Abstract
Porphyromonas gingivalis, one of the gram-negative organisms associated with periodontal disease, possesses potential virulence factors, including fimbriae, proteases, and major outer membrane proteins (OMPs). In this study, P. gingivalis ATCC 33277 was cultured in a chemostat under hemin excess and presumably peptide-limiting conditions to better understand the mechanisms of expression of the virulence factors upon environmental changes. At higher growth rates, the amounts of FimA and the 75-kDa protein, forming long and short fimbriae, respectively, increased significantly, whereas gingipains decreased in amount and activity. In a nutrient-limited medium, lesser amounts of the above two fimbrial proteins were observed, whereas clear differences were not found in the amounts of gingipains. In addition, two-dimensional electrophoresis revealed that proteins in cells were generally fewer in number during nutrient-limited growth. Under aeration, a considerable reduction in gingipain activity was found, whereas several proteins associated with intact cells significantly increased. However, the expression of major OMPs, such as RagA, RagB, and the OmpA-like proteins, was almost constant under all conditions tested. These results suggest that P. gingivalis may actively control expression of several virulence factors to survive in the widely fluctuating oral environment.
It has been reported that more than 80% of U.S. adults have overt gingivitis and about half of them have periodontitis (1). In periodontal diseases, teeth may be lost after inflammation spreads to the supporting periodontal tissues. Not only local events such as inflammation and trauma but also systemic and environmental factors are involved in establishment of periodontal diseases (9). The formation and maturation of plaque, especially subgingival plaque in periodontal pockets, are also involved (61). Although there are more than 600 species of bacteria in the mouth (30), specific species of gram-negative anaerobic bacteria, spirochetes, and motile rods are known to participate in formation of subgingival plaque (20, 60). Porphyromonas gingivalis, a gram-negative anaerobe, is considered to contribute to periodontitis, since it is detected frequently in patients with these diseases (56, 60).
Asaccharolytic P. gingivalis mainly obtains nutrients by degradation of proteins. This bacterium is known to have many potential virulence factors, including fimbriae, proteases, hemagglutinins, lipopolysaccharide, capsule polysaccharides, and major outer membrane proteins (26, 31). P. gingivalis expresses two types of fimbriae, long fimbriae composed of FimA (fimbrilin) (67), and short fimbriae composed of a 75-kDa protein (21, 51, 68). Many studies have shown that FimA has various functions, which include attachment to and invasion of gingival epithelial cells (28, 64), coaggregation with other species of bacteria (2), attachment to salivary components (4), activation of fibroblasts and macrophages (24), induction of inflammatory cytokines, and bone resorption (23).
Several proteins are often found in the outer membrane fraction, particularly RagA, RagB, Arg-gingipains (Rgps), Lys-gingipain (Kgp), and the OmpA-like proteins (41, 42). RagA is a TonB-dependent outer membrane receptor. RagB forms a complex with RagA, and the complex participates in uptake of unknown substances (11). Gingipains are highly active endo-type, trypsin-like, cysteine proteases (27, 47). Rgps and Kgp hydrolyze the peptide bonds containing Arg and Lys residues, respectively. Most gingipain activity is found on the cell surface associated with the outer membrane, and the remainder is released to the culture medium. Recently, the OmpA-like proteins were shown to form a heterotrimeric structure composed of two homologous proteins, Pgm6/7 (44).
Bacteria sense surrounding environmental factors and subsequently adapt their physiology, including the growth rate, accordingly (52). Therefore, it is important to investigate the effects of the growth rate and environment on expression of virulence factors. Batch culture is generally used for bacterial studies. However, since bacteria are cultured in a closed environment, culture factors such as consumption of nutrients, accumulation of metabolic products, and cell number and growth phase are continuously changing. Therefore, batch culture is not always suitable for studies on bacterial physiology.
Another culture system, continuous culture in a chemostat, may also be used (50). By adjusting the dilution rate in a chemostat, reproducible steady-state conditions at a certain exponential growth rate are obtained because the physiological state of bacteria, i.e., the number of cells, growth rate, and surrounding environment, is stable. The growth rate of bacteria and the dilution rate become equal in the steady state. Once the steady state is established, culture fluid under the particular condition can be readily obtained for analysis during the experiment. With these characteristics, this method is ideal for studying the physiology of bacteria.
It is important to investigate how P. gingivalis adapts to changes in the oral environment. In this study, we examined the effects of growth rate, medium, and aeration on expression of major virulence factors and cellular proteins in P. gingivalis ATCC 33277 in a chemostat.
MATERIALS AND METHODS
Bacterial strain, culture media, and culture conditions.
P. gingivalis ATCC 33277 was used throughout this study. The strain was grown in Trypticase soy broth (BBL; Becton Dickinson Microbiology Systems, Sparks, MD) supplemented with 0.25% yeast extract (Difco, Becton Dickinson Microbiology Systems), 2.5 μg/ml hemin, 5.0 μg/ml menadione, and 0.01% dithiothreitol (DTT) (sTSB broth) to examine the effects of growth rate and aeration. To examine effects of medium, sTSB was used as a nutrient-rich medium and synthetic Eagle's minimal essential medium (MEM; Nissui Pharmaceutical Co., Tokyo, Japan) supplemented with 0.3% bovine serum albumin (BSA; fraction V), 2.5 μg/ml hemin, 5.0 μg/ml menadione, and 0.01% DTT (0.3% BSA-MEM) was used as a nutrient-poor medium. A preliminary experiment in batch culture revealed that P. gingivalis could not grow in Eagle's MEM; however, it could grow after addition of BSA. P. gingivalis was precultured under anaerobic conditions at 37°C for 48 h using prereduced sTSB broth. The preculture was then inoculated into 300 ml of fresh prewarmed sTSB broth and incubated anaerobically at 37°C for 48 h.
Continuous culture was performed in a model BioFlo 3000 chemostat (New Brunswick Scientific, Edison, NJ) with a working volume of 600 ml. Three hundred milliliters of the bacterial culture in sTSB described above was added to 300 ml of prereduced fresh sTSB broth in the chemostat vessel. The mixture was allowed to grow under batch culture conditions at 37°C overnight under an atmosphere of N2. After growth, the medium reservoir pump was turned on and the medium flow was adjusted to give an appropriate dilution rate. Culture fluid in the vessel was continuously agitated at 50 rpm. Both the culture vessel and medium reservoir were continuously gassed with N2 (100 ml/min). The temperature was set at 37°C, and the pH was controlled at 7.4 by addition of 1 N HCl or 1 N NaOH.
The effects on the growth rate were examined in sTSB broth by sequential establishment of steady-state growth at dilution rates (D) of 0.05 (doubling time [td] of 13.9 h), 0.15 (td = 4.6 h), and 0.3 h−1 (td = 2.3 h). The effects of growth medium were determined at a constant dilution rate of 0.05 h−1. After the steady-state growth had been established in sTSB broth, the medium was then replaced with 0.3% BSA-MEM and cultured until steady-state conditions were established again. The effect of aeration was examined in sTSB broth at a medium dilution rate of 0.15 h−1. After the steady-state growth had been established under anaerobic conditions with continuous N2 flow, the gas was changed to compressed air (100 ml/min), and organisms were cultured until steady-state conditions were achieved.
Bacterial growth was monitored by measuring optical density at 600 nm (OD600). Anaerobic conditions were checked by monitoring redox potential (Eh). Usually Eh values were less than −400 mV. After at least eight vessel volumes of the medium were fed into and out of the vessel, a steady state was achieved, based on the constancy of the OD600. All results described below were confirmed by at least duplicate independent experiments. The cultures were checked daily for purity by Gram staining.
