Abstract
The in vivo kinetics in Saccharomyces cerevisiae CEN.PK 113-7D was evaluated during a 300-second transient period after applying a glucose pulse to an aerobic, carbon-limited chemostat culture. We quantified the responses of extracellular metabolites, intracellular intermediates in primary metabolism, intracellular free amino acids, and in vivo rates of O2 uptake and CO2 evolution. With these measurements, dynamic carbon, electron, and ATP balances were set up to identify major carbon, electron, and energy sinks during the postpulse period. There were three distinct metabolic phases during this time. In phase I (0 to 50 seconds after the pulse), the carbon/electron balances closed up to 85%. The accumulation of glycolytic and storage compounds accounted for 60% of the consumed glucose, caused an energy depletion, and may have led to a temporary decrease in the anabolic flux. In phase II (50 to 150 seconds), the fermentative metabolism gradually became the most important carbon/electron sink. In phase III (150 to 300 seconds), 29% of the carbon uptake was not identified in the measurements, and the ATP balance had a large surplus. These results indicate an increase in the anabolic flux, which is consistent with macroscopic balances of extracellular fluxes and the observed increase in CO2 evolution associated with nonfermentative metabolism. The identified metabolic processes involving major carbon, electron, and energy sinks must be taken into account in in vivo kinetic models based on short-term dynamic metabolome responses.
Mathematical models of in vivo enzyme kinetics in microorganisms are important for understanding metabolic control mechanisms operating on the level of the metabolome and can be used to assist the rational redesign of metabolic pathways to enhance desired functionalities of microbes (54). Kinetic parameters in this kind of model can be obtained by stimulus-response experiments, in which cells grown in a (quasi-)steady state are perturbed by an external stimulus and the dynamic responses of intra- and extracellular metabolites are monitored. The time window of observation is usually within tens to a few hundred seconds after the application of the stimulus, and the responses are usually attributed to rapid (allosteric) enzyme-metabolite interactions (38). Kinetic parameters can be estimated from the measured responses, based on a set of (dynamic) material balances (7, 30, 49), as follows: dx/dt = Sv (equation 1), where x is a vector of the metabolite concentrations, S is the stoichiometry matrix, and v is a vector of the reaction rates as a function of (yet) unknown kinetic parameters. The stimulus-response methodology is an ideal tool for obtaining kinetic information and has been applied to various microorganisms under different growth conditions (7, 26, 32, 38, 52).
For Saccharomyces cerevisiae, a frequently applied perturbation is the addition of a concentrated glucose solution, i.e., a glucose pulse, to a glucose-limited chemostat culture, thereby inducing a short-term Crabtree effect. Theobald et al. (38) first quantified for a 180-second postpulse period the responses of extracellular metabolites (glucose, ethanol, acetate, and glycerol) and intracellular intermediates of and cofactors involved in glycolysis. These measurements were used to establish a kinetic model focused on glycolysis (30). In addition, Vaseghi et al. (49, 50) quantified responses of the pentose phosphate pathway and the protein kinase A/phosphofructokinase 2 (PKA/PFK2) signal transduction cascade to such a glucose pulse to extend this kinetic model. Models developed so far incorporate detailed descriptions of metabolic subnetworks, such as glycolysis, for which metabolome responses have been quantified. Other metabolic processes, such as the tricarboxylic acid (TCA) cycle, storage carbon metabolism, and anabolism, are represented by lumped reactions with highly simplified kinetics due to a lack of experimental information.
The importance of various metabolic processes in a kinetic model can be evaluated by the relative contributions of metabolites involved in these processes to the overall material balance, which forms the basis for kinetic parameter estimation (equation 1). Under quasi-steady-state conditions, for example, material balances of extracellular metabolites are required for metabolic flux analysis (33). Previous work has evaluated dynamic carbon balances in a time frame of tens of minutes to hours (14, 26, 45), based on measured extracellular fluxes and an assumed intracellular quasi-steady state. Under dynamic conditions and within the time frame of interest for in vivo kinetic analysis, setting up material balances requires comprehensive quantification of extra- and intracellular metabolite concentrations as well as the in vivo O2 uptake rate and CO2 evolution rate.
The objectives of this study were (i) to quantify an extended range of metabolome responses following a glucose pulse to an aerobic, carbon-limited chemostat culture of S. cerevisiae, (ii) to construct complete carbon, electron, and energy balances during a 300-second postpulse period from these measurements, (iii) to identify major carbon/electron sinks and energy production/consumption processes in this period, and (iv) to determine if major unquantified metabolic processes remain. This information will increase our understanding of short-term physiological responses of S. cerevisiae to glucose excess and will assist in building more realistic and comprehensive kinetic models.
MATERIALS AND METHODS
Yeast strain and chemostat cultivation.
The haploid S. cerevisiae strain CEN.PK 113-7D was cultivated in an aerobic, carbon-limited chemostat with a working volume of 4 liters at a dilution rate of 0.051 h−1, as previously described (18). The medium used for chemostat cultivation was based on a previously described medium (18), with 0.15 M (27.1 g liter−1) glucose and 0.03 M (1.42 g liter−1) ethanol to obtain a biomass of ∼15 g dry weight liter−1 during steady state. The dissolved oxygen tension (DOT) in the fermentor and the O2 and CO2 contents in the fermentor off-gas were analyzed as previously described (55). The DOT was maintained above 70% in the steady state.
Rapid sampling, quenching, and metabolite extraction.
Culture supernatant for the analysis of extracellular metabolites was obtained by rapidly quenching the broth with stainless steel beads (4-mm diameter) at −20°C, followed by filtration, as described previously (21), for a sample volume of 2 ml. For analysis of intracellular metabolites, sampling and sample preparation were carried out as previously described (22). Briefly, ∼1 ml of broth was rapidly quenched in 5 ml of −40°C 60:40 (vol/vol) methanol-H2O. After centrifugation (2,000 × g, −20°C, 5 min), the supernatant was decanted, and the pellet was resuspended in 5 ml of −40°C 60:40 (vol/vol) methanol-H2O and centrifuged again. Intracellular metabolites were extracted from the pellet with boiling 75:25 (vol/vol) ethanol-H2O. The ethanol extracts were evaporated to dryness. The dried samples were resuspended in 0.5 ml deionized water and centrifuged (3,000 × g, 4°C, 5 min). The supernatant and pellet were separated and stored at −80°C until they were analyzed.
Determination of culture dry weight.
Culture samples (5 ml, in triplicate) were filtered through predried, tared nitrocellulose filters (pore size, 0.45 μm; Gelman Science, Ann Arbor, MI). Filters were washed twice with 5 ml demineralized water, dried at 70°C for 48 h, and weighed.
Extracellular metabolite analysis.
The amounts of glucose, acetate, and glycerol in the supernatant were measured with Enzytec kits (Scil Diagnostics GmbH, Martinsried, Germany). The concentration of ethanol in the supernatant was analyzed with an Enzymatic Bioanalysis kit (R-Biopharm AG, Darmstadt, Germany). Absorbance was read on an Agilent 8453 UV-visible spectroscopy system (Agilent Technologies Deutschland GmbH, Waldbronn, Germany).
MS analysis of intracellular metabolites.
The concentrations of glucose 6-phosphate (G6P), fructose 6-phosphate (F6P), fructose 1,6-bisphosphate (F1,6bP), 2-phosphoglycerate (2PG), 3-phosphoglycerate (3PG), and phosphoenol pyruvate (PEP) in the cell extract were analyzed by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) according to the method of van Dam et al. (40). The same analysis method and conditions were applied for the analysis of pyruvate (Pyr), citrate (Cit), isocitrate (iCit), α-ketoglutarate (αKG), succinate (Suc), fumarate (Fum), malate (Mal), glucose 1-phosphate (G1P), mannose 6-phosphate (M6P), trehalose 6-phosphate (T6P), fructose 2,6-bisphosphate (F2,6bP), and orthophosphate (Pi) in the cell extract. Neither 2PG and 3PG nor Cit and iCit could be resolved with this procedure, so only the sums of these compounds were determined. For F2,6bP, only the peak area was quantified instead of the absolute amount in the sample.
