Abstract
The phylogenetic diversity and seasonal dynamics of freshwater Actinobacteria populations in four limnologically different lakes of the Mecklenburg-Brandenburg Lake District (northeastern Germany) were investigated. Fluorescence in situ hybridization was used to determine the seasonal abundances and dynamics of total Actinobacteria (probe HGC69a) and the three actinobacterial subclusters acI, acI-A, and acI-B (probes AcI-852, AcI-840-1, and AcI-840-2). Seasonal means of total Actinobacteria abundances in the epilimnia of the lakes varied from 13 to 36%, with maximum values of 30 to 58%, of all DAPI (4′,6′-diamidino-2-phenylindole)-stained cells. Around 80% of total Actinobacteria belonged to the acI cluster. The two subclusters acI-A and acI-B accounted for 60 to 91% of the acI cluster and showed seasonal means of 49% (acI-B) and 23% (acI-A) in relation to the acI cluster. Total Actinobacteria and members of the clusters acI and acI-B showed distinct seasonal changes in their absolute abundances, with maxima in late spring and fall/winter. In eight clone libraries constructed from the lakes, a total of 76 actinobacterial 16S rRNA gene sequences were identified from a total of 177 clones. The majority of the Actinobacteria sequences belonged to the acI and acIV cluster. Several new clusters and subclusters were found (acSTL, scB1-4, and acIVA-D). The majority of all obtained 16S rRNA gene sequences are distinct from those of already-cultured freshwater Actinobacteria.
Actinobacteria are typically known as inhabitants of soils (10, 30). This class of gram-positive bacteria with a high genomic G+C content comprises a variety of described species and environmental isolates (11, 15), including biotechnologically important species, such as Corynebacterium glutamicum and Streptomyces griseus.
Detailed investigations of aquatic microbial communities during recent years have shown that members of the Actinobacteria also occur in marine and freshwater habitats (5, 9, 22, 29, 34). Freshwater Actinobacteria are globally distributed throughout a variety of limnetic systems, such as lakes and rivers, with very different limnological features (4, 9, 12, 14, 23, 39, 40, 45). Actinobacteria frequently belong to the dominant fraction of the heterotrophic bacterioplankton (33, 39, 40, 42). Their relatively small cell size and their cell wall structure suggest protection from protistan grazing (11, 16, 27). Actinobacteria often reach their maximal abundances in late fall and winter (4, 9). Nevertheless, the present knowledge on the occurrence and seasonal dynamics of freshwater Actinobacteria is rather scarce.
Phylogenetic analyses based solely on the comparison of 16S rRNA gene sequences revealed distinct phylogenetic lineages of freshwater Actinobacteria, which were clearly separated from actinobacterial sequences of other environments (41). These sequences were derived from globally distributed freshwater habitats and did not show ecosystem-specific lineages or clusters in the overall tree topology. The formation of phylogenetically distinct clusters and their in situ activity provide evidence that freshwater Actinobacteria are an autochthonous component of limnetic ecosystems and are not introduced from the catchment area.
Thus, it is not surprising that the majority of the recently determined actinobacterial sequences from freshwater belong to yet-uncultured bacteria. At present only a few isolates of freshwater Actinobacteria exist. These isolates are derived from a variety of aquatic habitats and are representative of only a few phylogenetic lineages of freshwater Actinobacteria (11, 21, 34, 43). Due to the rather small number of isolates and their affiliation with distinct phylogenetic lineages, almost nothing is known about their physiology and ecological role.
This study investigated actinobacterial populations of four lakes of the Mecklenburg-Brandenburg Lake District (northeastern Germany) with respect to their phylogenetic diversity and seasonal dynamics. Today, the recent development of new oligonucleotide probes for different clusters of freshwater Actinobacteria (42) enables high-resolution analyses of freshwater actinobacterial communities by fluorescence in situ hybridization (FISH). The occurrence of widely distributed or lake-specific actinobacterial lineages was determined by phylogenetic comparison of cloned and sequenced 16S rRNA gene fragments.
MATERIALS AND METHODS
Lake descriptions and sampling methods.
Four lakes from the same geographical location but with different limnological characteristics were selected for this study: Lake Stechlin, Lake Breiter Luzin, Lake Tiefwaren, and Lake Grosse Fuchskuhle. The lakes are located in the Mecklenburg-Brandenburg Lake District (northeastern Germany), which was formed after the last ice age (Weichselian stage). The main limnological characteristics of the lakes are given in Table 1.
TABLE 1.
Limnological characteristics of sampled lakes
| Parameter (unit) | Valuea for:
|
||||
|---|---|---|---|---|---|
| Lake Stechlin | Lake Grosse Fuchskuhleb
|
Lake Breiter Luzin | Lake Tiefwaren | ||
| Northeast basin | Southwest basin | ||||
| Geographical position | 53°10′N, 13°02′E | 53°10′N, 13°02′E | 53°20′N, 13°28′E | 53°31′N, 12°42′E | |
| Altitude above sea level (m) | 59.7 | 59.0 | 84.2 | 63.5 | |
| Maximum depth (m) | 69.5 | 5.6 | 58.5 | 24 | |
| Surface area (km2) | 4.3 | 0.02 | 3.57 | 1.4 | |
| Vol (106 m3) | 96.9 | 0.05 | 67.5 | 12.9 | |
| Catchment area (km2) | 26.0 | 0.005 | 14.0 | 17.5 | |
| Characteristics of catchment area | Mixed forest | Mixed forest, bog area | Forest and agriculture | Forest, small town | |
| Secchi depth (m) | 6.5-10.5 | 1.0-2.2 | 0.9-1.5 | 1.6-3.8 | 3.1-8.9 |
| Trophyc | Oligotrophic | Eutrophic | Dystrophic | Mesotrophic | Eutrophic |
| pH | 8.5 (0.2) | 6.5 (0.6) | 4.7 (0.2) | 8.5 (0.2) | 8.3 (0.2) |
| Alkalinity (mvale liter−1) | 2 (0.1) | NDd | ND | 2 (0.2) | 3 (0.2) |
| PO43− (μg l−1) | 2 (0.9) | 5 (2) | 7 (2) | 3 (4) | 2 (1) |
| DOC (mg liter−1) | 4.3 (1.1) | 10.3 (1.3) | 24.8 (5.1) | 5.8 (0.6) | 10.6 (5.3) |
| Bacterial numbers (106 ml−1) | 1.35 (0.58) | 2.74 (1.0) | 1.93 (0.69) | 1.93 (0.49) | 2.21 (0.47) |
| Primary production (μg C liter−1 day−1) | 31 (11) | 182 (156) | 512 (863) | 103 (44) | 114 (99) |
| Bacterial production (μg C liter−1 day−1) | 22 (15) | 39 (21) | 63 (101) | 51 (58) | 35 (37) |
Data for pH, alkalinity, PO43−, DOC, bacterial numbers, primary production, and bacterial production are average values, with standard deviations in parentheses, for epilimnetic samples from April to November 2003 and March 2004.