P. gingivalis cell fractionation.
All of the fractionation procedures were performed at 4°C. Bacterial cultures were immediately centrifuged at 10,000 × g for 20 min to separate the cell-free culture supernatant and cells. Cells were gently washed once to minimize the removal of fimbriae with 10 mM HEPES-NaOH (pH 7.4) containing 0.15 M NaCl and then resuspended in 10 mM HEPES-NaOH (pH 7.4) containing protease inhibitors (0.1 mM N-α-p-tosyl-l-lysine chloromethyl ketone, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1 mM leupeptine; HEPES buffer) to prevent proteolytic degradation. The cells were disrupted using a sonicator (Bioruptor UCD-200T; Cosmo Bio, Tokyo, Japan) and repeating 30-s bursts and 30-s intervals for 15 min. After the remaining unbroken cells were removed by centrifugation at 1,000 × g for 10 min, the supernatant was used as the whole-cell lysate fraction, which was subjected to ultracentrifugation at 100,000 × g for 60 min in a Beckman fixed-angle rotor (TLA-100.2; Palo Alto, CA) to separate the envelope and soluble fractions (43). The culture supernatant was first passed through a filter (0.22 μm) to remove residual cells. Then, proteins were precipitated with 80% saturation of ammonium sulfate, followed by centrifugation to collect the precipitate at 20,000 × g for 30 min. The precipitate was dissolved in a small volume of HEPES buffer and dialyzed extensively against the same buffer. This solution was used as the culture supernatant fraction.
Samples for two-dimensional electrophoresis were prepared from washed cells treated in 10% trichloroacetic acid for 20 min on ice. The cells were recovered by centrifugation at 20,000 × g for 20 min and washed twice with diethyl ether to remove trichloroacetic acid. After being dried at room temperature, the resulting material was used as the whole-cell sample. The amount of protein was estimated by the method of Bradford (8) with bovine serum albumin as the standard.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out in a 12% gel as described by Lugtenberg et al. (33). The samples were solubilized with a reduction mixture containing SDS and 2-mercaptoethanol at 100°C for 5 min. The gel was stained with Coomassie brilliant blue R-250 (CBB) to detect proteins and to compare the intensities of protein bands of the major virulence factors.
Antibodies.
Antisera raised in rabbits against RagA, RagB, and putative porin were used (43). Antisera against the fimbrilin monomer (FimA), fimbriae (fimbrilin polymer), and the 75-kDa protein were described previously (67, 68). Antiserum recognizing both Kgp and Rgps was kindly provided by K. Yamamoto (Department of Pharmacology, Graduate School of Dental Science, Kyushu University).
Western immunoblotting.
After SDS-PAGE, proteins in gels were electrophoretically transferred onto nitrocellulose membranes (63) and then immunostained with specific antibodies essentially as described previously (43). For detection of Rgps and Kgp, the ECL Plus Western blotting detection system (Amersham Biosciences) was used.
Enzyme assays.
Lys-X- and Arg-X-specific cysteine proteinase activities were determined by measuring the hydrolysis of the synthetic substrates N-p-tosyl-Gly-Pro-Lys p-nitroanilide (Sigma Chemical, St. Louis, MO) and N-α-benzoyl-l-Arg p-nitroanilide (Sigma Chemical), respectively (25). Briefly, a portion of the culture fluid or the cell-free supernatant was used to make a reaction mixture containing 50 mM Tris-HCl (pH 8.5) and 10 mM DTT. The reaction was started by addition of the substrate (final concentration of 0.2 mM) followed by incubation at 37°C for 10 min. The reaction was stopped by addition of 0.2 ml of 50% acetic acid. Release of cleaved p-nitroanilide was determined by measuring the OD405. Protease activities were divided by the OD600 values of the cell density to normalize all values per OD600 unit.
Two-dimensional gel electrophoresis (2-DE).
Isoelectric focusing (IEF) in the first dimension was carried out using an Ettan IPGphor II equipped with a cup-loading manifold as a sample addition apparatus (Amersham Biosciences). The whole-cell sample was dissolved in a solubilization solution (7 M urea, 2 M thiourea, 4% 3-[{3-cholamidopropyl} dimethylammonio]-1-propanesulfonic acid [CHAPS], 1 mM EDTA, 40 mM Tris base, and 0.2% tributyl phosphine). The dissolved whole-cell sample was applied to an Immobiline DryStrip (pH 4 to 7; 13 cm; Amersham Biosciences) swollen with a rehydration solution (7 M urea, 2 M thiourea, 4% CHAPS, 0.5% IPG buffer [pH 4 to 7; Amersham Biosciences], 1 mM EDTA, 50 mM DTT, and bromophenol blue). IEF was initially conducted at 100 V for 1 h, from 500 V to 5,000 V for 3 h, and at 5,000 V for 6 h. After IEF, the strips were equilibrated in a buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, and bromophenol blue) containing 1% DTT, followed by incubation in the same buffer except that DTT was replaced by 2.5% iodoacetamide. The equilibrated strips were embedded on top of 12% gels using molten agarose. After SDS-PAGE in the second dimension was performed, the gels were stained with CBB.
Image analysis.
Each CBB-stained one-dimensional SDS-PAGE gel and immunoblotted membrane was analyzed as a digitized image using NIH ImageJ version 1.3.4 software. The CBB-stained 2-DE gels were scanned with LabScan version 5.0 on an ImageScanner (Amersham Biosciences), and the images were processed with the software ImageMaster 2D Platinum version 5.0 (Amersham Biosciences).
Protein analysis by mass spectrometry.
Protein spots after 2-DE were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). After in-gel tryptic digestion, peptides were extracted, concentrated, and analyzed using a Voyage-DE STR BioSpectrometry work station (Applied Biosystems, Foster City, Calif.) in the reflector mode essentially as described elsewhere (16, 55). The identities of the proteins were deduced from MS peaks via the MS-Fit peptide mass fingerprinting methods in Mascot (http://www.matrixscience.com/) or ProteinProspector (http://prospector.ucsf.edu/).
RESULTS
Effects of growth rates.