The concentrations of the adenine nucleotides (AMP, ADP, and ATP) in the cell extract were quantified by LC-ESI-MS/MS. The nucleotides were separated by an ion-pairing reversed-phase high-performance liquid chromatography (HPLC) method adapted from a previously described method (8), using an XTerra MS C18 column (100 mm × 1 mm) equipped with a guard column (10 mm × 2.1 mm) (both from Waters, Milford, MA). The standard solution and sample injection volume was 10 μl; all standard solutions and samples were mixed 24:1 with a 2 M solution of the ion-pairing reagent tetrabutylammoniumacetate (Sigma-Aldrich, Steinheim, Germany) to obtain a final tetrabutylammoniumacetate concentration of 80 mM. Chromatography was performed at 20°C with an Alliance HT 2795 pump system (Waters), giving an isocratic elution flow of 0.1 ml min−1. The mobile phase was a 10 mM NH4H2PO4 solution that was adjusted to pH 6.4 with NH4OH, after which 2 mM tetrabutylammoniumhydroxide (Sigma-Aldrich) was added, giving a pH of 6.8, followed by the addition of 15% (vol/vol) acetonitrile solution. The postcolumn eluent was mixed with 0.1 ml min−1 80% (vol/vol) acetonitrile solution. The solutions were filtered (0.45 μm; Gelman Sciences) prior to use to remove particles and bubbles. The HPLC instrument was coupled to an MS/MS instrument (Quattro Ultima Pt; Micromass Ltd., Manchester, United Kingdom) via a flow splitter which sent a flow of 0.06 ml min−1 to the ESI instrument, which was operated in positive mode with a nebulizer gas (nitrogen) flow of 50 liter h−1 and a desolvation gas (nitrogen) flow of 500 liter h−1 at 300°C. The source block temperature was 120°C. The capillary voltage was set at 3.5 kV, and the cone voltage was set at 35 V. The [M + H]+ ions (m/z 348, 428, and 508 for AMP, ADP, and ATP, respectively) were fragmented by collision-induced dissociation with argon (collision energy, 24 V). The strongest daughter fragment of m/z 136 was monitored for each of them.
Intracellular trehalose analysis.
Intracellular trehalose was analyzed by quantitative 1H nuclear magnetic resonance. Analysis was performed at 360 MHz on a Bruker AMX 360 spectrometer (Bruker Analytik, Karlsruhe, Germany). To 0.5 ml of cell extract, an equal amount of a standard solution containing 5.6 g liter−1 maleic acid and 10 g liter−1 EDTA (natrium salt) was added. After lyophilization, the residue was dissolved in 100% D2O, and the 1H nuclear magnetic resonance spectrum was measured, with a relaxation delay of 30 seconds to ensure full relaxation of all hydrogen atoms between pulses. The integrals of the 1H protons of trehalose (doublet at 5.24 ppm) and the internal standard (singlet at 6.1 ppm) were measured, and the content of trehalose was calculated.
Intracellular glycogen analysis.
Intracellular glycogen was measured in the cell extracts and the insoluble residues as previously described (27). Glycogen in the cell extracts was directly hydrolyzed with amyloglucosidase (from Aspergillus niger; Sigma-Aldrich) without alkaline extraction.
Intracellular free amino acid analysis.
Intracellular concentrations of free amino acids were measured by HPLC using the AccQ-tag system, as previously described (53).
Estimation of extracellular Pi carryover to cell extracts.
About 0.25 ml of the 5 ml of quenching or washing solution remained in the test tube after decanting, which corresponds to a washing efficiency of 1 − (0.25/5)2, or 99.75%, and a carryover of 0.25% after the quenching and washing steps. A steady-state extracellular Pi concentration of 39.3 mM was calculated based on the biomass phosphate content (19) and the medium composition. Assuming a constant extracellular Pi concentration during the postpulse period, the carryover is no more than 8% of the measured Pi concentration in the cell extract.
Glucose pulse experiment.
S. cerevisiae was grown for 10 generations in an aerobic, carbon-limited chemostat, and 15 ml of a glucose solution (1.48 M, thus containing 22.2 mmol of glucose) was injected into the fermentor. Samples for extra- and intracellular metabolite analyses were taken during the steady state shortly before the glucose pulse and during the 300 seconds following the glucose pulse. During the postpulse period, the steady-state feed to the fermentor was continued, but no effluent was removed via the effluent pump since the chemostat was weight controlled and the fermentor weight decreased slightly due to sampling.
Steady-state metabolic flux analysis.
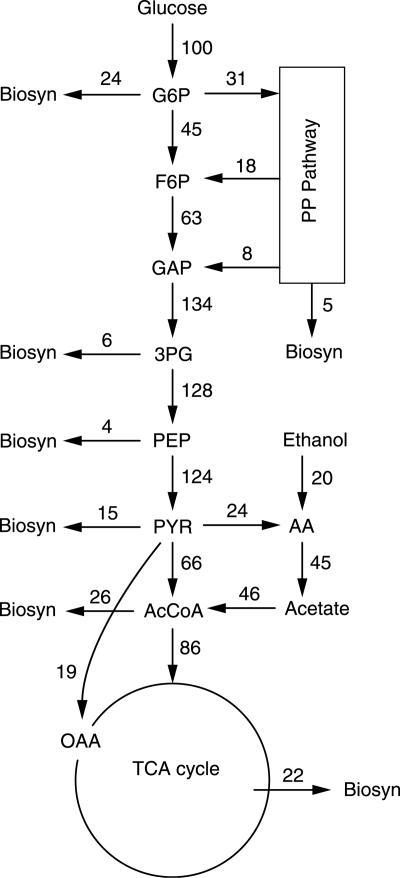
The measured extracellular fluxes in the steady state were used after reconciliation (41) to estimate intracellular fluxes by conventional metabolic flux analysis (33). The biomass composition determined for a growth rate of 0.05 h−1 was used (19), and 1 Cmol biomass (equivalent to the amount of biomass that contains 1 mol of carbon) corresponds to 26.15 g cell dry weight. The noncompartmented network of S. cerevisiae was based on previous work (34). We assumed that acetaldehyde dehydrogenase (AADH) is NADP+ specific and that isocitrate dehydrogenase is NAD+ specific. The inclusion of acetyl coenzyme A synthetase creates a parallel route for the synthesis of acetyl coenzyme A from pyruvate, which can proceed via either pyruvate dehydrogenase or the pyruvate bypass (Fig. 1). In the analyzed network, NADPH needed for growth was presumed to be regenerated by AADH and the pentose phosphate (PP) pathway; hence, intracellular fluxes can be calculated by specifying the PP pathway flux (or, equivalently, the PP split ratio). To estimate the PP split ratio, specific NADPH regeneration (mmol of NADPH g dry weight−1) via the PP pathway was assumed to decrease linearly with increasing NADPH regeneration by metabolizing ethanol via the NADP+-dependent AADH in a glucose-ethanol mixed-substrate feed when the ethanol fraction was below 0.4 Cmol of ethanol Cmol−1. Above this limit, the metabolic network would change due to the induction of gluconeogenic enzymes (9, 44). For the Saccharomyces strain used in this study, the PP split ratio was estimated to be 0.44 and 0.24 in two 13C-labeling studies (3, 48), for ethanol fractions of 0 and 0.28 Cmol ethanol Cmol−1, respectively, at a growth rate of 0.1 h−1. For the feed used in this study (0.06 Cmol ethanol Cmol−1), a PP split ratio of 0.31 was interpolated and used to estimate the intracellular fluxes.
FIG. 1.
Estimated intracellular fluxes in the reference steady state (normalized against a glucose influx of 0.013 mol Cmolx−1 h−1).
Estimation of qO2 and qCO2.
The in vivo O2 uptake rate (qO2) and the CO2 evolution rate (qCO2) during the postpulse period were calculated with a mass transport model as described previously (5). The model describes liquid/gas mass transfer in the fermentor, mixing/delay effects in the headspace and tubing system, and sensor dynamics. The parameters of this model were either derived from physical knowledge or computed by fitting the model to an identification data set. The qO2 and qCO2 values during the postpulse period were calculated from the off-gas and DOT measurements and the model by extended Kalman filtering. The cumulative O2 uptake and CO2 evolution rates were obtained by numerically integrating the obtained qO2 and qCO2 values.
Construction of cumulative carbon, electron, and ATP balances.
The cumulative carbon balance, in Cmol per Cmol of biomass (Cmol Cmolx−1), compares the cumulative uptake of carbon sources with the accumulation of known carbon sinks, calculated at each sampling time point relative to time zero, when the glucose pulse was given (14). During the postpulse period, glucose (added via both the pulse and the feed) was the sole carbon source, assuming that the small amount of ethanol in the feed was not metabolized. The known carbon sinks were secreted extracellular metabolites, intracellular metabolites, CO2, and flux towards anabolism. The anabolic flux was assumed to be equal to the biomass growth rate. Since possible changes in the growth rate during the postpulse period are difficult to quantify accurately, we assumed that it was the same as in steady state, i.e., 0.05 h−1. The adenine nucleotides were excluded from the carbon balances, assuming that the adenine moiety was conserved during the short time window. A constant total broth volume was also assumed, since within 300 seconds, effluent via sampling (∼0.1 liter) and influent via the feed pump (0.02 liter) were negligible compared to the total broth volume (4 liters). These assumptions were also made for the cumulative electron and ATP balances.
The cumulative electron balance (in mol Cmolx−1) was set up in a manner similar to that of the cumulative carbon balance, but with the degree of reduction of the metabolites (24), and it compares the cumulative uptake of electron sources with the accumulation of known electron sinks. During the postpulse period, glucose was the sole electron source. The known electron sinks include the carbon sinks and O2.