Data for “Geographical position” through “Characteristics of catchment area” apply to both the NE and SW basins.
Trophic status was determined according to guideline EUR 14563 EN of the Commission of the European Communities (28).
ND, not determined.
mval, milliequivalents.
In brief, the naturally acidic bog lake Grosse Fuchskuhle has been artificially divided into four compartments (southwest [SW], northwest, northeast [NE], and southeast) by large plastic curtains since 1990 (17, 18). In this study, only the NE and SW basins were investigated. The SW basin is the most acidic part (pH 4.7), since it is directly influenced by water run off from the adjacent bog area. In contrast, the NE basin is less influenced by humic acid input, resulting in an almost neutral pH (pH 6.5). The two basins differ significantly in their chemical and, hence, biological parameters (3, 13).
Lake Tiefwaren is located close to the village of Waren and has been under restoration by a combination of aluminate and calcium hydroxide precipitation (19) since 2001. Anthropogenic influences (e.g., agriculture and industry) in the late 1980s caused a hypertrophic state in this lake. Throughout the restoration, excess phosphorus was removed from the pelagic and fixed permanently in the sediment, resulting in its present eutrophic state.
Lake Stechlin and Lake Breiter Luzin are among the deepest lakes in the Mecklenburg-Brandenburg Lake District (69.5 m and 58.5 m, respectively). These lakes are characterized by high hypolimnetic oxygen concentrations (up to 60% O2 saturation). Lake Stechlin is one of the most oligotrophic lakes in northern Germany, whereas Lake Breiter Luzin is in a mesotrophic state due to extensive agriculture in its catchment area.
The trophic status of the lakes was determined following the guidelines of the Joint Research Centre of the Commission of the European Communities (28). In general, this protocol includes a variety of limnological parameters (e.g., Secchi depth, primary production, chlorophyll a, total phosphorous, total nitrogen, phytoplankton community composition and biomasses, and geomorphological features) for the trophic classification of a lake.
All lakes were sampled monthly between April 2003 and March 2004, except during ice coverage in December (Lake Breiter Luzin), January (all lakes), and February (Lake Grosse Fuchskuhle). Water samples of 1 liter were taken at the deepest point of each lake by using a Ruttner sampler. Depending on the thermal stratification, epilimnetic samples were obtained by taking subsamples at depths of 0, 5, and 10 m (April, May, and October through March) or 0 and 5 m (June through September) in Lake Stechlin, Lake Breiter Luzin, and Lake Tiefwaren. The subsamples were mixed in sterile glass flasks in equal volumes. The epilimnetic samples of the NE and SW basins of Lake Grosse Fuchskuhle were taken as mixed samples from depths of 0 and 2 m (April and October through March) or as surface samples (May through September). In addition, hypolimnetic samples (40 m) of Lake Stechlin were collected. Incubations for measurements of primary production and bacterial production were carried out under in situ conditions during sampling. Water samples for molecular and chemical analyses were taken to the lab in dark cooling boxes and processed for further analyses within 2 to 4 h after sampling.
Environmental variables.
The water samples were analyzed for basic physical, chemical, and biological parameters such as Secchi depth, pH, dissolved organic carbon (DOC), PO43− P, alkalinity, primary production (PP), bacterial protein production (BPP), and bacterial numbers. DOC was analyzed using a Shimadzu (Duisburg, Germany) TOC-5050 total organic carbon analyzer by standard methods as described by Wetzel and Likens (44). PO43− P was measured spectrophotometrically with a Tecator FIAstar 5010 analyzer (Rellingen, Germany) following standard protocols (38, 44) and the manufacturer's instructions. Alkalinity down to a pH of 4.3 was determined by titration with 1 N hydrochloric acid using a Metrohm 686 Titriprocessor (Filderstadt, Germany). Electrode measurements were performed for measuring pH (WTW pH 197; Weilheim, Germany), and Secchi depth was determined by the classical procedure using a Secchi disk (diameter, 0.25 m).
The total bacterial numbers, BPP, and PP were measured as follows. Bacterial numbers were determined by epifluorescence microscopy after staining of the bacterial cells with 4′,6′-diamidino-2-phenylindole (DAPI) (1 mg/100 ml) on a 0.2-μm Nuclepore polycarbonate membrane. Total BPP (>0.2 μm) and total PP (>0.6 μm) were determined by [14C]leucine incorporation (36) and H14CO3− uptake (see reference 44 and references therein), respectively.
Catalyzed reporter deposition FISH (CARD-FISH).
Fluorescence in situ hybridization was performed using horseradish peroxidase-labeled probes and tyramide signal amplification as described earlier (26, 42). To increase detection of gram-positive bacteria, a modified permeabilization protocol developed for freshwater bacterioplankton was used (32). Water samples of 10 ml were filtered through 0.2-μm Nuclepore polycarbonate filters (47-mm diameter) and fixed with 50% (vol/vol) ethanol. Filters were washed with 1× phosphate-buffered saline buffer and 70% (vol/vol) ethanol and stored at −20°C until further processing. For the quantification of total Actinobacteria and the three limnetic Actinobacteria clusters acI, acI-A, and acI-B, a set of four different probes was used: probe HGC69a as a universal probe for the class Actinobacteria (31), and probes AcI-852, AcI-840-1, and AcI-840-2 as specific probes for the acI lineage and the two subclusters acI-A and acI-B of this lineage, respectively (42). To improve the accessibility of the probe target site on the 16S rRNA, unlabeled helper oligonucleotides (AcI-852-H1, AcI-852-H2, AcI-840-H1, AcI-840-H2, and AcI-840-H3) were added to probes AcI-852, AcI-840-1, and AcI-840-2 (42). The hybridization efficiency was determined with the probe mix EUB I-III, which detects most bacteria, including Verrucomicrobia and Planctomycetes (6). The filters were counterstained with DAPI (1 mg/100 ml) and inspected with a Leica DR-MB epifluorescence microscope at a magnification of ×1,000. At least 10 randomly selected microscopic fields (each with 10,000 μm2) were counted for the enumeration of the FISH-stained bacteria. Based on these numbers, average values and the standard deviations were calculated. No replicate filters were tested.
