Abstract
A bacteriocin-producing strain, Lactococcus lactis QU 4, was isolated from corn. The bacteriocin, termed lactococcin Q, showed antibacterial activity only against L. lactis strains among a wide range of gram-positive indicator strains tested. Lactococcin Q was purified by acetone precipitation, cation exchange chromatography, and reverse-phase chromatography. Lactococcin Q consisted of two peptides, α and β, whose molecular masses were determined to be 4,260.43 Da and 4,018.36 Da, respectively. Amino acid and DNA sequencing analyses revealed that lactococcin Q was a novel two-peptide bacteriocin, homologous to lactococcin G. Comparative study using chemically synthesized lactococcin Q (Qα plus Qβ) and lactococcin G (Gα plus Gβ) clarified that hybrid combinations (Qα plus Gβ and Gα plus Qβ) as well as original combinations showed antibacterial activity, although each single peptide showed no significant activity. These four pairs of lactococcin peptides acted synergistically at a 1:1 molar ratio and exhibited identical antibacterial spectra but differed in MIC. The MIC of Qα plus Gβ was 32 times higher than that of Qα plus Qβ, suggesting that the difference in β peptides was important for the intensity of antibacterial activity.
Some strains of lactic acid bacteria (LAB) produce ribosomally synthesized antimicrobial peptides or proteins, termed bacteriocins, which generally inhibit the growth of closely related strains. Because of their potential use as food preservatives and pharmaceuticals, LAB bacteriocins have been widely explored and studied in recent years (3).
On the basis of their structures, LAB bacteriocins are classified mainly into two groups: lantibiotic (class I) and nonlantibiotic (class II) (13). Class I bacteriocins contain posttranslationally modified unusual amino acids, like lanthionine and dehydrated amino acids. In contrast, class II bacteriocins comprise only unmodified amino acid residues. Class II bacteriocins are further classified into three subclasses, IIa, IIb, and IIc, by their detailed characteristics (21, 22).
Most LAB bacteriocins were found as single active peptides, but some of them were found as two-peptide systems (9). Each two peptides are encoded by adjacent open reading frames (ORFs) in the same operon, transcribed, and synthesized together. They are fully active generally in consequence of the synergistic action of the two peptides, whereas each peptide alone has no activity or less activity. In all cases studied so far, the optimal combination ratio of the peptides is approximately 1:1. Two-peptide bacteriocins are found in both lantibiotics and unmodified bacteriocins. Unmodified two-peptide bacteriocins are classified into class IIb, although two-peptide lantibiotics have not been categorized into a defined class. Recently many class IIb bacteriocins have been discovered from LAB: e.g., ABP-118 (6), plantaricin NC8 (14), and enterocin 1071 (2, 7).
The first reported one was lactococcin G produced by Lactococcus lactis subsp. lactis LMG 2081 (23). Its activity, structure, and mode of action have been examined extensively. It was reported that lactococcin G was active against various LAB and clostridia and that the constituents, α and β peptides, acted synergistically at a 1:1 molar ratio. In addition, it was suggested that both peptides of lactococcin G formed amphiphilic α-helices and that their complex showed bactericidal activity by forming a transmembrane pore that conducted potassium ions (10, 17, 18). However, the recognition of target cells and partner peptides is still unclear.
Until now, only one homolog of lactococcin G, namely enterocin 1071, produced by Enterococcus faecalis BFE 1071 and E. faecalis FAIR-E 309, has been reported (2, 7). The primary structure of enterocin 1071, consisting of enterocin 1071A and 1071B, was reported firstly by Balla et al. (2) and corrected by Franz et al. (7). Enterocin 1071 inhibited the growth of the wide range of gram-positive bacteria, especially enterococcal strains. In addition, the peptides were presumed to have amphiphilic α-helices in their structures as in the case of lactococcin G.
We have screened a variety of plant sources in Japan and isolated various bacteriocin-producing LAB, including 11 strains from corn. Among the new corn isolates, five strains, including Lactococcus lactis QU 4, produced a bacteriocin with a specific antibacterial spectrum. In this study, we described purification, structural analysis, and characterization of a novel two-peptide bacteriocin, named lactococcin Q, produced by L. lactis QU 4.
MATERIALS AND METHODS
Bacterial strains and media.
The bacteriocin-producing strain, L. lactis QU 4 was isolated from fresh corn grown in Aso, Kumamoto, Japan. L. lactis QU 4 and the indicator strain, L. lactis subsp. lactis ATCC 19435T, were stored at −80°C in MRS medium (Oxoid, Basingstoke, United Kingdom) with 15% glycerol and propagated in MRS medium at 30°C for 18 h before use. The other indicator strains for determination of antibacterial spectra were propagated at appropriate temperatures (30 or 37°C) recommended by culture collections for 18 h before use. MRS medium was used to culture LAB strains, except Streptococcus sp. strains. Tryptic soy broth (Difco, Sparks, Md.) supplemented with 0.6% yeast extract (Nacalai Tesque, Kyoto, Japan) was used to culture Streptococcus strains and the other gram-positive indicator strains. Escherichia coli JM109 was grown in LB medium (1) at 37°C. Transformants of E. coli JM109 were selected on LB agar plates containing 50 mg liter−1 of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 40 mg liter−1 of isopropyl-β-d-thiogalactopyranoside, and 20 mg liter−1 of ampicillin.