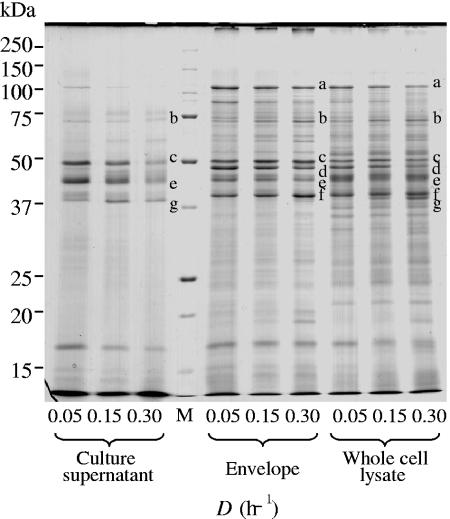
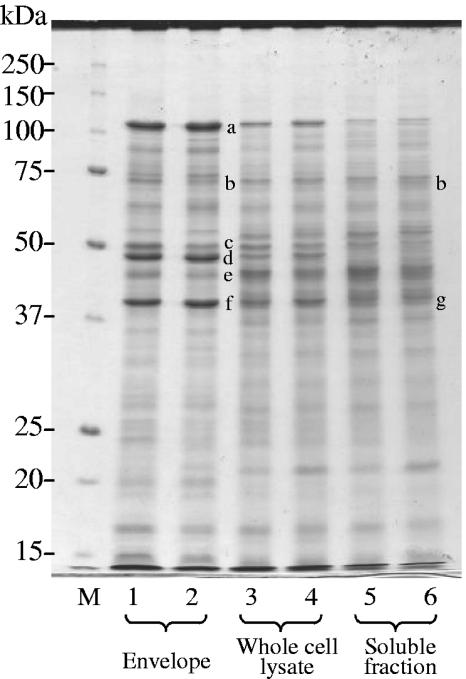
The growth rate of P. gingivalis was changed in sTSB broth by controlling dilution rates at 0.05, 0.15, and 0.30 h−1, for which the OD600 values in the culture under steady-state conditions were 1.4, 1.8, and 1.1, respectively. Effects of growth rates on expression of virulence factors were examined by SDS-PAGE. To investigate the localization of major virulence factors, the supernatant, the envelope, and whole-cell lysate fractions were used (Fig. 1). The amount of FimA (approximately 37 kDa) forming long fimbriae increased in the whole-cell lysate fraction as a function of the dilution rate. The 75-kDa protein forming short fimbriae also increased in the envelope and whole-cell fractions as the growth rate became faster. Similar results were obtained when dilution rates were decreased sequentially from 0.3 to 0.15 and 0.05 h−1 in reverse order (data not shown). Because estimation of the exact amounts of FimA and the 75-kDa protein in the CBB-stained gel was difficult because of faint bands, the amounts of them were compared by Western blotting as described below.
FIG. 1.
Comparison of SDS-PAGE patterns at various growth rates. Cells were grown in sTSB. Dilution rates were set at 0.05, 0.15, and 0.30 h−1. Samples for the culture supernatant fraction were prepared from the same volume of culture and loaded on the basis of the same volume. The same amount of protein (50 μg) was applied for the envelope and whole-cell lysate fractions. The gel was stained with CBB. Protein bands of major virulence factors are shown as follows: a, RagA; b, 75-kDa protein; c, Kgp; d, RagB, e, Rgps; f, OmpA-like proteins; g, FimA. Lane M, molecular mass marker.
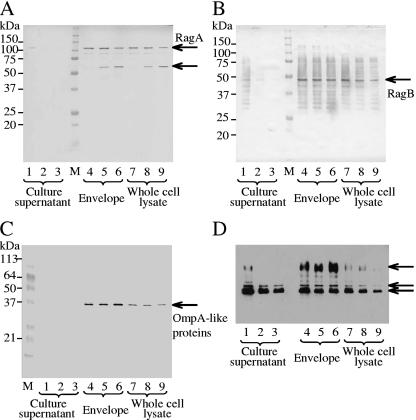
Expression of RagA and RagB in the envelope and whole-cell fractions was decreased at higher growth rates. The amounts of Rgps and Kgp in the envelope and whole-cell fractions did not clearly differ at the various dilution rates; however, those in the culture supernatant decreased as the growth rate increased. OmpA-like protein levels in the envelope and whole-cell fractions were almost constant, regardless of the dilution rate. It was noteworthy that only trace amounts of RagA, RagB, and OmpA-like proteins existed in the supernatant fraction. The above-mentioned major protein identification, based on mobility in SDS-PAGE, was carried out previously (41-43).
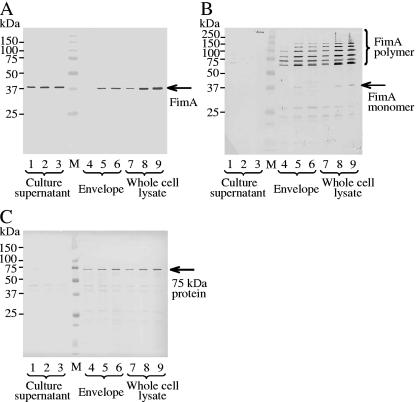
To confirm the results of SDS-PAGE, expression of major virulence factors was examined using specific antibodies against FimA (fimbrilin monomer), fimbriae (fimbrilin polymer), the 75-kDa protein, RagA, RagB, Rgps/Kgp, and the OmpA-like proteins (Fig. 2 and 3). Both FimA and the 75-kDa protein in the envelope and whole-cell lysate fractions clearly increased (more than twofold) at higher growth rates (Fig. 2A and C). An increase of FimA was also observed in the supernatant fraction. An antifimbria antibody was used to examine whether FimA was polymerized to fimbriae (Fig. 2B). In this case, samples were heated at 80°C for 20 min before SDS-PAGE. At each growth rate, high-molecular-weight forms of fimbriae were detected. The anti-RagA antibody revealed that a band near 110 kDa became weak (approximately 0.5-fold) with the increase of the growth rate, whereas a new band near 60 kDa, an apparent degraded product of RagA, became intense (Fig. 3A). Reaction against anti-RagB did not change in the envelope fraction but decreased in the whole-cell lysate fraction (Fig. 3B). A faster growth rate had little influence on expression of OmpA-like proteins in the envelope fraction, although the band appeared weaker at the highest growth rate (Fig. 3C). The amounts of Rgps and Kgp in the envelope fraction did not change; however, membrane-bound-type Rgps (mt-Rgps) somewhat increased at the dilution rate of 0.3 h−1 (Fig. 3D). In the whole-cell lysate and supernatant fractions, expression of Rgps, Kgp, and mt-Rgps decreased when the growth rate became higher. The intensities of mt-Rgps were stronger in the envelope fraction than in the whole-cell lysate fraction. Various mt-Rgp species having different lengths of carbohydrate chains, concentrated in the envelope fraction, were detected as fused, broad bands with the highly sensitive chemiluminescent method.
FIG. 2.
Comparison of amounts of FimA, fimbriae, and the 75-kDa protein at various growth rates by Western blotting. Blots were probed with anti-FimA antibody (1:3,000) (A), antifimbria antibody (1:3,000) (B), or anti-75-kDa protein antibody (1:8,000) (C). Cells were grown in sTSB. Dilution rates were set at 0.05, 0.15, and 0.30 h−1. The sample application for SDS-PAGE was essentially the same as that in Fig. 1, except that 20 μg of protein from the envelope fraction and the whole-cell lysate was used for detection of FimA and the 75-kDa protein and 30 μg was used for detection of fimbriae. For detection of fimbriae, samples were heated at 80°C for 20 min before SDS-PAGE. Lane M, molecular mass marker; lanes 1, 4, and 7, D = 0.05 h−1; lanes 2, 5, and 8, D = 0.15 h−1; lanes 3, 6, and 9, D = 0.30 h−1.
FIG. 3.
Comparison of amounts of RagA, RagB, OmpA-like proteins, and gingipains at various growth rates by Western blotting. Blots were probed with anti-RagA antibody (1:8,000) (A), anti-RagB antibody (1:8,000) (B), anti-OmpA-like protein antibody (1:8,000) (C), or anti-Rgp/Kgp antibody (1:2,000) (D). Cells were grown in sTSB. Dilution rates were set at 0.05, 0.15, and 0.30 h−1. The sample application was essentially the same as that in Fig. 1 and 2, except that 20 μg of protein from the envelope fraction and the whole-cell lysate was used for detection of RagA, RagB, and the OmpA-like proteins and 2 μg was used for detection of Rgp/Kgp. Lane M, molecular mass marker; lanes 1, 4, and 7, D = 0.05 h−1; lanes 2, 5, and 8, D = 0.15 h−1; lanes 3, 6, and 9, D = 0.30 h−1. Arrows in panel D indicate mt-Rgps, Kgp, and Rgps from top to bottom, respectively.