The cumulative ATP balance (in mol ATP equivalents Cmolx−1) (Table 1) was also set up in a manner similar to that of the cumulative carbon balance. The P/O ratio (the ratio of ATPs produced to the number of atmospheric oxygen atoms converted to water) was assumed to be constant during the postpulse period. Intermediates in the PP pathway and the TCA cycle, as well as free amino acids, were excluded from the ATP balance because they account for only a very small proportion (<3%) of the total carbon uptake.
TABLE 1.
Energy production or consumption associated with accumulation of metabolites
| ATP status | Metabolite | mol ATP equivalents mol−1 |
|---|---|---|
| Production | Ethanol | 1 |
| Acetate | 1a | |
| O2 | 2b | |
| Consumption | Glucose | 0a |
| Glycerol | 1 | |
| Biomass | 1.65b | |
| G6P | 1 | |
| F6P | 1 | |
| F1,6bP | 2 | |
| 2PG + 3PG | 0 | |
| PEP | 0 | |
| G1P | 1 | |
| T6P | 3 | |
| Trehalose | 3 | |
| M6P | 1 |
Assuming no ATP cost for the uptake of glucose and the secretion of acetate.
According to reference 51.
RESULTS
Changes in metabolic fluxes and intracellular metabolites following a glucose pulse.
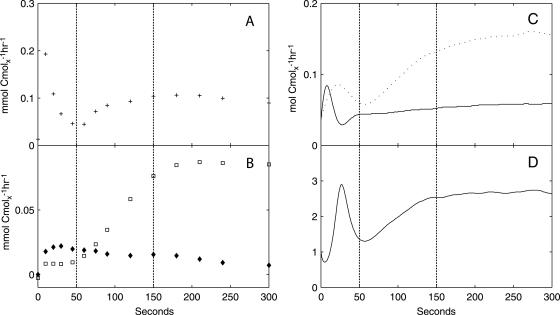
Three independent glucose pulse experiments yielded highly reproducible trends in metabolic responses. The average variations in the accumulation (in mCmol Cmolx−1) of extracellular carbon sinks (e.g., ethanol) and groups of intracellular carbon sinks (e.g., intracellular metabolites in glycolysis) (Table 2) relative to the steady-state levels were within 10% and 15%, respectively, in these experiments. Results from one representative experiment are presented here and were used for setting up material balances. Prior to the glucose pulse, the culture of S. cerevisiae was maintained in a reference steady state at a growth rate of 0.05 h−1 (Fig. 1; Table 2). Perturbation of this reference state by a glucose pulse at time zero, which increased the residual glucose concentration from 0.15 mM to 5.56 mM, led to dynamics in the biomass-specific glucose uptake rate (qglucose) (Fig. 2A), the secretion of ethanol, acetate (Fig. 2B), and a small amount of glycerol, and dynamic changes in the biomass-specific rates of O2 uptake and CO2 evolution (qO2 and qCO2, respectively) (Fig. 2C). The respiratory quotient (RQ) was >1 during the postpulse period, consistent with respiratory fermentative metabolism (Fig. 2D).
TABLE 2.
Steady-state concentrations of different classes of intracellular metabolites
| Metabolite | Concn (μmol Cmolx−1)a |
|---|---|
| Glycolytic intermediates | |
| G6P | 53 ± 2.6 |
| F6P | 9.9 ± 0.8 |
| F1,6bP | 7.1 ± 0.7 |
| 2PG + 3PG | 24 ± 1.2 |
| PEP | 25 ± 2.4 |
| Pyr | 2.2 ± 0.2 |
| TCA cycle intermediates | |
| Cit + iCit | 140 ± 5.1 |
| αKG | 2.2 ± 0.2 |
| Suc | 1.5 ± 0.3 |
| Fum | 1.4 ± 0.2 |
| Mal | 7.3 ± 0.8 |
| PP pathway intermediates | |
| 6PG | 6.1 ± 0.6 |
| Storage carbon metabolism | |
| G1P | 7.8 ± 2.2 |
| T6P | 18 ± 0.9 |
| Trehalose | 2.0 × 103 ± 38 |
| M6P | 22 ± 4.2 |
| Free amino acids | |
| Asp | 36 ± 0.4 |
| Ser | 8.3 ± 2.0 |
| Asn | 30 ± 1.4 |
| Glm | 362 ± 4.4 |
| Gln | 142 ± 1.6 |
| His | 36 ± 1.6 |
| Thr | 11 ± 0.1 |
| Ala | 55 ± 0.7 |
| Arg | 47 ± 0.7 |
| Pro | 7.9 ± 1.4 |
| Tyr | 6.8 ± 0.2 |
| Val | 20 ± 0.7 |
| Orn | 27 ± 0.7 |
| Lys | 23 ± 0.3 |
| Ile | 5.7 ± 0.1 |
| Leu | 35 ± 0.7 |
| Phe | 3.2 ± 0.3 |
| Adenine nucleotides | |
| ATP | 192 ± 0.3 |
| ADP | 45 ± 1.3 |
| AMP | 12 ± 1.5 |
Average metabolite concentrations with standard deviations were determined from duplicate analyses of three steady-state samples.
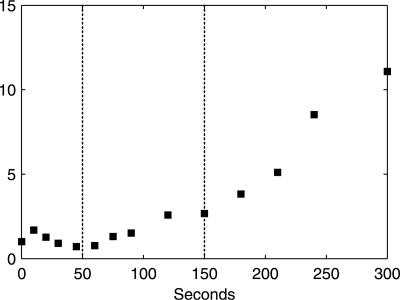
FIG. 2.
Estimated biomass-specific fluxes of extracellular metabolites. (A) qglucose; (B) qethanol (□) and qacetate (⧫); (C) qO2 (solid line) and qCO2 (dotted line); (D) RQ. The vertical dotted lines indicate the three metabolic phases.
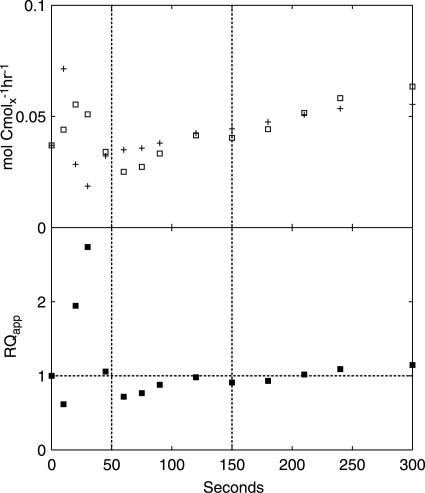
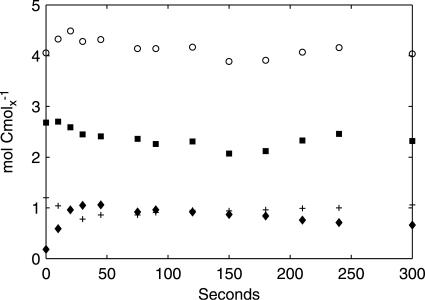
Nonfermentative CO2 evolution and O2 uptake provide a measure of changes in the flux towards respiratory, nonfermentative metabolism, i.e., the conversion of glucose to CO2 and biomass components coupled to NADH generation in lower glycolysis and the TCA cycle. The qO2 and qCO2 values associated with nonfermentative metabolism can be calculated, along with the apparent RQ, from estimated extracellular fluxes, as follows:
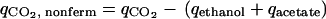 |
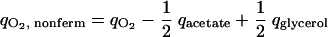 |
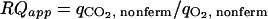 |
assuming an NADP+-dependent acetaldehyde dehydrogenase (Fig. 3). Notably, both qO2 and qCO2 are closely related to flux through the TCA cycle, which is a major source of NADH and nonfermentative CO2 and accounts for 69% and 82% of the steady-state qO2 and qCO2, respectively.
FIG. 3.
Estimated nonfermentative qO2 (+) and qCO2 (□) (upper panel) and apparent RQ (lower panel). The vertical dotted lines indicate the three metabolic phases.
We divided the 300-second postpulse period into three sequential metabolic phases that are characterized by distinct metabolic responses for a number of metabolites (see Fig. 4 to 7).
FIG. 4.