Identification of strain QU 4.
The sugar fermentation pattern of strain QU 4 was tested by the API 50 CHL system (bioMérieux, Marcy l'Etoile, France). The obtained pattern was analyzed by using the APILAB Plus software (bioMérieux). A partial 16S rRNA gene region of strain QU 4, corresponding to positions 8 to 519 of E. coli 16S rRNA gene, was analyzed as described previously (29).
Determination of bacteriocin activity.
Bacteriocin activity was determined by the spot-on-lawn method as described previously (5, 16) using L. lactis subsp. lactis ATCC 19435T as an indicator strain unless otherwise mentioned. Briefly, 10 microliters of a bacteriocin preparation were spotted onto a double-layered agar plate comprised of 5 ml of Lactobacilli Agar AOAC (Difco) inoculated with an overnight culture of an indicator strain as an upper layer and 10 ml of MRS medium with 1.2% agar as a lower layer. After overnight incubation at appropriate temperatures for the indicator strains, bacterial lawns were checked for inhibition zones.
Purification of lactococcin Q.
L. lactis QU 4 was grown at 30°C for 12 h in 250 ml of MRS medium, and the culture supernatant was obtained by centrifugation. Proteins including lactococcin Q in the supernatant were precipitated with 750 ml of acetone at −30°C overnight. The pellet obtained by centrifugation at 4°C was dried under vacuum and dissolved in 50 ml of 20 mM sodium phosphate buffer, pH 5.7 (buffer A). This solution was applied at a flow rate of 2 ml min−1 to a SP-Sepharose cation-exchange column (100-mm length, 15-mm internal diameter; Amersham Pharmacia Biotech, Uppsala, Sweden) equilibrated with buffer A. After a subsequent washing with 20 ml of buffer A, the active fraction containing lactococcin Q was eluted with a stepwise gradient consisting of 20 ml each of 0.5, 0.75, and 1 M NaCl in buffer A. The active fraction obtained was then applied to a reverse-phase column (Resource RPC 3-ml; Amersham Pharmacia Biotech) incorporated in LC-10A high-performance liquid chromatography (HPLC) system (Shimadzu, Kyoto, Japan) and equilibrated with 20% acetonitrile containing 0.1% trifluoroacetic acid (TFA). Lactococcin Q was eluted with a linear gradient ranging from 20 to 50% acetonitrile containing 0.1% TFA for 30 min at a flow rate of 1 ml min−1. Purified lactococcin Q was stored at −30°C. The antibacterial activity of each fraction in the purification steps was determined as described above.
Mass spectrometry and amino acid sequence analysis of lactococcin Q.
The molecular masses of the purified lactococcin Q peptides were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) using a Voyager DE-STR mass spectrometer (Applied Biosystems, Foster City, Calif.) in the linear positive-ion mode and α-cyano-4-hydroxycinnamic acid (Sigma, St. Louis, Mo.) as a matrix. The N-terminal amino acid sequences of the peptides were determined by Edman degradation using a 476A gas-phase automatic sequencer (Applied Biosystems).
Analysis of the gene encoding lactococcin Q.
DNA polymerases, restriction enzymes, and other DNA modifying enzymes were used as recommended by the manufacturers. The total genomic DNA of L. lactis QU 4 was extracted from the cells treated with lysozyme (Seikagaku, Tokyo, Japan) and labiase (Seikagaku) as described previously (1) and used as a template for PCR. All primers used to analyze the gene encoding lactococcin Q are listed in Table 1. Two degenerate primers, lcnQ1 and lcnQ2, designed from the obtained amino acid sequences of lactococcin Qα and Qβ, respectively, were used to amplify a fragment corresponding to a part of the gene encoding lactococcin Q. Degenerate PCR was performed using Taq DNA polymerase (Promega, Madison, Wis.) under the condition consisting of denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 51°C for 30 s, and polymerization at 72°C for 1 min. The PCR fragment was cloned using pGEM-T vector systems (Promega) into E. coli JM109 and sequenced with an ALF Express DNA sequencer (Amersham Pharmacia Biotech), according to the manufacturer's instructions. To obtain the region downstream of the lactococcin Q gene, cassette PCR was applied. Two primers, S-1 and S-2, were designed from the DNA sequence obtained by degenerate PCR and used for cassette PCR with a HindIII cassette (Takara Bio, Otsu, Japan) (Table 1) and primers C-1 and C-2 corresponding to the sequence of the HindIII cassette. The total genomic DNA of strain QU 4 was completely digested with HindIII (Fermentas, Hanover, Md.), and the generated DNA fragments were ligated to the HindIII cassette. They were used as templates for PCR (first PCR) using primers S-1 and C-1. PCR was performed with LA Taq DNA polymerase (Takara Bio) under the condition consisting of denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 2 min, and polymerization at 72°C for 1 min. The products of first PCR were then used as templates for nested PCR using primers S-2 and C-2 under the same conditions as were employed for the first PCR. The product of the nested PCR was cloned and sequenced as described above. To obtain the region upstream of the lactococcin Q gene, inverse PCR was applied. The total genomic DNA of strain QU 4 was completely digested with MboI (Fermentas) and self-ligated for use as the template. Inverse PCR, using primers S-2 and lcnQ3, designed from the sequence obtained by the degenerate PCR, was performed with KOD Dash DNA polymerase (Toyobo, Osaka, Japan) under the condition including denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and polymerization at 74°C for 3.5 min. The product obtained by inverse PCR was cloned and sequenced as described above. Based on the sequences obtained from the PCR products, new specific primers were synthesized and used to confirm the obtained DNA sequence, according to the procedure described above.