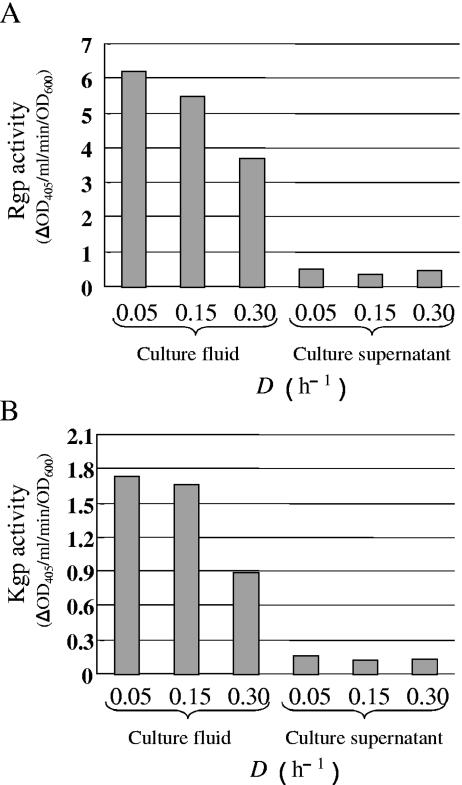
Rgp and Kgp enzyme activities at each growth rate were examined using specific artificial substrates. Culture fluid (containing intact cells plus the supernatant fluids) and cell-free culture supernatant in the steady state were used as samples. Rgp and Kgp activities in the culture fluid gradually decreased with the increase of the dilution rate (Fig. 4). Rgp and Kgp activities in the cell-free supernatant were more than five times lower than those in the culture fluid, indicating that most of their activities were associated with cells at all dilution rates.
FIG. 4.
Comparison of Rgp and Kgp activities at various growth rates. (A) Rgp activity of the culture fluid and cell-free culture supernatant; (B) Kgp activity of the culture fluid and cell-free culture supernatant. Cells were grown in sTSB. Dilution rates were set at 0.05, 0.15, and 0.30 h−1. Samples recovered after reaching the steady state were used for the enzyme assay. Duplicate enzyme assays were performed independently three times. A typical result is shown.
Effects of growth medium.
Continuous culture of P. gingivalis was performed at a fixed dilution rate of 0.05 h−1 using a nutrient-rich sTSB broth and nutrient-limited 0.3% BSA-MEM. The OD600 values in the culture vessels under the steady-state conditions were 1.4 in sTSB and 0.5 in 0.3% BSA-MEM.
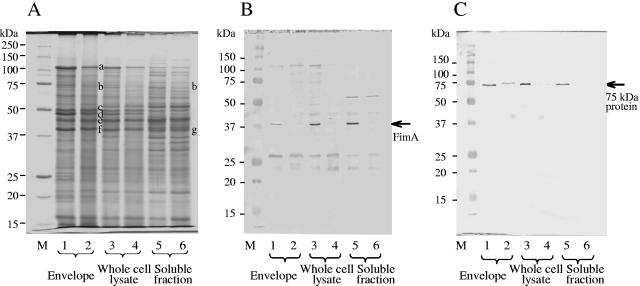
Protein profiles in each fraction were compared by SDS-PAGE (Fig. 5A). When changes in major outer membrane proteins were examined in the envelope and whole-cell lysate fractions, RagA and RagB decreased less than 0.5-fold in 0.3% BSA-MEM. In contrast, clear differences were not found in the amounts of Kgp, Rgps, and the OmpA-like proteins. These were confirmed by Western blotting (data not shown). The anti-FimA antibody detected a 37-kDa band in each cell fraction when the nutrient-rich sTSB medium was used; however, the band could not be detected when the nutrient-limited 0.3% BSA-MEM was used (Fig. 5B). Similarly, the anti-75-kDa protein antibody detected a band near 75 kDa in sTSB culture, whereas only a faint band was detected in 0.3% BSA-MEM culture (Fig. 5C).
FIG. 5.
Comparison of SDS-PAGE patterns and amounts of FimA and the 75-kDa protein by Western blotting in different culture media. sTSB was used as a nutrient-rich medium, and 0.3% BSA-MEM was the nutrient-poor medium. The dilution rate was set at 0.05 h−1. Fifty micrograms of sample was loaded per lane. (A) Stained with CBB; (B) probed with anti-FimA antibody (1:4,000); (C) probed with anti-75-kDa protein antibody (1:8,000). Lanes 1, 3, and 5, sTSB; lanes 2, 4, and 6, 0.3% BSA-MEM. Other symbols are the same as those indicated in Fig. 1.
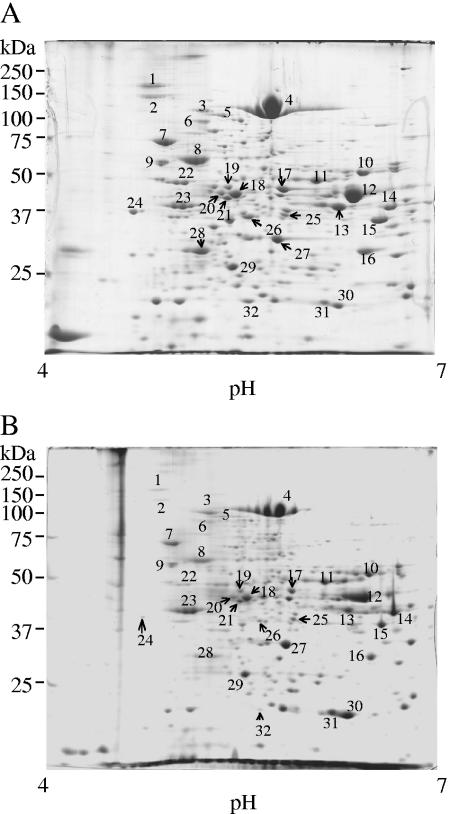
Whole-cell proteins in sTSB and 0.3% BSA-MEM were examined by 2-DE. The whole-cell samples were prepared from equal amounts of bacterial cells. Protein spot patterns after CBB staining are shown in Fig. 6. Generally, the number of detectable protein spots in 0.3% BSA-MEM was smaller than that in sTSB, and more than half of the stained spots were less intense. To identify proteins affected by the environmental change, major protein spots were excised and analyzed by MALDI-TOF MS after in-gel tryptic digestion. Altered expression of the identified proteins during growth in the nutrient-limited medium is shown in Table 1. The numbers of protein spots in Fig. 6 correspond to those in Table 1. Several spots remained unidentified, presumably because of strain variation between W83 (genome sequenced) and ATCC 33277 (genome sequence unavailable).
FIG. 6.
Comparison of 2-DE patterns of the whole-cell samples from different culture media. (A) sTSB as a nutrient-rich medium; (B) 0.3% BSA-MEM as a nutrient-limited medium. Growth medium was changed from sTSB to 0.3% BSA-MEM. The dilution rate was set at 0.05 h−1. Whole-cell samples prepared from the same cell numbers, measured on the basis of absorbance, were loaded. Gels were stained with CBB. The numbers refer to the proteins that have been identified (see Table 1).
TABLE 1.