16S rRNA gene clone libraries.
16S rRNA gene clone libraries were constructed for the May samples from Lake Stechlin (epilimnion and hypolimnion) and for the November samples from all lakes. Water samples of 300 ml were prefiltered through a 5.0-μm Nuclepore polycarbonate membrane, and subsequently genomic DNA of free-living bacteria was collected by filtering 100 to 150 ml of prefiltered sample onto a 0.2-μm Nuclepore polycarbonate filter (47-mm diameter). Filters were transferred into sterile Eppendorf tubes and kept frozen at −20°C until DNA extraction.
DNA was extracted using a protocol with phenol-chloroform-isoamylalcohol, sodium dodecyl sulfate, Tris-EDTA-sodium chloride-polyvinylpyrrolidone buffer, and zirconium beads. The filters were cut into sections and placed in 2-ml Eppendorf reaction tubes. Zirconium beads (150 mg), sodium dodecyl sulfate (1.5%), Tris-EDTA-sodium chloride-polyvinylpyrrolidone buffer (50 mM Tris, 20 mM EDTA, 100 mM NaCl, 20 mg/ml polyvinylpyrrolidone), and phenol-chloroform-isoamylalcohol (50%) were added to the filter sections and mixed for ca. 10 to 15 min on a Vortex mixer at maximum speed. The remaining proteins were removed by repeating the phenol-chloroform extraction step up to five times. After addition of Na acetate (3 M, pH 5.2) and isopropanol (2.5 volumes), DNA was precipitated at −20°C for 3 h. The DNA was washed two times with 70% ethanol and resuspended in 50 to 100 μl nuclease-free water.
The almost complete 16S rRNA genes were amplified using the primer pair 8F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-GGY TAC CTT GTT ACG ACT T-3′), which is specific for the domain Bacteria (24). The reaction mixtures for the PCR amplification contained 2 to 5 μl template DNA, 250 nM of each of the appropriate primers, 200 μM of each deoxyribonucleoside triphosphate, 2 mM MgCl2, 10 μl of 10× PCR buffer, and 2 U of BIOTAQ Red DNA polymerase (Bioline) in a total volume of 50 μl.
The PCRs were performed in a PT-200 gradient cycler (MJ Research) by using an initial denaturation step at 94°C (2 min), followed by 30 cycles of denaturation at 94°C (1 min), annealing at 54°C (1 min), and extension at 72°C (2 min). A final extension at 72°C (10 min) and subsequent cooling at 4°C completed the reaction. The amplified DNA was purified using the NucleoSpin Extract II PCR purification kit (Macherey-Nagel) and stored at −20°C.
Cloning of the almost complete 16S rRNA genes of free-living bacteria was done using the pGEM-T-Easy Vector System II (Promega) according to the manufacturer's protocol. A total of 20 to 30 clones of each clone library were picked and sequenced. Plasmids were isolated from the clones with the Wizard Plus SV Minipreps DNA purification system (Promega). Sequencing reactions were performed using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) and an ABI Prism 3100-Avant genetic analyzer (Applied Biosystems) according to the manufacturer's instructions. Three different primers were used to sequence the cloned 16S rRNA gene fragments: M13F (5′-GTT TTC CCA GTC ACG AC-3′), M13R (5′-CAG GAA ACA GCT ATG AC-3′), and 341F (5′-CCT ACG GGA GGC AGC AG-3′) (25). Partial sequences were assembled and corrected manually using the software Chromas, version 1.45 (Technelysium Pty Ltd).
Phylogenetic analysis.
Phylogenetic analyses were performed using the ARB software package (http://arb-home.de). The partial 16S rRNA gene sequences were imported into the ARB database of ca. 51,900 aligned reference sequences, including the closest related sequences determined by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The sequences were automatically aligned and corrected manually. For the calculation of the phylogenetic trees, only 16S rRNA gene sequences longer than 1,400 nucleotides were used. To exclude highly variable positions within the 16S rRNA gene sequences, a 50% base frequency filter was calculated. This filter only uses those sequence positions of an alignment for phylogenetic reconstructions, where 50% of the analyzed sequences have identical entries. Thus, the phylogenetic calculations become more robust and potential alignment errors are excluded. Phylogenetic analyses were performed by the maximum-likelihood algorithm using the fastDNAml program of the ARB package. The stability of the trees was tested by comparison of the results with those of other tree construction methods, such as neighbor joining or maximum parsimony. Partial sequences (shorter than 1,400 nucleotides) were added to the trees according to maximum-parsimony criteria and with the 50% base frequency filter. This tool does not correct for evolutionary distances and does not allow changes in the overall tree topology.
Nucleotide sequence accession numbers.
The partial sequences of 16S rRNA genes obtained in this study were deposited in GenBank with accession numbers DQ316321 to DQ316396.
RESULTS
Limnology of the sampled lakes.
The physical and chemical features of the four studied lakes reflect a wide range of environmental conditions (Table 1). The selected lakes varied in trophy, from oligotrophic (Lake Stechlin) to mesotrophic (Lake Breiter Luzin) to eutrophic (Lake Tiefwaren and the NE basin of Lake Grosse Fuchskuhle) to dystrophic (SW basin of Lake Grosse Fuchskuhle). Primary production and bacterial production ranged from 31 to 512 μg C liter−1 day−1 and from 22 to 63 μg C liter−1 day−1, respectively. Minor differences were observed in the concentrations of DOC and PO43− and in alkalinity (Table 1). In general, oligotrophic Lake Stechlin and the dystrophic SW basin of Lake Grosse Fuchskuhle showed the greatest differences in their limnological characteristics.
Abundances and seasonal dynamics of freshwater Actinobacteria populations.
The detailed results of the CARD-FISH analyses of all lakes with all probes used are given in Table 2. The hybridization efficiency determined by the probe mix EUB I-III was relatively high (77 to 100%). However, various samples from the epilimnion and hypolimnion of Lake Stechlin had lower hybridization efficiencies (35 to 69%) (Table 2).
TABLE 2.