TABLE 1.
Oligonucleotide primers and a cassette used in this study
| Name | Sequence |
|---|---|
| lcnQ1 | 5′-AT(T/C/A)TGGGG(T/C/A/G)GG(T/C)AT(T/C/A) GG-3′ |
| lcnQ2 | 5′-CCA(T/C)TT(A/G)TC(T/C)TT(A/G)TT(T/C/A/G) CC(T/C)TC-3′ |
| lcnQ3 | 5′-CCAATAAGCGGCTTTTCCTACTCCTT-3′ |
| S-1 | 5′-CAAGGAGTAGGAAAAGCCGCTTATTGGGTTGG-3′ |
| S-2 | 5′-GCCATGGGAAATATGAGCGATGTTAATCAGGCTTC-3′ |
| C-1 | 5′-GTACATATTGTCGTTAGAACGCGTAATACGACTCA-3′ |
| C-2 | 5′-CGTTAGAACGCGTAATACGACTCACTATAGGGAGA-3′ |
| HindIII cassette | 5′-HO GTACATATTGTCGTTAGAACGCGTAATACGACTCAC TATAGGGAGA-3′ |
| 3′-CATGTATAACAGCAATCTTGCGCATTATGCTGAGTGATATCCCTCTTCGA OH-5′ |
Computer analysis of DNA and amino acid sequences.
The obtained DNA and amino acid sequences were analyzed using GENETYX-WIN software (GENETYX, Tokyo, Japan). Database searches were performed using the BLAST program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/).
Peptide synthesis of lactococcins Q and G.
Lactococcin peptides, Qα, Qβ, Gα, and Gβ, were synthesized by a PSSM-8 automated peptide synthesizer (Shimadzu) as described previously (12). All the synthesized peptides were purified by reverse-phase HPLC, and their molecular masses were confirmed by MALDI-TOF MS as described above. Concentrations of the peptides were determined spectrophotometrically using molar adsorption coefficients at 280 nm (ɛ280) deduced from amino acid composition of the peptides.
Evaluation of antibacterial activity of synthetic lactococcin peptides.
Synthetic lactococcin peptides were dissolved in 0.05% TFA and diluted by 0.1% Tween 80 solution to appropriate concentrations. Single peptides and mixtures of α and β peptides at various molar ratios were tested for antibacterial activity as described above, and MIC was determined. For analysis of antibacterial spectra, preparations containing 500 nM (each) of synthetic α and β peptides were assayed. The supernatant of an overnight culture of L. lactis QU 4, adjusted to pH 6.0 with 5 M NaOH and filter sterilized, was also assayed as a crude lactococcin Q.
Nucleotide sequence accession number.
The DNA sequence presented in this article has been deposited in the DDBJ database with accession no. AB182406.
RESULTS
Identification of strain QU 4.
By the sugar fermentation pattern test using the API 50 CHL system, strain QU 4 was identified as Lactococcus lactis subsp. lactis. The 16S rRNA gene sequence of strain QU 4 showed 99% identity to those of the reference strains of L. lactis. Other characteristics of strain QU 4, such as being a gram positive diplococcus, being catalase negative, having growth at 10°C and no growth at 45°C, and producing homo l-lactic acid from glucose, also agreed well with the criteria of L. lactis. Considering these results, we concluded that strain QU 4 belonged to L. lactis.
Purification of lactococcin Q.
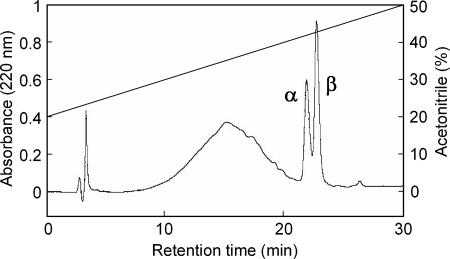
Lactococcin Q was purified by a three-step procedure including acetone precipitation, cation exchange, and reverse-phase HPLC. Most of the activity in the culture supernatant was recovered by acetone precipitation. In cation exchange chromatography, the bacteriocin activity was mainly recovered in a 0.75 M NaCl fraction. This fraction was applied subsequently to a reverse-phase HPLC, in which two peaks with antibacterial activity were obtained (Fig. 1). These antibacterial substances, termed peptides α and β, were eluted at retention times of 21.9 and 22.7 min, respectively. This implied that lactococcin Q consists of two peptides.
FIG. 1.