Proteins with altered expression during growth in nutrient-limited medium
| Spot no.a | Proteinb | TIGR locusb | Fold changec |
|---|---|---|---|
| 1 | Unidentified | 0.15 | |
| 2 | Unidentified | 0.09 | |
| 3 | Peptidase, M16 family | PG0196 | 1.75 |
| 4d | RagA protein | PG0185 | 0.90 |
| 5 | Pyruvate phosphate dikinase | PG1017 | 1.02 |
| 6 | Translation elongation factor G | PG1940 | 0.69 |
| 7 | DnaK protein | PG1208 | 0.77 |
| 8 | Chaperonin, 60 kDa | PG0520 | 0.41 |
| 9 | Trigger factor, putative | PG0762 | 0.80 |
| 10 | 4-Hydroxybutyryl-coenzyme A dehydratase | PG0692 | 0.76 |
| 11 | GTP-binding protein Era | PG2142 | 1.31 |
| 12 | Glutamate dehydrogenase, NAD specific | PG1232 | 1.02 |
| 13 | Unidentified | 0.99 | |
| 14 | Immunoreactive 42-kDa antigen PG33 | PG0694 | 1.61 |
| 15 | Phosphoserine aminotransferase | PG1278 | 0.88 |
| 16 | d-Isomer-specific 2-hydroxy acid dehydrogenase family protein | PG1279 | 0.86 |
| 17 | 3-Oxoacyl-(acyl carrier protein) synthase II | PG1764 | 1.14 |
| 18e | Arginine-specific cystein proteinase | PG0506 | 1.21 |
| Hemagglutinin protein HagE | PG2024 | ||
| 19 | Tetratrico peptide repeat domain protein | PG0449 | 0.84 |
| 20e | Arginine-specific cystein proteinase | PG0506 | 1.12 |
| Hemagglutinin protein HagE | PG2024 | ||
| 21 | Enolase | PG1824 | 1.15 |
| 22 | v-type ATPase, subunit B | PG1804 | 0.32 |
| 23 | Tetratrico peptide repeat domain protein | PG1385 | 2.15 |
| 24 | Hypothetical protein | PG0027 | 0.10 |
| 25 | Methionine gamma-lyase | PG0343 | 0.95 |
| 26 | Electron transfer flavoprotein, alpha subunit | PG0776 | 0.33 |
| 27 | Fructose-bisphosphate aldolase, class I | PG1755 | 1.94 |
| 28 | Electron transfer flavoprotein, beta subunit | PG1077 | 0.54 |
| 29 | Translation elongation factor Ts | PG0378 | 1.19 |
| 30 | Superoxide dismutase, Fe-Mn | PG1545 | 3.98 |
| 31 | Secretion activator protein, putative | PG0293 | 2.63 |
| 32 | Alkyl hydroperoxide reductase, C subunit | PG0618 | 0.07 |
Spot numbers are identical to those in Fig. 6.
Annotation and locus names are from The Institute for Genomic Research (TIGR; http://www.tigr.org/).
Fold change was calculated as the ratio of the spot intensity of a protein grown in the nutrient-limited medium to that of the same protein grown in the nutrient-rich medium.
This protein was identified by a comparison with the sequence of the RagA region of P. gingivalis ATCC 33277 performed at the Department of Microbiology, School of Dentistry, Aichi-Gakuin University (data not shown).
Either protein is possible.
Effect of aeration.
Continuous culture was performed in sTSB broth at the medium dilution rate of 0.15 h−1 under anaerobic and aerated conditions. Bacterial densities (OD600) in the steady state were 1.8 and 1.1 under anaerobic and aerated conditions, respectively.
When protein patterns of cellular components under each condition were examined by SDS-PAGE, few differences were observed in amounts of RagA, RagB, and the OmpA-like proteins. Expression of Rgps and Kgp was slightly lower (approximately 0.8-fold) under the aerated conditions (Fig. 7). Western blotting showed little difference in the amounts of FimA and the 75-kDa protein between the two conditions (data not shown).
FIG. 7.
Effect of aeration on SDS-PAGE pattern. Cells were grown in sTSB at a dilution rate of 0.15 h−1. Fractions derived from cells under anaerobic and aerobic conditions were examined by SDS-PAGE. The samples (50 μg each) were applied onto an SDS-PAGE gel, and the gel was stained with CBB. Lanes 1, 3, and 5, results under anaerobic conditions; lanes 2, 4, and 6, results under aerated conditions. Other symbols are the same as those indicated in Fig. 1.
Rgp activity in both the culture fluid and cell-free supernatant fractions under the anaerobic conditions was approximately eightfold higher than that under aeration (data not shown). Kgp activity in the culture fluid was also approximately 8-fold higher and that in the cell-free supernatant was about 2.5-fold higher under anaerobic conditions than when aerated, suggesting that the major proteinases could be inactivated rather than repressed by oxygen stress.
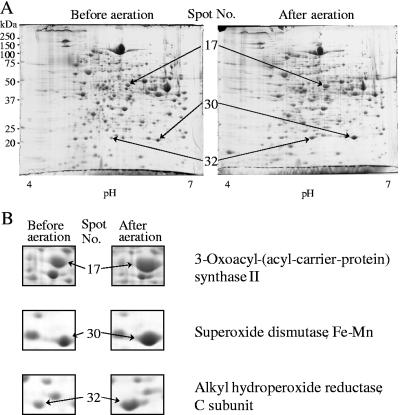
Figure 8A shows 2-DE patterns of whole-cell proteins after CBB staining. The intensities of many protein spots under both conditions were almost identical or slightly decreased under the aerobic conditions; however, several spots increased in intensity. Three spots that significantly increased more than 2.5-fold were chosen for further analysis by MALDI-TOF MS (Fig. 8B). The proteins were identified as 3-oxoacyl-(acyl-carrier-protein) synthase II (about 45 kDa; spot no. 17), superoxide dismutase, Fe-Mn (about 21 kDa; spot no. 30), and alkyl hydroperoxide reductase, C subunit (about 21 kDa; spot no. 32).
FIG. 8.
Effect of aeration on the 2-DE pattern of the whole-cell sample and identification of protein spots whose amounts were increased after aeration. (A) 2D-PAGE was performed using the whole-cell samples from anaerobic and aerated conditions. Cells were grown in sTSB at a dilution rate of 0.15 h−1. Samples were prepared from the same cell numbers, as described for Fig. 6. After electrophoresis, gels were stained with CBB. Numbers of protein spots were identical to those in Fig. 6. (B) Enlarged images and identification of protein spots which clearly increased after aeration.
DISCUSSION
P. gingivalis is the most extensively investigated bacterium related to periodontal disease. Many studies have been performed concerning the relationship between expression of virulence factors and environmental factors using batch culture (15, 41, 43). Previous pioneering studies (12-14, 35, 39, 40, 57-59) have reported application of continuous culture to P. gingivalis. However, effects on virulence factors such as fimbriae and major outer membrane proteins were not fully investigated.
Bacterial growth in the oral environment is much slower than that under conventional batch culture. It was reported that supragingival plaques rich in facultative anaerobes had a doubling time of 1 to 66 h (corresponding to dilution rates of 0.69 to 0.01 h−1) (22). Thus, it is reasonable to speculate that fastidious anaerobic bacteria in subgingival plaque would grow slower than supragingival plaque bacteria. The average growth rate of P. gingivalis in the mouth is considered to be much slower than the value at which bacterial wash-out would occur (36). Since the culture was washed out at the dilution rate of 0.35 h−1 in our preliminary experiment, 0.30 h−1 was considered the highest practical dilution rate, although the actual range of the growth rate is still unknown.