Results of FISH counts of total Actinobacteria and the actinobacterial freshwater clusters acI, acI-A, and acI-B
| Probe | Locationa | Mean (SD) % DAPI-stained cells
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| April | May | June | July | August | September | October | November | December | February | March | ||
| EUB I-III | BL | 90 (19) | 93 (11) | 97 (12) | 93 (11) | 99 (16) | 99 (14) | 100 (10) | 98 (7) | NDb | 84 (14) | 87 (12) |
| FU-NE | 90 (15) | 82 (9) | 92 (8) | 85 (27) | 90 (6) | 90 (9) | 90 (6) | 93 (7) | 92 (6) | ND | 89 (9) | |
| FU-SW | 92 (11) | 92 (8) | 96 (9) | 89 (19) | 91 (10) | 92 (12) | 94 (5) | 97 (11) | 92 (18) | ND | 92 (4) | |
| ST | 36 (11) | 87 (8) | 77 (17) | 98 (6) | 85 (19) | 98 (17) | 35 (3) | 55 (6) | 60 (7) | 69 (6) | 64 (8) | |
| ST-HL | 36 (5) | 44 (2) | 63 (3) | 68 (5) | 96 (9) | 57 (5) | 64 (12) | 69 (11) | 99 (9) | 77 (7) | 98 (9) | |
| TW | 96 (13) | 97 (5) | 77 (9) | 83 (9) | 88 (11) | 100 (6) | 85 (10) | 91 (11) | 87 (10) | 87 (11) | 87 (4) | |
| HGC69a (Actinobacteria) | BL | 13 (5) | 24 (4) | 42 (5) | 17 (2) | 27 (4) | 19 (3) | 35 (5) | 30 (5) | ND | 21 (4) | 24 (7) |
| FU-NE | 9 (1) | 8 (1) | 14 (1) | 4 (1) | 6 (1) | 11 (6) | 21 (2) | 30 (2) | 21 (2) | ND | 10 (2) | |
| FU-SW | 12 (1) | 20 (2) | 23 (4) | 16 (2) | 21 (3) | 34 (8) | 31 (7) | 31 (4) | 27 (8) | ND | 10 (2) | |
| ST | 15 (4) | 14 (4) | 36 (6) | 25 (3) | 47 (9) | 23 (12) | 21 (4) | 38 (10) | 35 (5) | 23 (4) | 40 (6) | |
| ST-HL | 12 (2) | 5 (1) | 18 (3) | 7 (2) | 12 (2) | 7 (1) | 22 (8) | 21 (3) | 16 (9) | 23 (3) | 20 (6) | |
| TW | 57 (6) | 19 (2) | 33 (9) | 31 (5) | 29 (6) | 29 (4) | 58 (12) | 40 (4) | 23 (6) | 27 (4) | 53 (7) | |
| AcI-852 (acI) | BL | 7 (2) | 19 (3) | 33 (3) | 15 (2) | 22 (3) | 15 (2) | 27 (3) | 22 (3) | ND | 14 (2) | 22 (2) |
| FU-NE | 6 (1) | 7 (1) | 11 (2) | 3 (1) | 4 (1) | 10 (3) | 18 (2) | 29 (2) | 19 (2) | ND | 7 (1) | |
| FU-SW | 10 (2) | 15 (3) | 20 (3) | 12 (2) | 16 (2) | 24 (3) | 22 (3) | 27 (2) | 18 (3) | ND | 7 (1) | |
| ST | 14 (7) | 14 (4) | 27 (3) | 20 (4) | 37 (9) | 22 (4) | 20 (3) | 30 (7) | 35 (3) | 16 (3) | 19 (5) | |
| ST-HL | 11 (1) | 5 (1) | 18 (2) | 7 (1) | 9 (1) | 5 (1) | 17 (3) | 18 (7) | 16 (7) | 23 (3) | 19 (4) | |
| TW | 43 (6) | 17 (1) | 28 (2) | 26 (2) | 21 (1) | 28 (3) | 47 (3) | 27 (7) | 19 (3) | 23 (2) | 47 (4) | |
| AcI-840-1 (acI-A) | BL | 3 (3) | 3 (1) | 4 (1) | 4 (2) | 8 (2) | 2 (0.5) | 1 (1) | 2 (1) | ND | 3 (1) | 4 (1) |
| FU-NE | 3 (1) | 2 (0.4) | 3 (1) | 0.7 (0.5) | 0.5 (0.4) | 0.7 (0.6) | 3 (1) | 7 (2) | 4 (1) | ND | 2 (1) | |
| FU-SW | 2 (1) | 3 (1) | 1 (1) | 2 (1) | 4 (2) | 5 (2) | 3 (0.5) | 5 (0.5) | 4 (1) | ND | 1 (1) | |
| ST | 7 (2) | 6 (2) | 10 (3) | 8 (1) | 13 (3) | 9 (2) | 7 (2) | 4 (1) | 4 (1) | 2 (1) | 3 (3) | |
| ST-HL | 3 (1) | 1 (0.3) | 4 (1) | 1 (0.3) | 4 (1) | 2 (1) | 9 (2) | 8 (2) | 6 (3) | 9 (2) | 6 (2) | |
| TW | 4 (1) | 4 (1) | 3 (0.5) | 4 (1) | 6 (1) | 4 (1) | 5 (1) | 6 (4) | 6 (1) | 6 (1) | 5 (1) | |
| AcI-840-2 (acI-B) | BL | 3 (3) | 14 (2) | 13 (3) | 9 (2) | 10 (3) | 10 (2) | 13 (1) | 12 (1) | ND | 7 (1) | 17 (2) |
| FU-NE | 2 (1) | 2 (1) | 3 (1) | 1 (0.8) | 1 (0.8) | 2 (2) | 5 (1) | 12 (1) | 10 (2) | ND | 5 (1) | |
| FU-SW | 4 (1) | 11 (3) | 13 (2) | 5 (1) | 11 (2) | 12 (2) | 14 (3) | 17 (2) | 11 (1) | ND | 5 (1) | |
| ST | 4 (2) | 6 (2) | 9 (2) | 11 (2) | 18 (3) | 12 (4) | 10 (4) | 12 (3) | 13 (3) | 10 (2) | 7 (4) | |
| ST-HL | 8 (2) | 3 (0.5) | 9 (2) | 4 (1) | 5 (2) | 2 (0.6) | 6 (1) | 9 (3) | 8 (4) | 11 (2) | 13 (2) | |
| TW | 18 (3) | 5 (1) | 12 (1) | 10 (1) | 9 (2) | 18 (3) | 16 (2) | 11 (2) | 12 (1) | 16 (1) | 15 (2) | |
BL, Lake Breiter Luzin; FU-NE, northeast basin of Lake Grosse Fuchskuhle; FU-SW, southwest basin of Lake Grosse Fuchskuhle; ST, Lake Stechlin (epilimnion); ST-HL, Lake Stechlin (hypolimnion); TW, Lake Tiefwaren.