Reverse-phase chromatography of lactococcin Q. The bacteriocin activity was detected in the fractions containing the peaks indicated by α and β. The peaks α and β correspond to peptides α and β of lactococcin Q, respectively.
Mass spectrometry and amino acid sequence analysis.
MALDI-TOF MS analysis showed that purified peptides α and β of lactococcin Q had molecular masses of 4,260.43 Da and 4,018.36 Da, respectively. Edman degradation of peptides α and β of lactococcin Q revealed the sequences of 39 and 35 amino acid residues, respectively. The following N-terminal sequences were obtained: for lactococcin Qα, SIWGDIGQGVGKAAYWVGKAMGNMSDVNQASRINRKKKH; and for lactococcin Qβ, KKWGWLAWVEPAGEFLKGFGKGAIKEGNKDKWKNI. The respective calculated masses of 4,260 Da and 4,018 Da agreed completely with those observed by MALDI-TOF MS. This proved that the amino acid sequencing of peptides α and β was complete.
Analysis of the gene encoding lactococcin Q.
The structural gene sequence of lactococcin Q was obtained by degenerate PCR and subsequent cassette and inverse PCR. Sequencing analysis revealed two ORFs, termed laqA and laqB, encoding lactococcins Qα and Qβ, respectively (Fig. 2). Putative promoter and terminator sequences and ribosome binding sites were identified (Fig. 2). As in the cases of lactococcin G (20) and enterocin 1071 (7), the ORF for peptide β was encoded adjacent to and downstream of that for peptide α, and the two ORFs constituted an operon. The two ORFs, laqA and laqB, encoded prepeptides consisting of 54 and 61 amino acids, respectively. The deduced prepeptides possessed double-glycine sites for the cleavage of the leader peptides. The deduced amino acid sequences of mature peptides were identical to those of lactococcins Qα and Qβ obtained by Edman degradation.
FIG. 2.
Nucleotide sequence of the region encoding lactococcin Qα and lactococcin Qβ of L. lactis QU 4 and the deduced amino acid sequences. The putative −35 and −10 promoter regions and ribosome binding sites (RBS) are underlined. The vertical arrows indicate the cleavage sites of the peptides. The reversed horizontal arrows indicate the putative transcriptional terminator.
The database search showed that lactococcin Q was a novel two-peptide bacteriocin, homologous to enterocin 1071 (2, 7) and especially to lactococcin G (Fig. 3) (20, 23). Peptides α and β of lactococcin Q showed the highest homologies with peptides α and β of lactococcin G, having six and three amino acids different in mature peptides, respectively. The leader peptides of lactococcin Q also showed high homology with those of lactococcin G, as in the case of the mature peptides (Fig. 3).
FIG. 3.
Alignment of leader peptides (a) and mature peptides (b) of lactococcin Q, lactococcin G, and enterocin 1071. Identical amino acid residues are boxed.
Antibacterial activity of synthetic lactococcins Q and G.
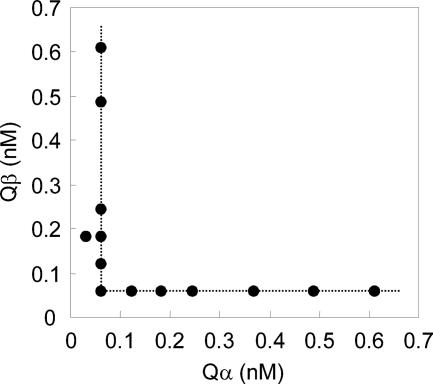
Lactococcins Qα and Qβ were chemically synthesized and tested for bacteriocin activity against the indicator strain, L. lactis subsp. lactis ATCC 19435T. Lactococcins Gα and Gβ were also synthesized and compared to lactococcin Q. None of the single peptides showed bacteriocin activity even at concentrations of 1 μM. All the peptides needed a complementary peptide to show antibacterial activity, as α peptide needed β peptide and vice versa (Table 2). Hybrid pairs Qα plus Gβ and Gα plus Qβ, as well as the lactococcin Q (Qα plus Qβ) and lactococcin G (Gα plus Gβ) original pairs, showed antibacterial activity. The Qα-plus-Qβ combination showed antibacterial activity at various molar ratios (Fig. 4). However, even when Qα or Qβ peptide existed in excess, intensities of antibacterial activity depended on the other peptide at a lower ratio. This indicated that they acted synergistically at a molar ratio of 1:1. All the other combinations also showed an equimolar ratio for the highest synergistic activities (data not shown). MICs for 1:1 mixtures of α and β peptides were determined (Table 2). The original combination of lactococcin Q, Qα plus Qβ, exhibited the lowest MIC of 0.0610 nM. However, Qα plus Gβ resulted in a 32-fold MIC, 1.95 nM.
TABLE 2.
Antibacterial activity of lactococcin peptides
| Lactococcin peptide | MIC (nM)a |
|---|---|
| Qα | >1,000 |
| Qβ | >1,000 |
| Gα | >1,000 |
| Gβ | >1,000 |
| Qα plus Gα | >500 |
| Qβ plus Gβ | >500 |
| Qα plus Qβ | 0.0610 |
| Qα plus Gβ | 1.95 |
| Gα plus Qβ | 0.122 |
| Gα plus Gβ | 0.488 |
L. lactis subsp. lactis ATCC 19435T was used as an indicator strain. Preparations containing 1,000 nM of single peptides were used as lactococcin peptides. Samples containing two peptides were prepared by mixing single-peptide preparations at an equimolar ratio, and their dilutions were assayed.