SDS-PAGE and Western blotting showed that FimA and the 75-kDa protein increased markedly in each fraction as the dilution rate increased (Fig. 1 and 2). FimA was also shown to exist as a form of fibrous fimbriae at all dilution rates (Fig. 2B). When P. gingivalis grows faster, expression of fimbriae may increase to attach and invade host cells; when the growth rate is slower, P. gingivalis may have reduced production of fimbriae. Although fimbriae are very important cellular components, high energy consumption is required for their production.
As the growth rate became higher, expression of the RagA protein (110 kDa) was reduced. Although an approximately 60-kDa band, an apparent degraded product of RagA, increased (Fig. 1 and 3A), the sum of the 110- and 60-kDa band intensities appeared to be reduced. Expression of RagB was also reduced in the whole-cell lysate fraction (Fig. 1 and 3B). It has been reported that ragA and ragB form a temperature-regulated operon (7); thus, it was no surprise that RagA and RagB varied simultaneously.
OmpA-like proteins were expressed in a large amount independent of the growth rate (Fig. 1 and 3C). Since the OmpA-like proteins possess eight-stranded β-barrels in the N-terminal region and the peptidoglycan binding region in the C-terminal region is similar to that of Escherichia coli OmpA, the proteins are mainly thought to play a role in stabilization of the outer membrane, maintaining the bacterial surface structure.
RgpA and RgpB as well as Kgp enzymes are strong trypsin-like cysteine proteases, and they are thought to cooperate in protein digestion for acquisition of nutrients, maturation of FimA and the 75-kDa protein, and processing of Rgps and Kgp themselves. Gingipains could directly disrupt periodontal tissues, such as collagen and fibronectin. Disruption of immunoglobulin, cytokines, and the complement system and inhibition of phagocytosis in polymorphonuclear leukocytes would disturb the immune system, enabling evasion of the host defense system (27, 47). Moreover, gingipains induce apoptosis of gingival fibroblasts and endothelial cells (5, 6). These functions are implicated in development of periodontal disease. The amounts of Rgps and Kgp were slightly reduced at the higher growth rate (Fig. 1 and 3D), whereas their activities in culture fluid were considerably reduced (Fig. 4). Marsh et al. reported that the faster the growth rate, the greater the Rgp activity (35). The reason for this discrepancy remains unclear, but it may be related to the different bacterial strains, culture media, and dilution rates used. It is thought that P. gingivalis will grow at a relatively slower rate when attached to gingival epithelium or when coexisting with various bacteria forming a biofilm. Under these conditions, increased gingipain activity may be of help to the invasion of cells or acquisition of nutrients. Western blotting showed that a large amount of mt-Rgps existed in the envelope fraction at a high-molecular-mass position at all three growth rates (Fig. 3D). Although the significance of mt-Rgps is still unknown, it is not clear why mt-Rgps were produced at a high level under all the conditions.
Since it is technically difficult to sterilize a high concentration of BSA solution by filtration, we used 0.3% BSA, a relatively low concentration, in the nutrient-poor medium. Continuous culture in this medium is considered to be in keeping with the relatively nutrient-limited condition of the oral cavity.
The amounts of RagA and RagB were lower in 0.3% BSA-MEM than in sTSB, based on comparison of the band intensities in SDS-PAGE. Nevertheless, these proteins were still produced at high levels similar to other major proteins (Fig. 5A). Expression of Rgps, Kgp, and OmpA-like proteins was almost unchanged in both media. Since major outer membrane proteins, including RagA and RagB, are considered important for maintaining bacterial cells, they would be produced in large quantities under any circumstances.
Western blotting showed that expression of FimA and the 75-kDa protein was decreased in 0.3% BSA-MEM (Fig. 5B and C). Since P. gingivalis is known to utilize dipeptides digested from proteins with its own proteinases (62), it is possible that production of FimA and the 75-kDa protein is negatively regulated by a shortage of essential dipeptides. Thus, during growth in medium containing a limited amount of peptides, production of FimA and the 75-kDa protein decreased, suggesting that a stringent control response might negatively regulate their expression.
Whole-cell components in sTSB and 0.3% BSA-MEM were compared by 2-DE (Fig. 6; Table 1). Although samples were prepared from the same number of cells to be compared quantitatively, there were fewer, less intense spots in 0.3% BSA-MEM. Since 0.3% BSA-MEM was nutrient poor, the amount of proteins in cells decreased. In contrast, some spots, such as that of superoxide dismutase, Fe-Mn (spot no. 30), increased in intensity.
Proteins were identified after peptide mass fingerprinting based on the database of P. gingivalis W83 on The Institute for Genomic Research website (http://www.tigr.org/). Since we used P. gingivalis ATCC 33277 in this study, a few spots were unidentified (Table 1). Most of the proteins identified were putative and hypothetical proteins whose functions in P. gingivalis are unknown. We intend to continue analysis of the protein spots that have not yet been examined by mass spectrometry.
Expression of virulence factors in cells under the anaerobic and aerated steady-state conditions was compared. Since the oral cavity is directly open to the external environment, P. gingivalis should occasionally be exposed to aerobic conditions. It has been reported that neutrophils in saliva, but not in peripheral blood, always produce active oxygen molecules (66). Active oxygen molecules produced by phagocytes in gingival epithelial cells (32) may have inhibitory effects on invading P. gingivalis. Therefore, it is of interest to examine the reaction of the strict anaerobe P. gingivalis to oxygen stress.
Expression of RagA, RagB, and the OmpA-like proteins did not show much difference between the anaerobic and aerated conditions. These major outer membrane proteins seemed to be expressed in quantity even under unfavorable conditions, such as aeration (Fig. 7). Expression of FimA was slightly reduced under the aerated conditions, while that of the 75-kDa protein was almost constant (data not shown). Thus, the aeration rate used in this study (100 ml/min) had little effect on expression of fimbriae.
When cellular proteins were compared by 2-DE, most protein spots were comparable to each other in cells under anaerobic and aerated conditions, though some were reduced under aerated conditions (Fig. 8). We focused on three spots that increased markedly, which were identified as 3-oxoacyl-(acyl-carrier-protein) synthase II, superoxide dismutase, Fe-Mn, and alkyl hydroperoxide reductase, C subunit. 3-Oxoacyl-(acyl-carrier-protein) synthase II is known as beta-ketoacyl-ACP synthase II (FabF) in fatty acid biosynthesis of E. coli (10, 17, 18). The homolog of FabF detected in P. gingivalis could be involved in the mechanism that modifies the bacterial membrane to “a state” more resistant to oxidative circumstances, although no report indicating a relation between bacterial membrane fluidity and air tolerance was found. There is a report that describes morphological changes in P. gingivalis cells under oxygen stress (12).
Superoxide dismutase (SOD) is an enzyme that converts superoxide to hydrogen peroxide (19, 37, 38). Since oxygen resistance in anaerobes depends on the existence of SOD, several studies characterized SOD in P. gingivalis (3, 34, 45, 46). The present study confirmed the induction of SOD by exposure to air (46).
Alkyl hydroperoxide reductase, C subunit (AhpC) induced here is known to contribute to tolerance to reactive oxygen with another F subunit (AhpF) (14, 29, 53). Their genes, ahpCF, are organized in an operon (14, 54). AhpCF was reported not only in facultative anaerobes (29, 48, 49) but also in strict anaerobes, i.e., in Bacteroides fragilis (54), as well as P. gingivalis (14). Although P. gingivalis does not have catalase, it is known to be fairly aerotolerant. AhpC could contribute to this activity together with SOD, also detected here, although AhpF was not identified. More work is needed to identify other components involved in oxygen tolerance.