ND, not determined.
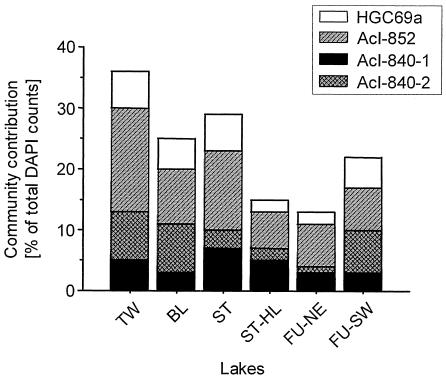
The seasonal means of the absolute bacterial numbers determined by DAPI staining varied from 1.35 × 106 to 2.74 × 106 cells ml−1 and generally reflect the limnological characteristics of the lakes (Table 1). The bacterioplankton was mainly dominated by members of the Actinobacteria. The proportions of cells hybridized by the universal actinobacterial probe HGC69a ranged in their seasonal means from 13 to 36%, with maximal abundances of 58 and 47% in the epilimnia of Lake Tiefwaren and Lake Stechlin, respectively (Fig. 1 and Table 2).
FIG. 1.
Seasonal means of abundances of total Actinobacteria (probe HGC69a) and members of the actinobacterial subgroups acI (AcI-852), acI-A (AcI-840-1), and acI-B (AcI-840-2) in relation to total bacterial numbers determined by direct microscopic counts.
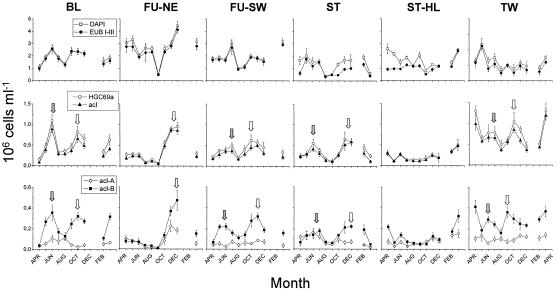
Actinobacteria populations conspicuously exhibited seasonal dynamics in their absolute abundances, as depicted in Fig. 2. Actinobacterial abundances determined as absolute numbers of cells hybridized with probe HGC69a showed a peak in all lakes except the NE basin of Lake Grosse Fuchskuhle in June and July. All lakes showed a peak between October and December. In Lake Tiefwaren two additional maxima were observed in April 2003 and March 2004, with absolute actinobacterial numbers of 1.3 × 106 and 1.33 × 106 cells ml−1, respectively. Cell numbers of Actinobacteria in the hypolimnion of Lake Stechlin ranged at a low level from 0.11 × 106 to 0.5 × 106 cells ml−1. Slightly elevated numbers were observed in winter and spring (April to June 2003 and February and March 2004). In general, lowest actinobacterial abundances were detected in summer around August and September.
FIG. 2.
Seasonal dynamics of the abundance of Actinobacteria determined by the oligonucleotide probes HGC69a (Actinobacteria), AcI-852 (acI), AcI-840-1 (acI-A), and AcI-840-2 (acI-B). The upper panels show the total bacterial numbers as enumerated by DAPI direct microscopic counts in comparison to Eubacteria determined by using the oligonucleotide probe mix EUB I-III. The arrows mark the peaks in abundances of Actinobacteria in late spring (gray arrows) and fall/winter (white arrows). Error bars represent standard deviations determined from absolute cell numbers in 10 independently counted microscopic fields. ST, Lake Stechlin; BL, Lake Breiter Luzin; TW, Lake Tiefwaren; FU, Lake Grosse Fuchskuhle; HL, hypolimnion.
The seasonal dynamic of Actinobacteria of the acI cluster was fairly similar to that of the total Actinobacteria populations, with peaks in spring and fall and lowest abundances in summer. The acI cluster represented the largest group within the Actinobacteria. Around 80% of total Actinobacteria belonged to the acI cluster, which accounted for 3 to 47% of all DAPI-stained cells (Fig. 1 and Table 2). The highest seasonal means of total Actinobacteria and the acI Actinobacteria group were found in the epilimnia of Lake Tiefwaren and Lake Stechlin (Fig. 1).
The proportion of cells hybridized with the probes AcI-840-1 (acI-A) and AcI-840-2 (acI-B) ranged between 0.5 and 10% and 1 and 18% of all DAPI-stained cells, respectively. In general, the fraction of the acI-B subcluster of all Actinobacteria was two to three times higher than that of the acI-A subcluster (Table 2). The observed seasonal means of acI-A varied between 3% (±1.7%) and 7% (±3.3%), and those of acI-B varied between 4% (±3.8%) and 13% (±4.1%) (Fig. 1). In relation to the seasonal means of acI, acI-B accounted for 49% and acI-A for 23%. Both subclusters together accounted for 60 to 91% (mean, 73%; n = 6) of the acI cluster. Nine to 40% (mean, 27%; n = 6) of the acI cluster were not detected by the probes AcI-840-1 and AcI-840-2 (Fig. 1).
Almost no seasonal variability was observed within the populations of acI-A Actinobacteria. Except for slight variations in spring and fall (e.g., in Lake Breiter Luzin, Lake Stechlin, and Lake Tiefwaren), the absolute cell numbers remained stable at a relatively low level (0.004 × 106 to 0.222 × 106 cells ml−1). Increased absolute cell numbers of acI-A Actinobacteria were observed only in November 2003 in the NE basin of Lake Grosse Fuchskuhle. However, more distinct seasonal dynamics were found for Actinobacteria of the acI-B subcluster. Their seasonal changes in absolute numbers were similar to those of the total Actinobacteria and members of the acI cluster.
Diversity and classification of freshwater bacterial 16S rRNA gene sequences.
Cloning and sequencing of 16S rRNA gene fragments of eight clone libraries from different samples of the four investigated lakes resulted in a total of 177 clones (Table 3). Phylogenetically, 76 clones (42.9%) belonged to the Actinobacteria, 31 clones (17.5%) to the Proteobacteria (α, β, and δ), 30 clones (16.9%) to the Bacteroidetes, 21 clones (11.9%) to the cyanobacteria/chloroplasts, and 19 clones (10.7%) to other bacterial groups (Table 3).