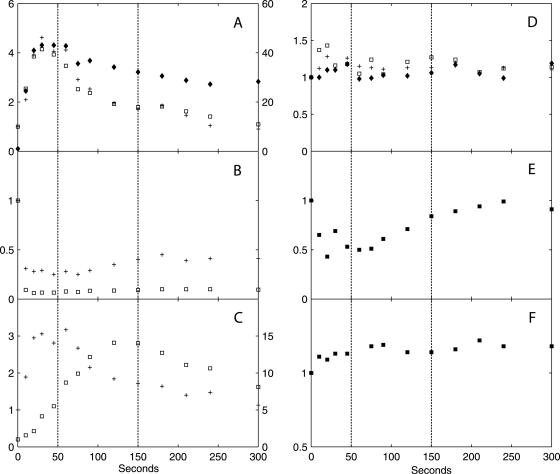
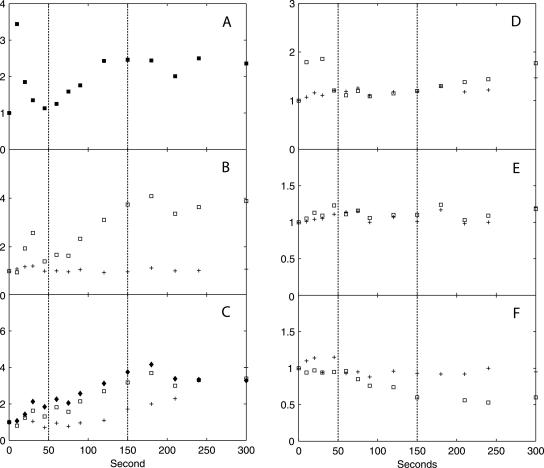
Dynamic responses of intermediates in glycolysis, the PP pathway, and storage carbon metabolism and thence-derived biosynthetic precursors, in x-fold changes relative to the reference steady state. (A) G6P (□, left axis), F6P (+, left axis), and F1,6bP (⧫, right axis); (B) 2PG + 3PG (+) and PEP (□); (C) G1P (+, left axis) and T6P (□, right axis); (D) His (+), Phe (□), and Tyr (⧫); (E) 6PG; (F) trehalose. The relative standard deviation of metabolite concentrations at each time point is within 10%, based on duplicate determinations. The vertical dotted lines indicate the three metabolic phases.
FIG. 7.
Dynamic response of F2,6bP (in x-fold changes relative to the reference steady state). The vertical dotted lines indicate the three metabolic phases.
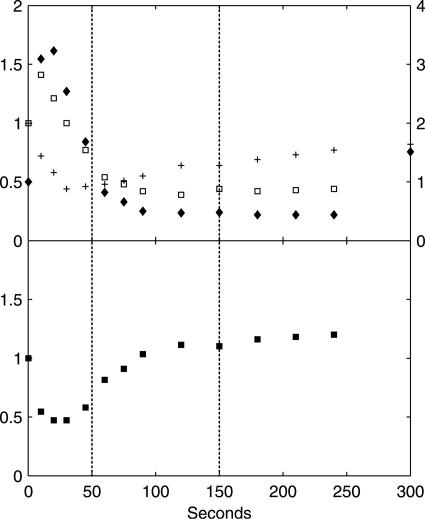
In phase I (0 to 50 seconds after the glucose pulse), highly dynamic changes occurred in metabolic fluxes and intracellular metabolite concentrations. qglucose increased to 17× the steady-state flux (qglucose0) at 10 seconds, followed by a sharp drop to ∼3× qglucose0 at 50 seconds. Similar dynamic patterns were found for qO2 and qCO2. Both nonfermentative qO2 and qCO2 decreased after an initial increase, indicating a similar change in flux through the TCA cycle. Ethanol production in this phase was not significant and was much lower than qacetate. Intracellular metabolite responses in phase I were characterized by accumulations in the hexose phosphate pool, T6P, and trehalose (relative to already high steady-state levels) (Table 2) and decreases in 6PG and the phosphorylated C3 pool, i.e., 2PG, 3PG, and PEP (Fig. 4). In addition, peaks in the concentrations of pyruvate and several TCA cycle intermediates were observed (Fig. 5). The ATP concentration and the energy charge, defined as (ATP + 1/2 ADP)/(ATP + ADP + AMP), sharply decreased (Fig. 6). The ADP and AMP concentrations both decreased after an initial increase.
FIG. 5.
Dynamic responses of pyruvate, TCA cycle intermediates, and thence-derived free amino acids, in x-fold changes relative to the reference steady state. (A) Pyr; (B) Cit + iCit (+) and αKG (□); (C) Suc (+), Fum (□), and Mal (⧫); (D) Ala (+) and Val (□); (E) Glm (+) and Gln (□); (F) Asn (+) and Asp (□). The relative standard deviation of metabolite concentrations at each time point is within 10%, based on duplicate determinations. The vertical dotted lines indicate the three metabolic phases.
FIG. 6.
(Upper panel) Dynamic responses of ATP (+, left axis), ADP (□, left axis), and AMP (⧫, right axis), in x-fold changes relative to the reference steady state. (Lower panel) Change in energy charge relative to the steady state. The energy charge in the steady state is 0.86, as calculated from the steady-state ATP, ADP, and AMP concentrations in Table 2. The relative standard deviation of metabolite concentrations at each time point is within 10%, based on duplicate determinations. The vertical dotted lines indicate the three metabolic phases.
In phase II (50 to 150 seconds after the pulse), gradual changes in the extracellular fluxes and intracellular metabolite concentrations were observed, and qglucose increased from 3× to 8× qglucose0. Fermentative metabolism began, as evidenced by strong increases in qethanol, qCO2, and the RQ. The ATP level and the energy charge recovered due to increased ATP generation in both fermentative metabolism, i.e., increased qethanol, and oxidative phosphorylation, i.e., increased qO2. In the cell, the concentrations of the hexose phosphates decreased, whereas those of the TCA cycle intermediates, T6P, and F2,6bP (Fig. 7) increased gradually.
In phase III (150 to 300 seconds after the pulse), the extracellular fluxes and metabolite concentrations remained rather constant. The magnitude of metabolite concentration changes, i.e., |dx/dt|, was negligible compared to the magnitude of extracellular fluxes, indicating that an intracellular quasi-steady state had been established. During phases II and III, nonfermentative qO2 and qCO2 continued to increase, up to 1.5× and 1.7× their steady-state values, respectively, indicating an increase in flux through the TCA cycle. The apparent RQ remained close to 1.
No significant changes were observed in intracellular free amino acid concentrations in all three phases (Fig. 4 and 5). The only exception was aspartate, with a 2× decrease in phase III relative to the reference steady state.
Cumulative carbon, electron, and ATP balances.
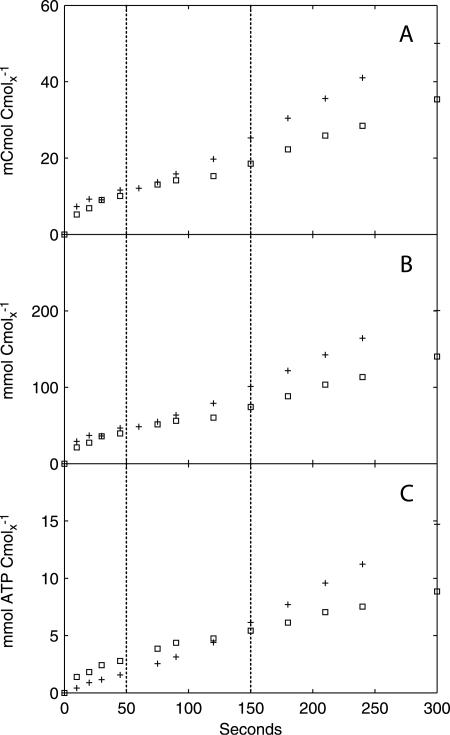
The available intra- and extracellular metabolite measurements were combined with the in vivo qO2 and qCO2 values to construct the cumulative carbon, electron, and ATP balances. The measured glycogen concentrations in the insoluble residues and the cell extracts were ∼3.1 mCmol Cmolx−1 and ∼0.5 mCmol Cmolx−1, respectively, which is much lower than the glycogen content of 102 mCmol Cmolx−1 reported for S. cerevisiae grown at 0.05 h−1 (13). The alkaline extraction seemed to be incomplete, presumably due to the use of insoluble residue obtained after boiling ethanol treatment rather than making the extraction directly from yeast cells, as previously described (27). Hence, we considered the glycogen measurements unreliable and excluded them from the balance calculations. The cumulative carbon, electron, and ATP balances (Fig. 8) were evaluated for each of the three metabolic regimens during the postpulse period.
FIG. 8.
Cumulative carbon (A), electron (B), and ATP (C) balances. Symbols: +, carbon/electron source and ATP production; □, carbon/electron sink and ATP consumption. The vertical dotted lines indicate the three metabolic phases.
For phase I (0 to 50 seconds), ∼85% of the measured carbon and electron uptake can be accounted for by the known carbon and electron sinks (Tables 3 and 4). More than 60% of the consumed glucose was found in (phosphorylated) intracellular metabolites in upper glycolysis and storage carbon metabolism, while 20% was converted to secreted compounds (ethanol, acetate, and CO2) and drained towards anabolism. The ATP balance (Table 5) shows that the ATP consumption exceeded its production, which is consistent with the sharp drop in the ATP level and energy charge. However, by the end of phase I, the resulting energy depletion (1.2 mmol ATP equivalents Cmolx−1) largely exceeded the observed total decrease in ATP (0.1 mmol ATP Cmolx−1). Since anabolism represents a major fraction of ATP consumption, but only a minor fraction of the carbon balance, the calculated difference between ATP consumption and production might indicate a temporary decrease of the anabolic flux (qanabolism).
