of P-type ATPases—significance of structural diversity. Biochemistry 34:15607-15613. [DOI] [PubMed] [Google Scholar]
- 30.Macaskie, L. E., R. M. Empson, A. K. Cheetham, C. P. Grey, and A. J. Skarnulis. 1992. Uranium bioaccumulation by a Citrobacter sp. as a result of enzymatically mediated growth of polycrystalline HUO2PO4. Science 257:782-784. [DOI] [PubMed] [Google Scholar]
- 31.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean, E. O. 1982. Soil pH and lime requirement, p. 199-209. In Methods of soil analysis, part 2. Chemical and microbiological properties. Agronomy Monograph 9, 2nd ed. ASA-SSSA, Madison, Wis.
- 33.Mergeay, M., S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. van der Lelie, and R. Wattiez. 2003. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27:385-410. [DOI] [PubMed] [Google Scholar]
- 34.Merroun, M. L., J. Raff, A. Rossberg, C. Hennig, T. Reich, and S. Selenska-Pobell. 2005. Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl. Environ. Microbiol. 71:5532-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, Y., T. Itoh, H. Matsuda, and T. Gojobori. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36:760-766. [DOI] [PubMed] [Google Scholar]
- 37.Nemergut, D. R., A. P. Martin, and S. K. Schmidt. 2004. Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution. Appl. Environ. Microbiol. 70:1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nevin, K. P., and D. R. Lovley. 2000. Potential for nonenzymatic reduction of Fe(III) via electron shuttling in subsurface sediments. Environ. Sci. Technol. 34:2472-2478. [Google Scholar]
- 39.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 40.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 41.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment co-contaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nucifora, G., L. Chu, T. K. Misra, and S. Silver. 1989. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. USA 86:3544-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Sullivan, D., R. P. Ross, D. P. Twomey, G. F. Fitzgerald, C. Hill, and A. Coffey. 2001. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility functions to a food-grade selectable marker. Appl. Environ. Microbiol. 67:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Payne, R. B., D. A. Gentry, B. J. Rapp-Giles, L. Casalot, and J. D. Wall. 2002. Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl. Environ. Microbiol. 68:3129-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson, A. J., K. D. Bruce, A. M. Osborn, D. A. Ritchie, and P. Strike. 1996. Distribution of class II transposase and resolvase genes in soil bacteria and their association with mer genes. Appl. Environ. Microbiol. 62:2961-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perriere, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 47.Petrie, L., N. N. North, S. L. Dollhopf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powers, E. M. 1995. Efficacy of the Ryu nonstaining KOH technique for rapidly determining Gram reactions of food-borne and waterborne bacteria and yeasts. Appl. Environ. Microbiol. 61:3756-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renninger, N., R. Knopp, H. Nitsche, D. S. Clark, and J. D. Keasling. 2004. Uranyl precipitation by Pseudomonas aeruginosa via controlled polyphosphate metabolism. Appl. Environ. Microbiol. 70:7404-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rensing, C., M. Ghosh, and B. P. Rosen. 1999. Families of soft-metal-ion-transporting ATPases. J. Bacteriol. 181:5891-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reyes, N. S., M. E. Frischer, and P. A. Sobecky. 1999. Characterization of mercury resistance mechanisms in marine sediment microbial communities. FEMS Microbiol. Ecol. 30:273-284. [DOI] [PubMed] [Google Scholar]
- 52.Sani, R. K., B. A. Peyton, and M. Jandhyala. 2003. Toxicity of lead in aqueous medium to Desulfovibrio desulfuricanis G20. Environ. Toxicol. Chem. 22:252-260. [PubMed] [Google Scholar]
- 53.Smith, W. L., and G. M. Gadd. 2000. Reduction and precipitation of chromate by mixed culture sulphate-reducing bacterial biofilms. J. Appl. Microbiol. 88:983-991. [DOI] [PubMed] [Google Scholar]
- 54.Sorensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki, Y., and J. F. Banfield. 2004. Resistance to, and accumulation of, uranium by bacteria from a uranium-contaminated site. Geomicrobiol. J. 21:113-121. [Google Scholar]
- 56.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 SEQUENCES, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. (Erratum, 177: 187-188.) [DOI] [PubMed] [Google Scholar]
- 57.Thomas, C. M., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuruta, T. 2002. Removal and recovery of uranyl ion using various microorganisms. J. Biosci. Bioeng. 94:23-28. [DOI] [PubMed] [Google Scholar]
- 60.Utgikar, V. P., H. H. Tabak, J. R. Haines, and R. Govind. 2003. Quantification of toxic and inhibitory impact of copper and zinc on mixed cultures of sulfate-reducing bacteria. Biotechnol. Bioeng. 82:306-312. [DOI] [PubMed] [Google Scholar]
- 61.van Waasbergen, L. G., D. L. Balkwill, F. H. Crocker, B. N. Bjornstad, and R. V. Miller. 2000. Genetic diversity among Arthrobacter species collected across a heterogeneous series of terrestrial deep-subsurface sediments as determined on the basis of 16S rRNA and recA gene sequences. Appl. Environ. Microbiol. 66:3454-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viamajala, S., B. M. Peyton, R. K. Sani, W. A. Apel, and J. N. Petersen. 2004. Toxic effects of chromium(VI) on anaerobic and aerobic growth of Shewanella oneidensis MR-1. Biotechnol. Prog. 20:87-95. [DOI] [PubMed] [Google Scholar]
- 63.Wade, R., and T. J. DiChristina. 2000. Isolation of U(VI) reduction-deficient mutants of Shewanella putrefaciens. FEMS Microbiol. Lett. 184:143-148. [DOI] [PubMed] [Google Scholar]