TABLE 3.
Phylogenetic classification of 16S rRNA gene sequences derived from eight clone libraries
| Class and cluster | No. of clones froma:
|
Total no. of clones (n = 177) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| STb (n = 29) | ST (n = 22) | ST-HLb (n = 13) | ST-HL (n = 20) | FU-NE (n = 20) | FU-SW (n = 20) | BL (n = 28) | TW (n = 25) | ||
| Proteobacteria (α, β, and δ) | 3 | 4 | 1 | 6 | 3 | 6 | 8 | 31 | |
| Actinobacteria | 7 | 13 | 4 | 8 | 11 | 7 | 16 | 10 | 76 |
| acI-A | 3 | 6 | 1 | 1 | 3 | 14 | |||
| acI-B | 2 | 1 | 2 | 5 | 6 | 7 | 4 | 27 | |
| acII | 1 | 1 | |||||||
| acIV | 3 | 6 | 1 | 4 | 8 | 3 | 25 | ||
| acSTL | 1 | 2 | 1 | 4 | |||||
| Other | 2 | 1 | 2 | 5 | |||||
| Bacteroidetes | 3 | 4 | 4 | 5 | 2 | 9 | 3 | 30 | |
| Cyanobacteria/chloroplasts | 15 | 1 | 1 | 1 | 1 | 2 | 21 | ||
| Other | 1 | 4 | 5 | 1 | 4 | 4 | 19 | ||
Except as indicated, samples were from November 2003. BL, Lake Breiter Luzin; FU-NE, northeast basin of Lake Grosse Fuchskuhle; FU-SW, southwest basin of Lake Grosse Fuchskuhle; ST, Lake Stechlin (epilimnion); ST-HL, Lake Stechlin (hypolimnion); TW, Lake Tiefwaren.
Samples were from May 2003.
Almost all of the clone libraries were dominated by 16S rRNA gene sequences of the class Actinobacteria (24.1 to 59.1%). Of the 76 Actinobacteria clones analyzed, 20 (26.3%) originated from the epilimnion and 12 (15.8%) from the hypolimnion of Lake Stechlin, 11 (14.5%) from the NE basin of Lake Grosse Fuchskuhle, 7 (9.2%) from the SW basin of Lake Grosse Fuchskuhle, 16 (21.1%) from Lake Breiter Luzin, and 10 (13.1%) from Lake Tiefwaren (Table 3).
Percentages of Proteobacteria in the epilimnetic samples varied from 10.3 to 32%. Almost no Proteobacteria were found in clone libraries of hypolimnetic samples from Lake Stechlin.
Clones of the Bacteroidetes were highly abundant in the SW basin of Lake Grosse Fuchskuhle (45%) and the hypolimnetic samples of Lake Stechlin (25 and 30.8%). The proportion of cyanobacteria/chloroplasts was generally low (0 to 7.7%), except in the epilimnetic May sample of Lake Stechlin (51.7%). The hypolimnetic samples of Lake Stechlin yielded the highest numbers of clones of other bacterial groups, such as Verrucomicrobia, Planctomycetes, or candidate division OP10 (data not shown).
Phylogenetic inferences for freshwater Actinobacteria.
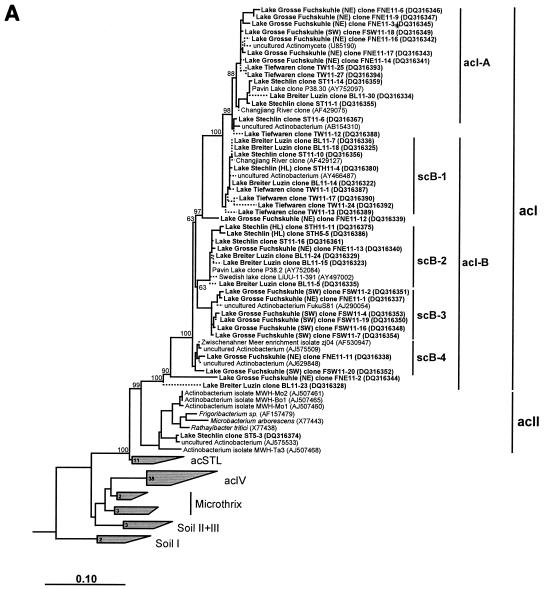
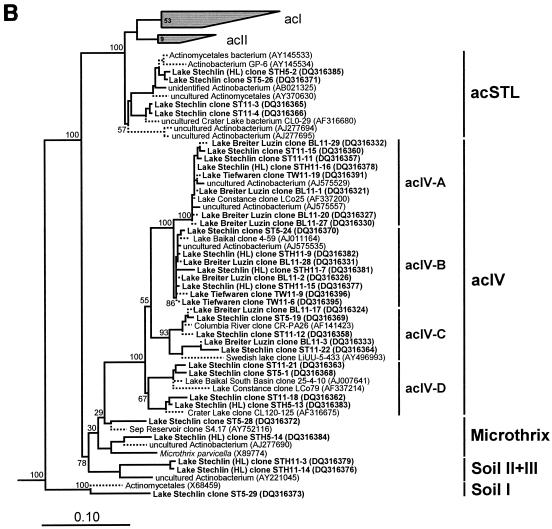
The majority of the clones belonged to the two clusters acI and acIV, proposed by Warnecke et al. (41). All other clones of Actinobacteria were related to the clusters acII, Soil I to Soil III, Microthrix, and the cluster newly proposed here, acSTL (Fig. 3A and B).
FIG.3.
Phylogenetic trees of freshwater actinobacterial 16S rRNA gene sequences. Solid lines indicate sequences that were included in the primary analyses (sequences of >1,400 nucleotides), whereas dotted lines indicate partial sequences (<1,400 nucleotides). Sequences produced during this study are shown in boldface. GenBank accession numbers are given in parentheses. The scale bar correspond to 10 base substitutions per 100 nucleotide positions. Bootstrap values at the main branching points are given. (A) Detailed view of phylogenetic relationships within the acI cluster. (B) Phylogeny of other actinobacterial freshwater clusters.