TABLE 3.
Relative contributions of different carbon sinks to cumulative glucose uptake relative to time zero
| Carbon source or sink | Contribution (%) to cumulative glucose uptake or uptake value (mCmol Cmolx−1)
|
||
|---|---|---|---|
| 45 s | 150 s | 300 s | |
| Cumulative glucose uptake | 12 | 25 | 50 |
| Carbon sinks | |||
| Extracellular sinks | |||
| Ethanol | 2.0 | 12 | 19 |
| Acetate | 4.3 | 5.7 | 4.7 |
| CO2 | 7.8 | 14 | 20 |
| Glycerol | 0.0 | 0.8 | 2.0 |
| Anabolisma | 5.4 | 8.3 | 8.3 |
| Intracellular sinks | |||
| Glycolytic intermediates | 27 | 6.7 | 2.4 |
| Storage carbon | 36 | 25 | 12 |
| TCA cycle intermediates | 0.1 | 0.4 | 0.4 |
| Free amino acids | 5.4 | 0.8 | 1.7 |
| Unquantified sinks | 13 | 27 | 29 |
A constant qanabolism was assumed, corresponding to a growth rate of 0.05 h−1.
TABLE 4.
Relative contributions of different electron sinks to cumulative electron uptake relative to time zero
| Electron source or sink | Contribution (%) to cumulative electron uptake or uptake value (mmol Cmolx−1)
|
||
|---|---|---|---|
| 45 s | 150 s | 300 s | |
| Cumulative electron uptake | 47 | 101 | 200 |
| Electron sinks | |||
| Extracellular sinks | |||
| Ethanol | 3.0 | 17 | 29 |
| Acetate | 4.3 | 5.7 | 4.7 |
| Glycerol | 0.0 | 0.9 | 2.4 |
| Anabolisma | 5.7 | 8.7 | 8.8 |
| O2 | 5.2 | 7.8 | 8.7 |
| Intracellular sinks | |||
| Glycolytic intermediates | 27 | 6.7 | 2.4 |
| Storage carbon | 37 | 25 | 12 |
| TCA cycle intermediates | 0.2 | 0.3 | 0.3 |
| Free amino acids | 5.0 | 0.8 | 1.7 |
| Unquantified sinks | 15 | 27 | 30 |
A constant qanabolism was assumed, corresponding to a growth rate of 0.05 h−1.
TABLE 5.
Cumulative energy generation and consumption relative to time zero
| Energy status | Process | Energy (mmol ATP equivalents Cmolx−1)
|
||
|---|---|---|---|---|
| 45 s | 150 s | 300 s | ||
| Generation | Acetate secretion | 0.3 | 0.7 | 1.2 |
| Ethanol secretion | 0.1 | 1.5 | 4.8 | |
| Oxidative phosphorylation | 1.2 | 4.0 | 8.7 | |
| Total | 1.6 | 6.2 | 14.7 | |
| Consumption | Anabolisma | 1.0 | 3.3 | 6.6 |
| Glycerol secretion | 0.0 | 0.1 | 0.4 | |
| Accumulation of phosphorylated compounds and storage material | 1.8 | 2.1 | 1.8 | |
| Total | 2.8 | 5.4 | 8.9 | |
A constant qanabolism was assumed, corresponding to a growth rate of 0.05 h−1.
In summary, in phase I intense dynamics in glycolysis lead to elevated pools of phosphorylated compounds and storage material, leading to a large energy drain and possible decrease in qanabolism. Only 2% of the consumed glucose was converted to ethanol. This result suggests that in biomass-directed industrial applications of S. cerevisiae, undesired ethanol formation due to inhomogeneous distribution and mixing in large-scale fermentors (29) could be reduced if the mixing time, i.e., the maximum time that yeast cells encounter a local glucose excess, is reduced to <50 seconds.
In phase II (50 to 150 seconds), a gap between the carbon/electron sources and sinks emerged at about 100 seconds. At 150 seconds, 26% of the consumed glucose had been channeled into unquantified sinks, and another 26% was involved in fermentative metabolism, which was the main carbon and electron sink at the end of phase II. With the onset of fermentative metabolism, qO2 increased and ATP generation accelerated. The ATP balance shows a slight ATP excess at 150 seconds, which corresponds to the measured increase in the ATP concentration and the recovery of the energy charge.
In phase III (150 to 300 seconds), the difference between the carbon/electron sources and sinks continued to increase. At 300 seconds, 29% of the consumed glucose was channeled into unquantified sinks. Fermentative metabolism was the largest carbon and electron sink, with ethanol, acetate, and the related CO2 evolution accounting for 36% of the total glucose uptake at the end of phase III. The ATP balance has a large energy surplus, which indicates that the unquantified carbon/electron sinks might be related to an unidentified energy-consuming process.
Estimation of qanabolism in phase III.
The amount of carbon/electrons and energy withdrawn by anabolic processes is constant in the balance calculations due to an assumed constant growth rate of 0.05 h−1 during the postpulse period. An increase in qanabolism could potentially account for the missing carbon and electrons and the surplus ATP equivalents. However, such an increase would produce a practically undetectable change in the biomass dry weight, since even assuming that all of the carbon that had not been accounted for was converted to biomass, it would increase the dry weight only ∼7.6 mCmolx liter−1 (0.2 g dry weight liter−1), relative to a steady-state biomass concentration of 573.6 mCmolx liter−1 (15 g dry weight liter−1).
To quantify possible changes in qanabolism in phase III, we used a reaction network model, which contains six macroscopic processes (Table 6) and assumes a quasi-steady state in phase III. This type of model has been used successfully to describe catabolic and anabolic processes in postpulse periods lasting several hours (12). The quasi-steady-state fluxes were estimated from metabolite concentration profiles at the end of phase III (Table 6). We used qethanol, qacetate, qglycerol, qO2, and two metabolite balances, i.e., CO2 and NADH (assuming a constant intracellular NADH concentration), to calculate the remaining two unknown fluxes, qanabolism and qcatabolism. The calculated qanabolism corresponds to a growth rate of 0.14 h−1, relative to a steady-state growth rate of 0.05 h−1. Thus, an increased qanabolism is consistent with the measured extracellular fluxes and the known macroscopic stoichiometry.
TABLE 6.
Stoichiometry of macroscopic model and estimated fluxes at the end of phase III
| Flux | mol Cmolx−1 h−1 | Macroscopic process |
|---|---|---|
| qethanol | 0.081 | 1/2 Glucose → ethanol + CO2 + ATP |
| qacetate | 0.008 | 1/2 Glucose → acetate + CO2 + ATP + NADH + NADPHc |
| qglycerol | 0.009 | 1/2 Glucose + ATP + NADH → glycerol |
| qanabolisma | 0.2 Glucose + 1.65 ATP → biomass + 0.24 CO2 + 0.38 NADH | |
| qcatabolism | Glucose → 6 CO2 + 12 NADH + 2 ATP | |
| qO2b | −0.061 | O2 + 2 NADH → 2 ATP |
| qglucosed | −0.096 | |
| qCO2d | 0.157 |
The stoichiometry of the anabolic reaction was adapted from reference 51.
An effective P/O ratio of 1 was assumed, as reported previously (51).
For simplicity, NADPH was not balanced.
qglucose and qCO2 represent the sums of fluxes through all glucose-consuming and CO2-producing processes, respectively.
The measured glucose uptake rate and the ATP balance were not utilized in the flux calculation and can be used as an independent check of the model. Two different qanabolism values were compared; one corresponds to an accelerated growth rate of 0.14 h−1, and the other corresponds to a steady-state growth rate of 0.05 h−1 (Table 7). Assuming increased qanabolism, both the carbon and electron recovery improved by 16% to 85% at the end of phase III. In addition, the increase in qanabolism can be energetically sustained, since ATP production now closely matches its consumption, while a qanabolism value corresponding to an unchanged growth rate would result in a large ATP surplus. Thus, an increased rather than unchanged qanabolism is consistent with the ATP balance; moreover, this increase is consistent with increased flux through nonfermentative respiratory metabolism in phases II and III, as indicated by increased qO2, nonferm and qCO2, nonferm.
TABLE 7.