FIG. 4.
Synergistic activities of α and β peptides of lactococcin Q. Various molar ratios of mixtures of α and β peptides were assayed. Minimum concentrations of the peptides in the mixtures exhibiting antibacterial activity were plotted. Even when α or β peptide existed in excess, intensities of antibacterial activity depended on the other peptide at a lower ratio. This indicated that synthetic lactococcin Qα and Qβ acted at a 1:1 molar ratio.
Antibacterial spectra of lactococcins Q and G.
Crude lactococcin Q (culture supernatant) and synthetic lactococcins Q and G (containing each 500 nM of α and β peptide) were tested for antibacterial activity against various indicator strains. All samples showed similar narrow antibacterial spectra (Table 3). Lactococcins Q and G inhibited only L. lactis strains, including nisin-producing strains (NCDO 497, a nisin A producer; ATCC 7962, JCM 7638, and QU 1, nisin Z producers; and 61-14, a nisin Q producer). The lactococcin Q producer, L. lactis QU 4, showed immunity against lactococcin G as well as against lactococcin Q. The other indicator strains were highly resistant to lactococcins. The only exception was Lactobacillus sakei subsp. sakei JCM 1157T, which was weakly sensitive to lactococcin peptides. However, the MICs were significantly higher, which ranged from 500 nM to 1 μM. Hybrid pairs of synthetic peptides, Qα plus Gβ and Gα plus Qβ, showed identical antibacterial spectra to those of synthetic lactococcins Q and G.
TABLE 3.
Antibacterial spectra of lactococcins Q and G
| Indicator species | Synthetic peptidec
|
|||||
|---|---|---|---|---|---|---|
| Straina | Crude lactococcin Qb | Lactococcin Q (Qα plus Qβ) | Lactococcin G (Gα plus Gβ) | Qα plus Gβ | Gα plus Qβ | |
| Lactococcus lactis subsp. lactis | ATCC 19435T | +d | + | + | + | + |
| ATCC 7962 | + | + | + | + | + | |
| IL-1403 | + | + | + | + | + | |
| JCM 7638 | + | + | + | + | + | |
| NCDO 497 | + | + | + | + | + | |
| 61-14e | + | + | + | + | + | |
| QU 1 | + | + | + | + | + | |
| QU 4f | − | − | − | − | − | |
| Lactococcus lactis subsp. cremoris | ATCC 19257T | + | + | + | + | + |
| Lactococcus lactis subsp. hordniae | JCM 1180T | + | + | + | + | + |
| Lactococcus garvieae | NCFB 2155T | − | − | − | − | − |
| Lactococcus raffinolactis | JCM 5706T | − | − | − | − | − |
| Streptococcus bovis | JCM 5802T | − | − | − | − | − |
| Streptococcus mutans | JCM 5705T | − | − | − | − | − |
| Streptococcus salivarius | JCM 5707T | − | − | − | − | − |
| Enterococcus faecalis | JCM 5803T | − | − | − | − | − |
| Enterococcus faecium | JCM 5804T | − | − | − | − | − |
| Enterococcus hirae | ATCC 10541 | − | − | − | − | − |
| Pediococcus pentosaceus | JCM 5885 | − | − | − | − | − |
| JCM 5890T | − | − | − | − | − | |
| Pediococcus acidilactici | JCM 8797T | − | − | − | − | − |
| Leuconostoc mesenteroides subsp. mesenteroides | JCM 6124T | − | − | − | − | − |
| Lactobacillus alimentarius | JCM 1095T | − | − | − | − | − |
| Lactobacillus brevis | JCM 1059T | − | − | − | − | − |
| Lactobacillus casei subsp. casei | JCM 1134T | − | − | − | − | − |
| Lactobacillus collinoides | JCM 1123T | − | − | − | − | − |
| Lactobacillus coryniformis subsp. coryniformis | JCM 1164T | − | − | − | − | − |
| Lactobacillus kimchii | JCM 10707T | − | − | − | − | − |
| Lactobacillus plantarum | JCM 1149T | − | − | − | − | − |
| Lactobacillus sakei subsp. sakei | JCM 1157T | − | w | w | w | w |
| Lactobacillus salivarius subsp. salivarius | JCM 1231T | − | − | − | − | − |
| Staphylococcus aureus subsp. aureus | ATCC 12600T | − | − | − | − | − |
| Micrococcus luteus | IFO 12708 | − | − | − | − | − |
| Bacillus subtilis | JCM 1465T | − | − | − | − | − |
| Bacillus cereus | JCM 2152T | − | − | − | − | − |
| Bacillus circulans | JCM 2504T | − | − | − | − | − |
| Bacillus coagulans | JCM 2257T | − | − | − | − | − |
| Listeria innocua | ATCC 33090T | − | − | − | − | − |
| Clostridium sporogenes | JCM 1416T | − | − | − | − | − |
| Escherichia coli | JM109 | − | − | − | − | − |
Abbreviations: ATCC, American Type Culture Collection, Rockville, Md.; IFO, Institute for Fermentation, Osaka, Japan; JCM, Japan Collection of Microorganisms, Wako, Japan; NCDO, National Collection of Dairy Organisms, Reading, United Kingdom; NCFB, National Collection of Food Bacteria, Reading, United Kingdom; QU, Laboratory of Microbial Technology, Kyushu University, Fukuoka, Japan.