The acI cluster was dominated by 16S rRNA sequences of the acI-B subcluster (Table 3). This subcluster showed a highly diverse branching pattern which leads to the formation of the four new groups scB-1, scB-2, scB-3, and scB-4 (Fig. 3 A). Although the group scB-1 was phylogenetically more closely related to the acI-A cluster, it was affiliated with the acI-B cluster due to the availability of the target sequence for the oligonucleotide probe AcI-840-2 on their 16S rRNA molecules. The groups scB-1 and scB-2 are comprised of sequences from Lake Stechlin (epilimnion and hypolimnion), Lake Breiter Luzin, and Lake Tiefwaren. Clones of Lake Breiter Luzin were equally distributed over both groups, whereas clones of Lake Stechlin occurred mainly in scB-2 and clones of Lake Tiefwaren occurred mainly in scB-1. The two groups scB-3 and scB-4 contained high numbers of clones from both basins of Lake Grosse Fuchskuhle.
The branching pattern of the acI-A subcluster was less diverse than that of the acI-B subcluster. The acI-A subcluster is closely related to the proposed scB-1 group of the acI-B subcluster and contains sequences from all four investigated lakes. Nevertheless, clones of the NE basin of Lake Grosse Fuchskuhle are the most numerous in this cluster (Fig. 3A).
The acIV cluster is clearly separated from all other Actinobacteria clusters and includes sequences from Lake Stechlin, Lake Breiter Luzin, and Lake Tiefwaren. No sequences from Lake Grosse Fuchskuhle were affiliated with this cluster. The phylogenetic calculations resulted in the four subclusters acIV-A to acIV-D (Fig. 3B), as described by Warnecke et al. (41). The subclusters acIV-A and acIV-B contain sequences from all three above-mentioned lakes, whereas the subcluster acIV-C includes sequences from Lake Stechlin and Lake Breiter Luzin and acIV-D includes sequences only from Lake Stechlin.
Four sequences from Lake Stechlin were affiliated with a newly proposed cluster, acSTL, which is closely related to the already-known clusters acII and acI (Fig. 3 B). The separation and stability of this cluster were consistent throughout the three phylogenetic reconstructions methods, i.e., maximum parsimony, neighbor joining, and maximum likelihood.
DISCUSSION
Members of the class Actinobacteria (37) are regarded as typical inhabitants of soil environments (10, 30). Recent studies on the composition of freshwater bacterial communities found, however, that Actinobacteria are also common in a variety of freshwater habitats (7, 9, 14, 39, 40, 41, 42, 45). In addition, the phylogeny of freshwater Actinobacteria is highly diverse and exhibits several clusters and subgroups (41). Due to the small numbers of currently existing isolates of limnetic Actinobacteria, very little is yet known about their physiology and ecological roles in these habitats.
Occurrence and seasonal variability of freshwater Actinobacteria.
As illustrated by earlier investigations, the proportion of Actinobacteria can vary considerably in different freshwater habitats (4, 9, 23, 40). The high abundance of Actinobacteria in Lake Stechlin, Lake Breiter Luzin, and Lake Tiefwaren indicates a dominance of this bacterial group in the epilimnia of these lakes. However, in the two compartments of Lake Grosse Fuchskuhle, numbers of Actinobacteria were relatively low. A previous study on Lake Grosse Fuchskuhle had shown that the bacterial communities of both the NE and SW compartments were dominated by Betaproteobacteria, with relative abundances of 29 and 42%, respectively (4). The dominance of Betaproteobacteria in freshwater habitats has been shown in a variety of other investigations (1, 14, 22, 35, 40). However, other studies revealed the Actinobacteria as the dominant fraction of freshwater bacterial communities (33, 39, 40, 42). Obviously, the dominance of particular bacterial phylogenetic groups depends on the respective ecosystem.
Since our study was focused mainly on Actinobacteria, we did not attempt an exhaustive community analysis by FISH. We therefore cannot draw any general conclusions about the phylogenetic diversity of the entire bacterioplankton communities in the lakes. However, several studies in the last few years determined that both Betaproteobacteria and Actinobacteria belong to the dominant fractions of freshwater bacterioplankton communities (1, 14, 39, 40, 45). Studies on abundances and biomasses of Actinobacteria and Betaproteobacteria in Lake Gossenköllesee and Lake Grosse Fuchskuhle suggested that the groups inhabit separate functional niches (4, 9).
Members of the acI cluster were the most prominent phylogenetic group of Actinobacteria in all studied lakes (Fig. 1 and Table 2). The classification into the two subclusters acI-A and acI-B revealed a higher fraction of acI-B than of acI-A Actinobacteria. However, the sum of these subclusters indicates that there is a substantial fraction of 9 to 40% (mean, 27%; n = 6) of acI Actinobacteria which is not detected by the two oligonucleotide probes AcI-840-1 and AcI-840-2. The fraction of nondetectable acI Actinobacteria was highly variable throughout the lakes and was not linked to any of the measured limnological features. To study the entire diversity of the acI cluster, the development of additional oligonucleotide probes is necessary (see also reference 42).
The Actinobacteria populations of the four investigated lakes exhibited common seasonal dynamics, with maxima in spring and fall. In general, the results were in accordance with earlier studies on Lake Grosse Fuchskuhle and Lake Gossenköllesee (4, 9). The comparative results of this study provide evidence that there is a common seasonal dynamic within freshwater actinobacterial populations which seems to be independent of basic limnological features (e.g., trophy).
Distinct phylogenetic diversity within freshwater actinobacterial communities.
Cloning and sequencing of 16S rRNA gene fragments of eight clone libraries yielded 177 clones, which were mainly related to the Proteobacteria, Actinobacteria, Bacteroidetes, and cyanobacteria/chloroplasts. We are aware of the fact that the numbers of sequenced clones (13 to 29 clones per library) were too low to draw any general conclusion on the abundances of phylogenetic groups. However, 16S rRNA gene sequences of our clone libraries were phylogenetically affiliated to already-known bacterial freshwater clusters (9, 45), and the phylogenetic reconstructions of the freshwater Actinobacteria are consistent with previous studies (9, 41, 45). The dominance of Actinobacteria of the acI cluster and the high percentage of clones from the acI-B cluster in our clone libraries were similar to results from CARD-FISH analysis (reference 42 and this study). We therefore assume that the analyzed clones can generally be considered to particularly represent the actinobacterial communities in the four investigated lakes.