Comparison of carbon, electron, and ATP balances in phase III under two assumptionsa
| Parameter | Carbon balance (Cmol Cmolx−1 h−1)
|
Electron balance (mol Cmolx−1 h−1)
|
ATP balance (mol ATP equivalents Cmolx−1 h−1)
|
|||
|---|---|---|---|---|---|---|
| A | B | A | B | A | B | |
| Source comparison | 0.58 | 0.58 | 2.3 | 2.3 | 0.22 | 0.22 |
| Sink comparison | 0.49 | 0.40 | 2.0 | 1.6 | 0.22 | 0.08 |
| Recovery (%) | 85 | 69 | 85 | 69 | 99 | 35 |
Assumption A, increased qanabolism, corresponding to a growth rate of 0.14 h−1; assumption B, qanabolism corresponding to a steady-state growth rate of 0.05 h−1.
Intracellular free and bound phosphate.
Intracellular phosphate exists in the free ion form (Pi) or can be bound in phosphorylated pathway intermediates, nucleoside phosphates, and pyro- and polyphosphate. We constructed a balance for the sum of free and bound intracellular phosphates with measured concentrations of adenine nucleotides, phosphorylated intermediates, and intracellular Pi (Fig. 9). Increased concentrations of phosphorylated intermediates are compensated for by decreased concentrations of adenine nucleotides and Pi, leading to a relatively constant amount of the measured free and bound phosphate in the cell during the postpulse period. This implies that the hydrolysis of pyro- or polyphosphate is not significant during the postpulse period for energy generation.
FIG. 9.
Balance of intracellular free and bound phosphate. The graph shows total free and bound phosphates present in measured intracellular metabolites (○), Pi (▪), phosphate bound in adenine nucleotides (+), and phosphate bound in pathway intermediates (⧫).
DISCUSSION
Comparison with previous glucose pulse experiments.
Responses to glucose excess have been measured for different strains of S. cerevisiae grown in glucose-limited chemostats (38, 45, 52). The time window of observation can be several hundred seconds to several hours. The measured responses of extracellular metabolites, intracellular glycolytic intermediates, and adenine nucleotides in this study are qualitatively comparable with previous observations of short-term glucose excess (5, 38, 52). The rapid increase in glucose influx in phase I, from 0.013 to 0.22 mol Cmolx−1 h−1 10 seconds after the pulse, is further consistent with a calculated maximal flux of 0.21 mol Cmolx−1 h−1 based on the Michaelis-Menten kinetic parameters estimated from zero-trans experiments (10).
A number of metabolic responses observed in this study were different from those observed in similar glucose pulse experiments with S. cerevisiae. In phase I, an initial decrease of the 6PG concentration was observed, although an increase has been reported previously (49). The difference could be due to differences in the S. cerevisiae strain, growth rate, or carbon source used in the studies. Notably, glucose was the sole carbon source used for the previous study (49), while a small amount of ethanol was supplied in this study, which might yield additional NADPH via the NADP+-dependent acetaldehyde dehydrogenase and hence could result in a lower steady-state PP split ratio. The drop of the 6PG concentration in our experiment might be due to an inhibition of G6P dehydrogenase by excess NADPH from NADP+-dependent acetaldehyde dehydrogenase that accompanied the sudden increase in qacetate (Fig. 2).
In phase I, a considerable amount of carbon was directed toward storage carbon metabolism. This result differs from previous reports of degradation of trehalose and glycogen by derepressed and chemostat-grown S. cerevisiae exposed to glucose excess, based on observations over a period of hours (42, 47). The difference may be due to a mutation in adenylate cyclase in the yeast strain used in this study, which largely abolishes the glucose-induced increase in cyclic AMP (cAMP) concentration (43). We measured a rather constant cAMP concentration in our experiment (M. T. A. P. Kresnowati, unpublished results), in contrast with strong increases observed for a different S. cerevisiae strain exposed to glucose excess (50). Since neutral trehalase (the major trehalose-degrading enzyme) is activated through a cAMP-dependent protein kinase A (39), S. cerevisiae strains carrying this mutation exhibit a glucose-induced accumulation of trehalose and, to a lesser extent, glycogen (43).
In phase II, F2,6bP began to accumulate, as observed by Vaseghi et al. (50) in a similar glucose pulse experiment with chemostat-grown S. cerevisiae CBS 7336. However, in their experiment, the cAMP concentration also increased, which activated the PKA, which in turn led to the phosphorylation of PFK2 by PKA. The absence of cAMP accumulation by the S. cerevisiae strain used in this study implies a PKA-independent regulation of phosphofructokinase 2 activity by, for example, other protein kinases (11) or allosteric mechanisms (15). The observed increases in F2,6bP concentration seen previously (50) and in this study suggest that within the short time window of observation (i.e., 300 seconds), protein phosphorylation needs to be taken into account in kinetic models, in addition to enzyme-metabolite interactions.
Reconciliation with known kinetic mechanisms.
A number of kinetic mechanisms known from in vitro studies have been shown to be consistent with metabolic responses observed in glucose pulse experiments; for example, the activation of pyruvate kinase by F1,6bP in vitro (23) serves as a possible explanation for the rapid decrease in PEP concentration upon an increase in F1,6bP concentration (38). In the following discussion, we evaluate the consistency of known in vitro kinetic mechanisms of primary metabolism of S. cerevisiae with the estimated metabolic fluxes and measured metabolite concentrations obtained in this study.
During primary metabolism, the reversible reactions catalyzed by phosphoglucoisomerase, aldolase, enolase, and fumarase are close to equilibrium in glucose-limited chemostat cultures of S. cerevisiae, as confirmed by measured mass isotopomer distributions of the reactants involved (48). With a glucose excess, G6P and F6P as well as fumarate and malate show highly correlated concentration changes, indicating that the reversible reactions catalyzed by phosphoglucoisomerase and fumarase are close to equilibrium. The reversible reaction catalyzed by enolase, however, is displaced from equilibrium, as can be seen from the more pronounced decrease of the concentration of PEP than that of 2PG plus 3PG.
The decrease of qglucose in phase I after the rapid initial increase indicates that glucose transport is subject to significant inhibition. Candidate metabolites that inhibit the facilitated diffusion of glucose are glucose (36) and G6P (2, 31), with the latter showing a strong increase after the pulse. We cannot exclude, however, the inhibitory effect of intracellular glucose, as it was not measured.
The intermediates in upper glycolysis (G6P, F6P, and F1,6bP) accumulated in phase I even after the drop in qglucose, which implies that the in vivo fluxes through the enzymes that metabolize these intermediates, i.e., hexokinase (HK), PFK, and the enzymes in lower glycolysis, must have decreased. Several known kinetic mechanisms can explain this seeming paradox. First, a strong decrease in the ATP concentration and/or the energy charge can limit the fluxes through HK and PFK (38). HK might be further inhibited by elevated levels of T6P (Fig. 4) (4, 16). In addition, decreased flux through PFK can be due to hyperaccumulation of the product F1,6bP (37) as well as to decreased levels of the allosteric activator AMP (after about 20 seconds, Fig. 6) (25). During phase I, the concentration of F2,6bP, a strong activator of PFK (1), remained close to the steady-state level. In phase II, the concentrations of G6P, F6P, and F1,6bP decreased, while qglucose increased, which implies that the in vivo fluxes through HK, PFK, and lower glycolysis must have increased. The stimulatory effects leading to the increased fluxes could be a recovered energy charge in phase II and elevated levels of F2,6bP (Fig. 7).
Pyruvate is partitioned between the TCA cycle and the fermentative pathway leading to ethanol and acetate. In phase I, qacetate is larger than qethanol, which could be explained by a much lower affinity of alcohol dehydrogenase than acetaldehyde dehydrogenase for acetaldehyde (28). The acetaldehyde dehydrogenase could be saturated with acetaldehyde, leading to the rather constant qacetate observed during the postpulse period. Furthermore, we observed that changes in pyruvate concentration paralleled the estimated nonfermentative qCO2 (which closely correlates with flux through the TCA cycle) and were similar to changes observed in most TCA cycle intermediates (Fig. 5). The in vitro Km estimated for pyruvate oxidation by intact mitochondria (0.3 mM) (46) is much higher than the pyruvate concentrations we observed, which suggests that the TCA cycle flux can respond quickly to changes in its precursor concentration.
The concentration increases of the more reduced fumarate and malate metabolites were accompanied by a concentration decrease of the more oxidized aspartate, while the sum of these C4 metabolites remained almost constant during the postpulse period (not shown). These results suggest that the fluxes into and out of the C4 pool are well balanced and that the redistribution of the pool components towards a more reduced status might be triggered by an increased free NADH/NAD+ ratio. This result is qualitatively consistent with an estimated increase in the cytosolic free NADH/NAD+ ratio by Theobald et al. (38).
Implications from carbon, electron, and energy balances.