Filter-sterilized culture supernatant of L. lactis QU 4 (pH 6) was used as crude lactococcin Q.
Each 500 nM of synthetic α and β peptide was contained.
+, sensitive; w, weakly sensitive; −, resistant to lactococcin.
Laboratory collection.
Lactococcin Q-producing strain.
DISCUSSION
In this paper, we have described purification, structural analysis, and characterization of a novel two-peptide bacteriocin, termed lactococcin Q, produced by L. lactis QU 4 isolated from corn. There have been only a few reports of the isolation of bacteriocin-producing LAB from corn-associated sources, such as corn steep liquor (4) and fermented corn products (25).
We isolated 11 strains of bacteriocin-producing LAB from a corn sample. Whereas five strains of the isolates showed specific antibacterial spectra similar to that of lactococcin Q, the other isolates showed broader antibacterial spectra, one of which was a nisin Z producer. Lactococcin Q and nisin Z were active against each other's producer, implying that they compete in corn microbiota. The remaining isolates showed unique antibacterial spectra, and further characterization is under investigation.
Amino acid and DNA sequence analyses revealed that lactococcin Q consisted of two peptides, termed lactococcins Qα and Qβ. Lactococcins Qα and Qβ contained 39 and 35 amino acids, respectively, but neither lanthionines nor dehydrated amino acids. MALDI-TOF MS analysis showed that each purified peptide was cross-contaminated in very small amounts (data not shown). Although amino acid sequence analysis was not affected, this indicated that further characterization of lactococcin Qα and Qβ using peptides purified through the procedures described above might lead to errors. An antibacterial activity assay using chemically synthesized lactococcin Qα and Qβ showed that two peptides exhibited full activity synergistically at a molar ratio of 1:1, whereas each peptide alone showed no activity. These characteristics are in accordance with those of class IIb bacteriocins of LAB, suggesting that lactococcin Q belongs to class IIb bacteriocin.
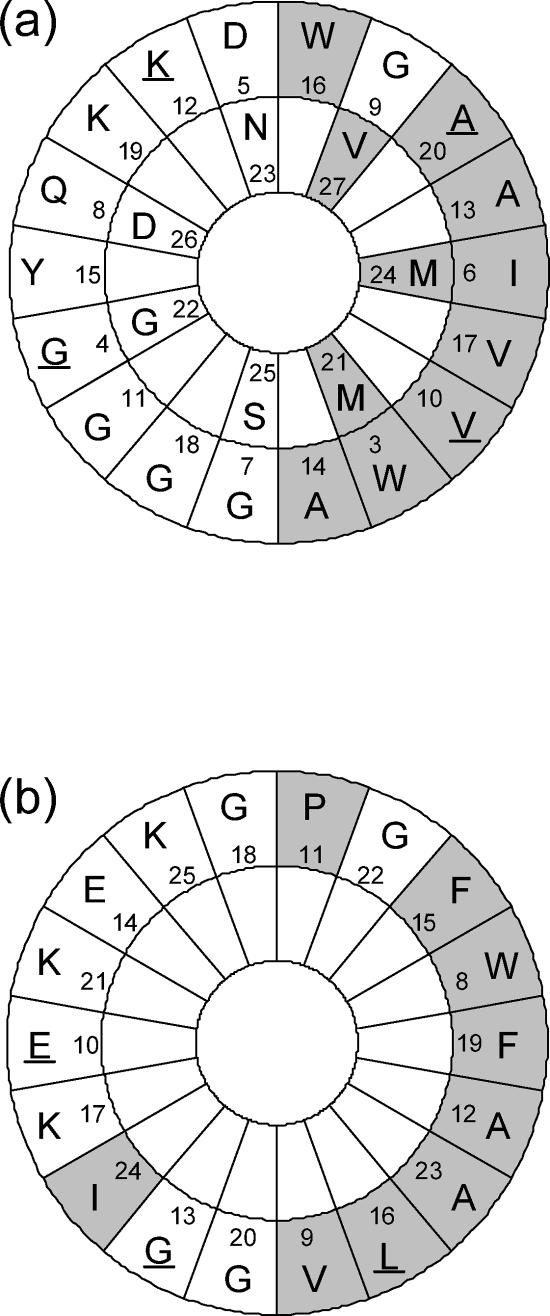
A database search showed that lactococcin Q was a novel class IIb bacteriocin with high homology to lactococcin G (Fig. 3). Based on the amino acid sequences, amphiphilic α-helical structures of lactococcin G and enterocin 1071 were predicted by Edmundson α-helical wheel representation (2, 23, 26). Amphiphilic α-helical structures are often found in pore-forming antimicrobial peptides (24). The analysis applied to lactococcin Q is shown in Fig. 5. It was presumed that each peptide of lactococcin Q formed an amphiphilic α-helix in the same positions as lactococcin G (23). In the amphiphilic region of α peptide, four residues were different between lactococcins Q and G, and in that of β peptide, three residues were different. In all the substitutions, the polarities of residues were not changed; hydrophobic and hydrophilic residues were changed to the corresponding ones. This supported the idea that lactococcins Q and G possessed similar secondary structures and therefore similar modes of action.