In this study, we examined the effects of growth rate, nutrition, and oxygen on the production of virulence factors in P. gingivalis using continuous culture in a chemostat. We showed that the amount of P. gingivalis fimbriae increased at higher growth rates and was reduced with limited nutrients. Although the production of specific proteins increased under oxygen stress, the expression of major outer membrane proteins was largely unaffected by environmental conditions. Complex factors, such as numerous bacterial species, salivary components, and gingival exudates, exist in the oral cavity. Nevertheless, this study provides important clues to the physiological character of P. gingivalis. As P. gingivalis ATCC 33277 constantly produces a large amount of fimbriae even after long successive culture, this strain is often used for fimbrial study. So far, previous studies reported that fimbriae were regulated by temperature with this strain (3, 65). Thus, this is the first report showing that the expression of fimbriae is influenced by other environmental factors, i.e., growth rate and nutrients.
Acknowledgments
Part of this work is taken from the thesis submitted by Takashi Masuda to the Graduate Faculty, School of Dentistry, Aichi-Gakuin University, in partial fulfillment of the requirements for his doctoral degree.
We thank M. Homma (Graduate School of Science, Nagoya University) for instruction on mass fingerprinting. We also thank Y. Nakano and H. Fukamachi (Graduate School of Dental Science, Kyushu University) for technical advice on 2-DE analysis.
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (15591957 to F.Y. and 15591958 to Y.M.) and the AGU High-Tech Research Center Project for Private Universities matching fund subsidy from the Ministry of Education, Culture, Sports, Science and Technology, Japan, 2003-2007.
REFERENCES
- 1.Albandar, J. M. 2002. Periodontal diseases in North America. Periodontology 2000 29:31-69. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., T. Fujiwara, H. Nagata, M. Kuboniwa, A. Sharma, H. T. Sojar, R. J. Genco, S. Hamada, and S. Shizukuishi. 1997. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J. Dent. Res. 76:852-857. [DOI] [PubMed] [Google Scholar]
- 3.Amano, A., A. Sharma, H. T. Sojar, H. K. Kuramitsu, and R. J. Genco. 1994. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect. Immun. 62:4682-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano, A., S. Shizukuishi, H. Horie, S. Kimura, I. Morisaki, and S. Hamada. 1998. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoprotein in parotid saliva via a domain shared by major salivary components. Infect. Immun. 66:2072-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba, A., N. Abe, T. Kadowaki, H. Nakanishi, M. Oishi, T. Asao, and K. Yamamoto. 2001. Arg-gingipain is responsible for the degradation of cell adhesion molecules of human gingival fibroblasts and their death induced by Porphyromonas gingivalis. Biol. Chem. 382:817-824. [DOI] [PubMed] [Google Scholar]
- 6.Baba, A., T. Kadowaki, T. Asao, and K. Yamamoto. 2002. Roles for Arg- and Lys-gingipains in the disruption of cytokine responses and loss of viability of human endothelial cells by Porphyromonas gingivalis infection. Biol. Chem. 383:1223-1230. [DOI] [PubMed] [Google Scholar]
- 7.Bonass, W. A., P. D. Marsh, R. S. Percival, J. Aduse-Opoke, S. A. Hanley, D. A. Devine, and M. A. Curtis. 2000. Identification of ragAB as a temperature-regulated operon of Porphyromonas gingivalis W50 using differential display of randomly primed RNA. Infect. Immun. 68:4012-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, N. G., and R. S. Hirsch. 1995. Personal risk factors for generalized periodontitis. J. Clin. Periodontol. 22:136-145. [DOI] [PubMed] [Google Scholar]
- 10.Cronan, J. E., Jr., and C. O. Rock. 1996. Biosynthesis of membrane lipid, p. 612-636. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 11.Curtis, M. A., S. A. Hanley, and J. Aduse-Opoke. 1999. The rag locus of Porphyromonas gingivalis: novel pathogenicity island. J. Periodont. Res. 34:400-405. [DOI] [PubMed] [Google Scholar]
- 12.Diaz, P. I., and A. H. Rogers. 2004. The effect of oxygen on the growth and physiology of Porphyromonas gingivalis. Oral Microbiol. Immunol. 19:88-94. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, P. I., P. S. Zilm, and A. H. Rogers. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology 148:467-472. [DOI] [PubMed] [Google Scholar]
- 14.Diaz, P. I., P. S. Zilm, V. Wasinger, G. L. Corthals, and A. H. Rogers. 2004. Studies on NADH oxidase and alkyl hydroperoxide reductase produced by Porphyromonas gingivalis. Oral Microbiol. Immunol. 19:137-143. [DOI] [PubMed] [Google Scholar]
- 15.Forng, R. Y., C. Champagne, W. Simpson, and C. A. Genco. 2000. Environmental cues and gene expression in Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Oral Dis. 6:351-365. [DOI] [PubMed] [Google Scholar]
- 16.Fountoulakis, M., and H. Langen. 1997. Identification of proteins by matrix-assisted laser desorption ionization-mass spectrometry following in-gel digestion in low-salt, nonvolatile buffer and simplified peptide recovery. Anal. Biochem. 250:153-156. [DOI] [PubMed] [Google Scholar]
- 17.Garwin, J. L., A. L. Klages, and J. E. Cronan, Jr. 1980. Structural, enzymatic, and genetic studies of β-ketoacyl-acyl carrier protein synthases I and II of Escherichia coli. J. Biol. Chem. 255:11949-11956. [PubMed] [Google Scholar]
- 18.Garwin, J. L., A. L. Klages, and J. E. Cronan, Jr. 1980. β-Ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J. Biol. Chem. 255:3263-3265. [PubMed] [Google Scholar]
- 19.Gregory, E. M., and I. Fridovich. 1973. Oxygen toxicity and the superoxide dismutase. J. Bacteriol. 114:1193-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontology 2000 5:78-111. [DOI] [PubMed] [Google Scholar]
- 21.Hamada, N., H. T. Sojar, M. I. Cho, and R. J. Genco. 1996. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect. Immun. 64:4788-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton, I. R., A. S. McKee, and G. H. Bowden. 1989. Growth and metabolic properties of Bacteroides intermedius in anaerobic continuous culture. Oral Microbiol. Immunol. 4:89-97. [DOI] [PubMed] [Google Scholar]
- 23.Hanazawa, S., Y. Kawata, Y. Murakami, K. Naganuma, S. Amano, Y. Miyata, and S. Kitano. 1995. Porphyromonas gingivalis fimbria-stimulated bone resorption in vitro is inhibited by a tyrosine kinase inhibitor. Infect. Immun. 63:2374-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanazawa, S., K. Hirose, Y. Ohmori, S. Amano, and S. Kitano. 1988. Bacteroides gingivalis fimbriae stimulate production of thymocyte-activating factor by human gingival fibroblasts. Infect. Immun. 56:272-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa, Y., S. Nishiyama, K. Nishikawa, T. Kadowaki, K. Yamamoto, T. Noguchi, and F. Yoshimura. 2003. A novel type of two-component regulatory system affecting gingipains in Porphyromonas gingivalis. Microbiol. Immunol. 47:849-858. [DOI] [PubMed] [Google Scholar]
- 26.Holt, S. C., L. Kesavalu, S. Walkers, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontology 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 27.Imamura, T. 2003. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74:111-118. [DOI] [PubMed] [Google Scholar]
- 28.Isogai, H., E. Isogai, F. Yoshimura, T. Suzuki, W. Kagota, and K. Takano. 1988. Specific inhibition of adherence of an oral strain of Bacteroides gingivalis 381 to epithelial cells by monoclonal antibodies against the bacterial fimbriae. Arch. Oral Biol. 33:479-485. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson, F. S., R. W. Morgan, M. F. Christman, and B. N. Ames. 1989. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage: purification and properties. J. Biol. Chem. 264:1488-1496. [PubMed] [Google Scholar]
- 30.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamont, R. J., and H. F. Jenkinson. 2000. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15:341-349. [DOI] [PubMed] [Google Scholar]
- 32.Listgarten, M. A. 1972. Normal development, structure, physiology and repair of gingival epithelium. Oral Sci. Rev. 1:3-67. [PubMed] [Google Scholar]
- 33.Lugtenberg, B., J. Meijers, R. Peters, P. van der Hoek, and L. van Alphen. 1975. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 58:254-258. [DOI] [PubMed] [Google Scholar]
- 34.Lynch, M. C., and H. K. Kuramitsu. 1999. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect. Immun. 67:3367-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh, P. D., A. S. McDermid, A. S. McKee, and A. Baskerville. 1994. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology 140:861-865. [DOI] [PubMed] [Google Scholar]
- 36.Marsh, P. D., A. S. McKee, and A. S. McDermid. 1993. Continuous culture studies, p. 105-126. In H. N. Shah, D. Mayrand, and R. J. Genco (ed.), Biology of the species Porphyromonas gingivalis. CRC Press, Boca Raton, Fla.