The carbon, electron, and energy balances suggest that qanabolism increased during phase III. The onset of anabolic processes does not seem to be related to concentration changes in free amino acids as direct precursors, as their average levels in cells were not significantly affected by the glucose pulse. In addition, during a prolonged chemostat cultivation of S. cerevisiae CEN.PK 113-7D, the growth rate remained constant for up to 90 generations, while average concentrations of free amino acids in the cells decreased severalfold (20). A possible signal for increased qanabolism might be a net increase in cellular energy. Although the energy charge is only slightly higher in phase III than in the steady state, the individual ratios of ATP/ADP and ATP/AMP are nearly twice their steady-state values. An increased qanabolism also implies an excess of ribosomal proteins at the 0.05 h−1 growth rate, as the amount of ribosomes for protein synthesis probably remains constant during the relatively short postpulse period. This excess capacity indicates a low ribosomal efficiency, which has been observed previously for low specific growth rates in both prokaryotes (17) and eukaryotes (6, 35).
The unmeasured carbon sinks account for ∼15% of the glucose uptake at the end of phase III (assuming increased qanabolism) (Table 7). The carbon, electron, and ATP balances suggest that these as yet unmeasured carbon sinks have an average degree of reduction of 4.17 per Cmol and do not involve intensively energy-consuming pathways. One such carbon sink could be glycogen (degree of reduction per Cmol = 4.0), which has a rather low energy requirement (0.33 mol of ATP mol of glycogen−1, compared to 1.65 mol of ATP Cmolx−1 for biomass). Given the high glycogen content in yeast cells, i.e., 102 mCmol Cmolx−1 (13), complete conversion of the remaining unquantified carbon sinks to glycogen would increase the cellular glycogen content only 7%, which would require a highly accurate method of glycogen quantification for correct assessment.
In summary, this study provides a comprehensive picture of the extra- and intracellular dynamic responses of S. cerevisiae within 300 seconds of a glucose pulse. The constructed carbon and electron balances close >70% in the 300-second postpulse period and are thus consistent with these measurements representing the major sinks. The discrepancy in the ATP balance in phases I and III, as well as the missing carbon and electron sinks in phase III, suggests that significant changes in the anabolic flux can occur during short transient periods. Our findings emphasize the importance of including storage and anabolic processes in kinetic models, in addition to glycolysis and fermentative metabolism, which have already been included (30, 49). Future research is needed to validate the estimated increase in qanabolism and to quantify possible changes in glycogen content.
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research (NWO) and by DSM.
REFERENCES
- 1.Avigad, G. 1981. Stimulation of yeast phosphofructokinase activity by fructose 2,6-bisphosphate. Biochem. Biophys. Res. Commun. 102:985-991. [DOI] [PubMed] [Google Scholar]
- 2.Azam, F., and A. Kotyk. 1969. Glucose-6-phosphate as regulator of monosaccharide transport in baker's yeast. FEBS Lett. 2:333-335. [DOI] [PubMed] [Google Scholar]
- 3.Bellaver, L. H., N. M. de Carvalho, J. Abrahao-Neto, and A. K. Gombert. 2004. Ethanol formation and enzyme activities around glucose-6-phosphate in Kluyveromyces marxianus CBS 6556 exposed to glucose or lactose excess. FEMS Yeast Res. 4:691-698. [DOI] [PubMed] [Google Scholar]
- 4.Blazquez, M. A., R. Lagunas, C. Gancedo, and J. M. Gancedo. 1993. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 329:51-54. [DOI] [PubMed] [Google Scholar]
- 5.Bloemen, H. J. J., L. Wu, W. M. van Gulik, J. J. Heijnen, and M. H. G. Verhaegen. 2003. Reconstruction of the O2 uptake rate and CO2 evolution rate on a time scale of seconds. AIChE J. 49:1895-1908. [Google Scholar]
- 6.Bushell, M. E., and A. T. Bull. 1999. Sporulation at minimum specific growth rate in Aspergillus nidulans chemostat culture predicted using protein synthesis efficiency estimations. J. Basic Microbiol. 39:293-298. [DOI] [PubMed] [Google Scholar]
- 7.Chassagnole, C., N. Noisommit-Rizzi, J. W. Schmid, K. Mauch, and M. Reuss. 2002. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng. 79:53-73. [DOI] [PubMed] [Google Scholar]
- 8.Claire, R. L., III. 2000. Positive ion electrospray ionization tandem mass spectrometry coupled to ion-pairing high-performance liquid chromatography with a phosphate buffer for the quantitative analysis of intracellular nucleotides. Rapid Commun. Mass Spectrom. 14:1625-1634. [DOI] [PubMed] [Google Scholar]
- 9.de Jong-Gubbels, P., P. Vanrolleghem, S. Heijnen, J. P. van Dijken, and J. T. Pronk. 1995. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast 11:407-418. [DOI] [PubMed] [Google Scholar]
- 10.Diderich, J. A., M. Schepper, P. van Hoek, M. A. Luttik, J. P. van Dijken, J. T. Pronk, P. Klaassen, H. F. Boelens, M. J. de Mattos, K. van Dam, and A. L. Kruckeberg. 1999. Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 274:15350-15359. [DOI] [PubMed] [Google Scholar]
- 11.Dihazi, H., R. Kessler, and K. Eschrich. 2003. Glucose-induced stimulation of the Ras-cAMP pathway in yeast leads to multiple phosphorylations and activation of 6-phosphofructo-2-kinase. Biochemistry 42:6275-6282. [DOI] [PubMed] [Google Scholar]
- 12.Duboc, P., U. von Stockar, and J. Villadsen. 1998. Simple generic model for dynamic experiments with Saccharomyces cerevisiae in continuous culture: decoupling between anabolism and catabolism. Biotechnol. Bioeng. 60:180-189. [DOI] [PubMed] [Google Scholar]
- 13.Guillou, V., L. Plourde-Owobi, J. L. Parrou, G. Goma, and J. Francois. 2004. Role of reserve carbohydrates in the growth dynamics of Saccharomyces cerevisiae. FEMS Yeast Res. 4:773-787. [DOI] [PubMed] [Google Scholar]
- 14.Herwig, C., I. Marison, and U. von Stockar. 2001. On-line stoichiometry and identification of metabolic state under dynamic process conditions. Biotechnol. Bioeng. 75:345-354. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, E., A. Bedri, R. Kessler, M. Kretschmer, and W. Schellenberger. 1989. 6-Phosphofructo-2-kinase and fructose-2,6-bisphosphatase from Saccharomyces cerevisiae. Adv. Enzyme Regul. 28:283-306. [DOI] [PubMed] [Google Scholar]
- 16.Hohmann, S., W. Bell, M. J. Neves, D. Valckx, and J. M. Thevelein. 1996. Evidence for trehalose-6-phosphate-dependent and -independent mechanisms in the control of sugar influx into yeast glycolysis. Mol. Microbiol. 20:981-991. [DOI] [PubMed] [Google Scholar]
- 17.Ingraham, J. L., O. Maaloee, and F. C. Neidhardt. 1983. Growth of the bacterial cell. Sinauer, Sunderland, Mass.
- 18.Lange, H. C., M. Eman, G. van Zuijlen, D. Visser, J. C. van Dam, J. Frank, M. J. de Mattos, and J. J. Heijnen. 2001. Improved rapid sampling for in vivo kinetics of intracellular metabolites in Saccharomyces cerevisiae. Biotechnol. Bioeng. 75:406-415. [DOI] [PubMed] [Google Scholar]
- 19.Lange, H. C., and J. J. Heijnen. 2001. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75:334-344. [DOI] [PubMed] [Google Scholar]
- 20.Mashego, M. R., M. L. Jansen, A. Hassane, J. L. Vinke, W. M. van Gulik, J. L. Pronk, and J. J. Heijnen. 2005. Changes in the metabolome of Saccharomyces cerevisiae associated with long-term aerobic glucose limited chemostat cultivation. FEMS Yeast Res. 5:419-430. [DOI] [PubMed] [Google Scholar]
- 21.Mashego, M. R., W. M. van Gulik, J. L. Vinke, and J. J. Heijnen. 2003. Critical evaluation of sampling techniques for residual glucose determination in carbon-limited chemostat culture of Saccharomyces cerevisiae. Biotechnol. Bioeng. 83:395-399. [DOI] [PubMed] [Google Scholar]
- 22.Mashego, M. R., L. Wu, J. C. Van Dam, C. Ras, J. L. Vinke, W. A. Van Winden, W. M. Van Gulik, and J. J. Heijnen. 2004. MIRACLE: mass isotopomer ratio analysis of U-13C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol. Bioeng. 85:620-628. [DOI] [PubMed] [Google Scholar]
- 23.Murcott, T. H., H. Gutfreund, and H. Muirhead. 1992. The cooperative binding of fructose-1,6-bisphosphate to yeast pyruvate kinase. EMBO J. 11:3811-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, J., and J. Villadsen. 1994. Bioreaction engineering principles. Kluwer Academic Publishers, New York, N.Y.