FIG. 5.
Edmundson α-helical wheel representation of the amphiphilic regions in lactococcin Qα (a) and lactococcin Qβ (b). For lactococcin Qα, the amphiphilic region starts with residue 3 and ends with residue 27, and for lactococcin Qβ, it starts with residue 8 and ends with residue 25, as in the cases of lactococcin Gα and lactococcin Gβ, respectively. The shaded areas indicate nonpolar residues, and the nonshaded areas indicate polar residues. The residues different from those of lactococcin G are underlined.
Antibacterial activities of lactococcins Q and G were compared in detail by using synthetic peptides. Lactococcins Gα and Gβ showed activity synergistically at a molar ratio of 1:1 as described previously (17). In addition, the other combinations of α and β peptides, Qα plus Gβ and Gα plus Qβ, also showed antibacterial activity at an equimolar ratio. This is the first case in which hybrid combinations between two-peptide bacteriocins provided synergistic activity.
MICs of lactococcin peptides were considerably different by combinations of α and β peptides (Table 2). These differences were probably caused by β peptides. The antibacterial activity of Qα plus Qβ was 32 times as high as that of Qα plus Gβ, whereas those of Qα plus Qβ and Gα plus Qβ were comparable. This suggested that the contribution of lactococcin Qβ for antibacterial activity was greater than that of lactococcin Gβ. Three amino acid residues that are different between lactococcins Qβ and Gβ might play important roles for the intensity of antibacterial activity.
Lactococcin Q showed antibacterial activity only against L. lactis strains (Table 3). Among LAB bacteriocins so far reported, lactococcin A (11), lactococcin B (19, 27, 28), lactococcin M (19, 27, 28), lactococcin 972 (15), LsbA (8), and LsbB (8) showed narrow and specific antibacterial spectra similar to lactococcin Q. They inhibited only the growth of lactococci and very few additional strains belonging to other genera. However, there is no structural homology between lactococcin Q and each of them.
Antibacterial spectra of lactococcin G and hybrid pairs were identical to lactococcin Q (Table 3). Their antibacterial activities were limited to the species of L. lactis. This suggested that different amino acid residues between lactococcins Q and G were not determinants of specificity of antibacterial activity. Another homologue, enterocin 1071, showed the activity against strains of various genera of gram-positive bacteria, including enterococci, but not against L. lactis (2). Amino acid residues that differ between lactococcins and enterocin 1071 might be involved in the specific recognition of target cells.
Recognition of target cells and partner peptides in a two-peptide bacteriocin system is still unknown. Further comparative studies on two-peptide bacteriocins are needed to solve the question of these types of recognition. This study is a good foothold to determine which parts of the peptides play important roles for the recognition. In addition, the results in this study implied that properties of two-peptide bacteriocins could be changed by substitution of partner peptides as well as amino acid residues. Other combinations of the peptides are worth further trying and consideration. Changing partner peptides of two-peptide bacteriocins may be an advantage over single-peptide bacteriocins for improvement of the properties.
Acknowledgments
We thank T. Horii and H. Nagasawa, Graduate School of Agricultural and Life Sciences, the University of Tokyo, for amino acid sequence analysis.
This work was partially supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, an AIST research grant, the Urakami Foundation for Food and Food Culture, the Hokuto Foundation for Bioscience, and the Iijima Memorial Foundation for the Promotion of Food Science and Technology.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley and Sons, Inc., Hoboken, N.J.