- 37.McCord, J. M. 1974. Free radicals and inflammation: protection of synovial fluid by superoxide dismutase. Science 185:529-531. [DOI] [PubMed] [Google Scholar]
- 38.McCord, J. M., B. B. Keele, Jr., and I. Fridovich. 1971. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc. Natl. Acad. Sci. USA 68:1024-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDermid, A. S., A. S. McKee, and P. D. Marsh. 1988. Effect of environmental pH on enzyme activity and growth of Bacteroides gingivalis W50. Infect. Immun. 56:1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKee, A. S., A. S. McDernid, A. Baskerville, A. B. Dowsett, D. C. Ellwood, and P. D. Marsh. 1986. Effect of hemin on the physiology and virulence of Bacteroides gingivalis W50. Infect. Immun. 52:349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami, Y., M. Imai, Y. Mukai, S. Ichihara, H. Nakamura, and F. Yoshimura. 2004. Effects of various culture environments of major outer membrane proteins from Porphyromonas gingivalis. FEMS Microbiol. Lett. 230:159-165. [DOI] [PubMed] [Google Scholar]
- 42.Murakami, Y., M. Imai, H. Nakamura, and F. Yoshimura. 2002. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 110:157-162. [DOI] [PubMed] [Google Scholar]
- 43.Murakami, Y., T. Masuda, M. Imai, J. Iwami, H. Nakamura, T. Noguchi, and F. Yoshimura. 2004. Analysis of major virulence factors in Porphyromonas gingivalis under various culture temperatures using specific antibodies. Microbiol. Immunol. 48:561-569. [DOI] [PubMed] [Google Scholar]
- 44.Nagano, K., E. K. Read, Y. Murakami, T. Masuda, T. Noguchi, and F. Yoshimura. 2005. Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis. J. Bacteriol. 187:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayama, K. 1990. The superoxide dismutase-encoding gene of the obligately anaerobic bacterium Bacteroides gingivalis. Gene 96:149-150. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama, K. 1994. Rapid viability loss on exposure to air in a superoxide dismutase-deficient mutant of Porphyromonas gingivalis. J. Bacteriol. 176:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama, K. 2003. Molecular genetics of Porphyromonas gingivalis: gingipains and other virulence factors. Curr. Protein Pept. Sci. 4:389-395. [DOI] [PubMed] [Google Scholar]
- 48.Niimura, Y., Y. Nishiyama, D. Saito, H. Tsuji, M. Hidaka, T. Miyaji, T. Watanabe, and V. Massey. 2000. A hydrogen peroxide-forming NADH oxidase that functions as an alkyl hydroperoxide reductase in Amphibacillus xylanus. J. Bacteriol. 182:5046-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishiyama, Y., V. Massey, K. Takeda, S. Kawasaki, J. Sato, T. Watanabe, and Y. Niimura. 2001. Hydrogen peroxide-forming NADH oxidase belonging to the peroxiredoxin oxidoreductase family: existence and physiological role in bacteria. J. Bacteriol. 183:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novick, A., and L. Szilard. 1950. Description of the chemostat. Science 112:715-716. [DOI] [PubMed] [Google Scholar]
- 51.Park, Y., M. R. Simionato, K. Sekiya, Y. Murakami, D. James, W. Chen, M. Hackett, F. Yoshimura, D. R. Demuth, and R. J. Lamont. 2005. The short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 53.Poole, L. B., C. M. Reynolds, Z. A. Wood, P. A. Karplus, H. R. Ellis, and M. Li Calzi. 2000. AhpF and other NADH: peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267:6126-6133. [DOI] [PubMed] [Google Scholar]
- 54.Rocha, E. R., and C. J. Smith. 1999. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 181:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 56.Slots, J., L. Bragd, M. Wikstrom, and G. Dahlen. 1986. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J. Clin. Periodontol. 13:570-577. [DOI] [PubMed] [Google Scholar]
- 57.Smalley, J. W., A. J. Birss, A. S. McKee, and P. D. Marsh. 1996. Haemin binding as a factor in the virulence of Porphyromonas gingivalis. FEMS Microbiol. Lett. 141:65-70. [DOI] [PubMed] [Google Scholar]
- 58.Smalley, J. W., A. J. Birss, A. S. McKee, and P. D. Marsh. 1998. Hemin regulation of hemoglobin binding by Porphyromonas gingivalis. Curr. Microbiol. 36:102-106. [DOI] [PubMed] [Google Scholar]
- 59.Smalley, J. W., A. J. Birss, R. Percival, and P. D. Marsh. 2000. Temperature elevation regulates iron protoporphyrin IX and hemoglobin binding by Porphyromonas gingivalis. Curr. Microbiol. 41:328-335. [DOI] [PubMed] [Google Scholar]
- 60.Socransky, S. S., A. D. Haffajee, M. A. Caging, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 61.Socransky, S. S., A. D. Haffajee, J. M. Goodson, and J. Lindhe. 1984. New concepts of destructive periodontal disease. J. Clin. Periodontol. 11:21-32. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi, N., and T. Sato. 2001. Preferential utilization of dipeptides by Porphyromonas gingivalis. J. Dent. Res. 80:1425-1429. [DOI] [PubMed] [Google Scholar]
- 63.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie, H., S. Cai, and R. J. Lamont. 1997. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect. Immun. 65:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto, M., K. Saeki, and K. Utsumi. 1991. Isolation of human salivary polymorphonuclear leukocytes and their stimulation-coupled responses. Arch. Biochem. Biophys. 289:76-82. [DOI] [PubMed] [Google Scholar]
- 67.Yoshimura, F., K. Takahashi, Y. Nodasaka, and T. Suzuki. 1984. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J. Bacteriol. 160:949-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshimura, F., K. Watanabe, T. Takasawa, M. Kawanami, and H. Kato. 1989. Purification and properties of a 75-kilodalton major protein, an immunodominant surface antigen, from the oral anaerobe Bacteroides gingivalis. Infect. Immun. 57:3646-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]