- 25.Nissler, K., A. Otto, W. Schellenberger, and E. Hofmann. 1983. Similarity of activation of yeast phosphofructokinase by AMP and fructose-2,6-bisphosphate. Biochem. Biophys. Res. Commun. 111:294-300. [DOI] [PubMed] [Google Scholar]
- 26.Ostergaard, S., L. Olsson, and J. Nielsen. 2001. In vivo dynamics of galactose metabolism in Saccharomyces cerevisiae: metabolic fluxes and metabolite levels. Biotechnol. Bioeng. 73:412-425. [DOI] [PubMed] [Google Scholar]
- 27.Parrou, J. L., and J. Francois. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248:186-188. [DOI] [PubMed] [Google Scholar]
- 28.Postma, E., C. Verduyn, W. A. Scheffers, and J. P. Van Dijken. 1989. Enzymic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 55:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuss, M., and R. K. Bajpai. 1991. Stirred tank modelling, p. 299-348. In H.-J. Rehm and G. Reed (ed.) Biotechnology, vol. 4. Measuring and control, 2nd ed. VCH Verlag, Weinheim, Germany. [Google Scholar]
- 30.Rizzi, M., M. Baltes, U. Theobald, and M. Reuss. 1997. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae. II. Mathematical model. Biotechnol. Bioeng. 55:592-608. [DOI] [PubMed] [Google Scholar]
- 31.Rizzi, M., U. Theobald, E. Querfurth, T. Rohrhirsh, M. Baltes, and M. Reuss. 1996. In vivo investigations of glucose transport in Saccharomyces cerevisiae. Biotechnol. Bioeng. 49:316-327. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz, M., E. Hirsch, J. Bongaerts, and R. Takors. 2002. Pulse experiments as a prerequisite for the quantification of in vivo enzyme kinetics in aromatic amino acid pathway of Escherichia coli. Biotechnol. Prog. 18:935-941. [DOI] [PubMed] [Google Scholar]
- 33.Stephanopoulos, G., A. A. Aristidou, and J. Nielsen. 1998. Metabolic engineering: principles and methodologies. Academic Press, San Diego, Calif.
- 34.Stuckrath, I., H. C. Lange, P. Kotter, W. M. van Gulik, K. D. Entian, and J. J. Heijnen. 2002. Characterization of null mutants of the glyoxylate cycle and gluconeogenic enzymes in S. cerevisiae through metabolic network modeling verified by chemostat cultivation. Biotechnol. Bioeng. 77:61-72. [DOI] [PubMed] [Google Scholar]
- 35.Sturani, E., M. G. Costantini, E. Martegani, and L. Alberghina. 1979. Level and turnover of polyadenylate-containing ribonucleic acid in Neurospora crassa in different steady states of growth. Eur. J. Biochem. 99:1-7. [DOI] [PubMed] [Google Scholar]
- 36.Teusink, B., J. A. Diderich, H. V. Westerhoff, K. van Dam, and M. C. Walsh. 1998. Intracellular glucose concentration in derepressed yeast cells consuming glucose is high enough to reduce the glucose transport rate by 50%. J. Bacteriol. 180:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teusink, B., J. Passarge, C. A. Reijenga, E. Esgalhado, C. C. van der Weijden, M. Schepper, M. C. Walsh, B. M. Bakker, K. van Dam, H. V. Westerhoff, and J. L. Snoep. 2000. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 267:5313-5329. [DOI] [PubMed] [Google Scholar]
- 38.Theobald, U., W. Mailinger, M. Baltes, M. Reuss, and M. Rizzi. 1997. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae. I. Experimental observations. Biotechnol. Bioeng. 55:305-316. [DOI] [PubMed] [Google Scholar]
- 39.Uno, I., K. Matsumoto, K. Adachi, and T. Ishikawa. 1983. Genetic and biochemical evidence that trehalase is a substrate of cAMP-dependent protein kinase in yeast. J. Biol. Chem. 258:10867-10872. [PubMed] [Google Scholar]
- 40.van Dam, J. C., M. Eman, J. Frank, H. C. Lange, G. W. K. van Dedem, and J. J. Heijnen. 2002. Analysis of glycolytic intermediates in Saccharomyces cerevisiae using anion exchange chromatography and electrospray ionization with tandem mass spectrometric detection. Anal. Chim. Acta 460:209-218. [Google Scholar]
- 41.Van der Heijden, R. T. J. M., J. J. Heijnen, C. Hellinga, B. Romein, and K. C. A. M. Luyben. 1994. Linear constraint relations in biochemical reaction systems. II. Diagnosis and estimation of gross errors. Biotechnol. Bioeng. 43:11-20. [DOI] [PubMed] [Google Scholar]
- 42.van der Plaat, J. B. 1974. Cyclic 3′,5′-adenosine monophosphate stimulates trehalose degradation in baker's yeast. Biochem. Biophys. Res. Commun. 56:580-587. [DOI] [PubMed] [Google Scholar]
- 43.Vanhalewyn, M., F. Dumortier, G. Debast, S. Colombo, P. Ma, J. Winderickx, P. Van Dijck, and J. M. Thevelein. 1999. A mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1K1876M, specifically affects glucose- and acidification-induced cAMP signaling and not the basal cAMP level. Mol. Microbiol. 33:363-376. [DOI] [PubMed] [Google Scholar]
- 44.Vanrolleghem, P. A., P. de Jong-Gubbels, W. M. van Gulik, J. T. Pronk, J. P. van Dijken, and S. Heijnen. 1996. Validation of a metabolic network for Saccharomyces cerevisiae using mixed substrate studies. Biotechnol. Prog. 12:434-448. [DOI] [PubMed] [Google Scholar]
- 45.Van Urk, H., P. R. Mak, W. A. Scheffers, and J. P. van Dijken. 1988. Metabolic responses of Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621 upon transition from glucose limitation to glucose excess. Yeast 4:283-291. [DOI] [PubMed] [Google Scholar]
- 46.Van Urk, H., D. Schipper, G. J. Breedveld, P. R. Mak, W. A. Scheffers, and P. Van Dijck. 1989. Localization and kinetics of pyruvate-metabolizing enzymes in relation to aerobic alcoholic fermentation in Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621. Biochim. Biophys. Acta 992:78-86. [DOI] [PubMed] [Google Scholar]
- 47.Van Urk, H., W. S. L. Voll, W. A. Scheffers, and J. P. van Dijken. 1990. Transient-state analysis of metabolic fluxes in crabtree-positive and crabtree-negative yeasts. Appl. Environ. Microbiol. 56:281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Winden, W. A., J. C. van Dam, C. Ras, R. J. Kleijn, J. L. Vinke, W. M. van Gulik, and J. J. Heijnen. 2005. Metabolic-flux analysis of Saccharomyces cerevisiae CEN. PK113-7D based on mass isotopomer measurements of 13C-labeled primary metabolites. FEMS Yeast Res. 5:559-568. [DOI] [PubMed] [Google Scholar]
- 49.Vaseghi, S., A. Baumeister, M. Rizzi, and M. Reuss. 1999. In vivo dynamics of the pentose phosphate pathway in Saccharomyces cerevisiae. Metab. Eng. 1:128-140. [DOI] [PubMed] [Google Scholar]
- 50.Vaseghi, S., F. Macherhammer, S. Zibek, and M. Reuss. 2001. Signal transduction dynamics of the protein kinase-A/phosphofructokinase-2 system in Saccharomyces cerevisiae. Metab. Eng. 3:163-172. [DOI] [PubMed] [Google Scholar]
- 51.Verduyn, C., A. H. Stouthamer, W. A. Scheffers, and J. P. van Dijken. 1991. A theoretical evaluation of growth yields of yeasts. Antonie Leeuwenhoek 59:49-63. [DOI] [PubMed] [Google Scholar]
- 52.Visser, D., G. A. van Zuylen, J. C. van Dam, M. R. Eman, A. Proll, C. Ras, L. Wu, W. M. van Gulik, and J. J. Heijnen. 2004. Analysis of in vivo kinetics of glycolysis in aerobic Saccharomyces cerevisiae by application of glucose and ethanol pulses. Biotechnol. Bioeng. 88:157-167. [DOI] [PubMed] [Google Scholar]
- 53.Vriezen, N., B. Romein, K. C. A. M. Luyben, and J. P. van Dijken. 1997. Effects of glutamine supply on growth and metabolism of mammalian cells in chemostat culture. Biotechnol. Bioeng. 54:271-286. [DOI] [PubMed] [Google Scholar]
- 54.Wiechert, W. 2002. Modeling and simulation: tools for metabolic engineering. J. Biotechnol. 94:37-63. [DOI] [PubMed] [Google Scholar]
- 55.Wu, L., H. C. Lange, W. M. Van Gulik, and J. J. Heijnen. 2003. Determination of in vivo oxygen uptake and carbon dioxide evolution rates from off-gas measurements under highly dynamic conditions. Biotechnol. Bioeng. 81:448-458. [DOI] [PubMed] [Google Scholar]