- 2.Balla, E., L. M. T. Dicks, M. Du Toit, M. J. Van Der Merwe, and W. H. Holzapfel. 2000. Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Appl. Environ. Microbiol. 66:1298-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobial for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 4.Contreras, B. G. L., L. De Vuyst, B. Devreese, K. Busanyova, J. Raymaeckers, F. Bosman, E. Sablon, and E. J. Vandamme. 1997. Isolation, purification, and amino acid sequence of lactobin A, one of the two bacteriocins produced by Lactobacillus amylovorus LMG P-13139. Appl. Environ. Microbiol. 63:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ennahar, S., Y. Asou, T. Zendo, K. Sonomoto, and A. Ishizaki. 2001. Biochemical and genetic evidence for production of enterocins A and B by Enterococcus faecium WHE 81. Int. J. Food Microbiol. 70:291-301. [DOI] [PubMed] [Google Scholar]
- 6.Flynn, S., D. van Sinderen, G. M. Thornton, H. Holo, I. F. Nes, and J. K. Collins. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973-984. [DOI] [PubMed] [Google Scholar]
- 7.Franz, C. M. A. P., A. Grube, A. Herrmann, H. Abriouel, J. Stärke, A. Lombardi, B. Tauscher, and W. H. Holzapfel. 2002. Biochemical and genetic characterization of the two-peptide bacteriocin enterocin 1071 produced by Enterococcus faecalis FAIR-E 309. Appl. Environ. Microbiol. 68:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gajic, O., G. Buist, M. Kojic, L. Topisirovic, O. P. Kuipers, and J. Kok. 2003. Novel mechanism of bacteriocin secretion and immunity carried out by lactococcal multidrug resistance proteins. J. Biol. Chem. 278:34291-34298. [DOI] [PubMed] [Google Scholar]
- 9.Garneau, S., N. I. Martin, and J. C. Vederas. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577-592. [DOI] [PubMed] [Google Scholar]
- 10.Hauge, H. H., J. Nissen-Meyer, I. F. Nes, and V. G. H. Eijsink. 1998. Amphiphilic α-helices are important structural motiefs in the α and β peptides that constitute the bacteriocin lactococcin G: enhancement of helix formation upon α-β interaction. Eur. J. Biochem. 251:565-572. [DOI] [PubMed] [Google Scholar]
- 11.Holo, H., Ø. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishida, A., Y. Shigeri, Y. Tatsu, K. Uegaki, I. Kameshita, S. Okuno, T. Kitani, N. Yumoto, and H. Fujisawa. 1998. Critical amino acid residues of AIP, a highly specific inhibitory peptide of calmodulin-dependent protein kinase II. FEBS Lett. 427:115-118. [DOI] [PubMed] [Google Scholar]
- 13.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-86. [DOI] [PubMed] [Google Scholar]
- 14.Maldonado, A., J. L. Ruiz-Barba, and R. Jiménez-Díaz. 2003. Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 69:383-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez, B., J. E. Suárez, and A. Rodríguez. 1996. Lactococcin 972: a homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 142:2393-2398. [DOI] [PubMed] [Google Scholar]
- 16.Mayr-Harting, A., A. J. Hedges, and R. C. W. Berkeley. 1972. Methods for studying bacteriocins. Methods Microbiol. 7:315-422. [Google Scholar]
- 17.Moll, G., T. Ubbink-Kok, H. Hildeng-Hauge, J. Nissen-Meyer, I. F. Nes, W. N. Konings, and A. J. M. Driessen. 1996. Lactococcin G is a potassium ion-conducting, two-component bacteriocin. J. Bacteriol. 178:600-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moll, G., H. Hildeng-Hauge, J. Nissen-Meyer, I. F. Nes, W. N. Konings, and A. J. M. Driessen. 1998. Mechanistic properties of the two-component bacteriocin lactococcin G. J. Bacteriol. 180:96-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan, S., R. P. Ross, and C. Hill. 1995. Bacteriolytic activity caused by the presence of a novel lactococcal plasmid encoding lactococcin A, B, and M. Appl. Environ. Microbiol. 61:2995-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nes, I. F., L. S. Håvarstein, and H. Holo. 1995. Genetics of nonlantibiotic bacteriocins, p. 645-651. In J. J. Ferretti, M. S. Gilmore, T. R. Klaenhammer, and F. Brown (ed.), Genetics of streptococci, enterococci and lactococci. Developments in biological standardisation, vol. 85. Karger, Basel, Switzerland. [PubMed] [Google Scholar]
- 21.Nes, I. F., D. B. Diep, L. S. Håvarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 22.Nes, I. F., and H. Holo. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 55:50-61. [DOI] [PubMed] [Google Scholar]
- 23.Nissen-Meyer, J., H. Holo, L. S. Håvarstein, K. Sletten, and I. F. Nes. 1992. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J. Bacteriol. 174:5686-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powers, J.-P., and R. E. W. Hancock. 2003. The relationship between peptide structure and antibacterial activity. Peptides 24:1681-1691. [DOI] [PubMed] [Google Scholar]
- 25.Sanni, A. I., A. A. Onilude, S. T. Ogunbanwo, and S. I. Smith. 1999. Antagonistic activity of bacteriocin produced by Lactobacillus species from ogi, an indigenous fermented food. J. Basic Microbiol. 39:189-195. [PubMed] [Google Scholar]
- 26.Schiffer, M., and A. B. Edmundson. 1967. Use of helical wheels to represent the structure of proteins and to identify segments with helical potential. Biophys. J. 7:121-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Belkum, M. J., B. J. Hayema, R. E. Jeeninga, J. Kok, and G. Venema. 1991. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl. Environ. Microbiol. 57:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Belkum, M. J., J. Kok, and G. Venema. 1992. Cloning, sequencing, and expression in Escherichia coli of lcnB, a third bacteriocin determinant from the lactococcal bacteriocin plasmid p9B4-6. Appl. Environ. Microbiol. 58:572-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zendo, T., N. Eungruttanagorn, S. Fujioka, Y. Tashiro, K. Nomura, Y. Sera, G. Kobayashi, J. Nakayama, A. Ishizaki, and K. Sonomoto. 2005. Identification and production of a bacteriocin from Enterococcus mundtii QU 2 isolated from soybean. J. Appl. Microbiol. 99:1181-1190. [DOI] [PubMed] [Google Scholar]