Phylogenetically, the acI cluster is clearly separated from other actinobacterial sequences. However, the separation into the two subclusters acI-A and acI-B is less obvious in our study than in the phylogenetic calculations of Warnecke et al. (41, 42). Further calculations with different sets of reference sequences and tree construction methods (e.g., maximum likelihood, maximum parsimony, and neighbor joining) as well as the high bootstrap values indicated the stability of the obtained phylogenetic relationships.
Three sequences from the epilimnion and hypolimnion of Lake Stechlin were phylogenetically affiliated to the clusters Soil I to Soil III. The input of allochthonous bacteria can amount to up to 70% of the internal production of bacterioplankton in the epilimnion of a lake (2). The composition of bacterioplankton communities also depends on the inlets and hydraulic retention times of the water bodies of the lakes. Bacterial communities of lakes with short hydraulic retention times are more strongly influenced by allochthonous bacteria than those of lakes with longer hydraulic retention times (20). The formation of specific phylogenetic freshwater clusters within the Actinobacteria and their in situ activity in lakes, however, indicates that this bacterial group is an autochthonous component of limnetic habitats (41, 42). Nevertheless, the introduction of Actinobacteria from other environments cannot be excluded unambiguously.
The overall phylogenetic tree of the freshwater Actinobacteria is highly diverse and contains sequences from different habitats, such as lakes and rivers, with a large variety of limnological features (9, 11, 23, 33, 39, 45). The tree consists predominantly of 16S rRNA gene sequences of yet-uncultured Actinobacteria. The few existing isolates of freshwater Actinobacteria belong to the acII cluster and to the newly proposed acSTL cluster. The isolates are derived from totally different habitats, such as lakes, the German Wadden Sea, paddy soil, or a phenol-degrading bioreactor (11, 21, 34, 43). Recently, the isolation of a member of the acI cluster was reported (8). Due to the lack of detailed characterizations of these isolates, almost nothing is known about their physiology and hence their ecological role.
In summary, our results show an overall seasonality within freshwater actinobacterial communities. The high proportions of Actinobacteria partly indicate a dominance of these bacteria in the investigated lakes. The 16S rRNA gene sequences obtained were highly diverse, and the phylogenetic reconstructions revealed several new clusters and subclusters. However, the majority of the retrieved sequences are distinct from already-cultured freshwater Actinobacteria. Therefore, the physiology of freshwater Actinobacteria is still unknown. Thus, the isolation of freshwater Actinobacteria, in particular those of the acI cluster, should be the aim of future studies.
Acknowledgments
We thank Elke Mach for technical assistance during sampling and for the measurement of various limnological parameters. We thank Rainer Koschel for providing data on chemistry, and we warmly acknowledge Kirsten Pohlmann for her many fruitful comments on the manuscript. We also thank Falk Warnecke and Jakob Pernthaler for an introduction to CARD-FISH and for helpful discussions about freshwater Actinobacteria during the last years. We appreciate the helpful comments of two anonymous reviewers on an earlier version of the manuscript.
This study was supported by a grant from the Studienstiftung des deutschen Volkes to M.A. and by the Leibniz Foundation.
REFERENCES
- 1.Bahr, M., J. E. Hobbie, and M. L. Sogin. 1999. Bacterial diversity in an arctic lake—a freshwater SAR11 cluster. Aquat. Microb. Ecol. 11:271-277. [Google Scholar]
- 2.Bergström, A.-K., and M. Jansson. 2000. Bacterioplankton production in humic lake Örträsket in relation to input of bacterial cells and input of allochthonous organic carbon. Microb. Ecol. 39:101-115. [DOI] [PubMed] [Google Scholar]
- 3.Bittl, T., and H. D. Babenzien. 1996. Microbial activities in an artificially divided acidic lake. Arch. Hydrobiol. Adv. Limnol. 48:113-121. [Google Scholar]
- 4.Burkert, U., F. Warnecke, H. D. Babenzien, E. Zwirnmann, and J. Pernthaler. 2003. Members of a readily enriched β-proteobacterial clade are common in surface waters of a humic lake. Appl. Environ. Microbiol. 68:6550-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daims, H., A. Bruhl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 7.Eiler, A., and S. Bertilsson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 8.Gich, F., K. Schubert, A. Bruns, H. Hoffelner, and J. Overmann. 2005. Specific detection, isolation, and characterization of selected, previously uncultured members of the freshwater bacterioplankton community. Appl. Environ. Microbiol. 71:5908-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodfellow, M., and S. T. Williams. 1983. Ecology of Actinomycetes. Annu. Rev. Microbiol. 37:189-216. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, M. W., H. Lünsdorf, Q. L. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haukka, K., E. Heikkinen, T. Kairesalo, H. Karjalainen, and K. Sivonen. 2005. Effect of humic material on the bacterioplankton community composition in boreal lakes and mesocosms. Environ. Microbiol. 7:620-630. [DOI] [PubMed] [Google Scholar]
- 13.Hehmann, A., L. Krienitz, and R. Koschel. 2001. Long-term phytoplankton changes in an artificially divided, top-down manipulated humic lake. Hydrobiologia 448:83-96. [Google Scholar]
- 14.Hiorns, W. D., B. A. Methé, S. A. Nierzwicki-Bauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iizuka, T., S. Yamanaka, T. Nishiyama, and A. Hiraishi. 1998. Isolation and phylogenetic analysis of aerobic copiotrophic ultramicrobacteria from urban soil. J. Gen. Appl. Microbiol. 44:75-84. [DOI] [PubMed] [Google Scholar]
- 16.Jezbera, J., K. Horňák, and K. Šimek. 2005. Food selection by bacteriovorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol. Ecol. 52:351-363. [DOI] [PubMed] [Google Scholar]
- 17.Kasprzak, P. 1993. The use of an artificially divided bog lake in food web studies. Verh. Int. Verein. Limnol. 25:652-656. [Google Scholar]
- 18.Koschel, R. 1995. Manipulation of whole lake ecosystems and long term limnological observations of the Brandenburg-Mecklenburg lake district. Int. Rev. Gesamten. Hydrobiol. 80:1-12. [Google Scholar]
- 19.Koschel, R., P. Casper, T. Gonsiorczyk, R. Rossberg, and G. Wauer. Hypolimnetic Al- and CaCO3-treatments and aeration for restoration of a stratified eutrophic hardwater lake (Lake Tiefwarensee, Mecklenburg-Vorpommern, Germany). Verh. Int. Verein. Limnol., in press.
- 20.Lindström, E. S., and A.-K. Bergström. 2004. Influence of inlet bacteria on bacterioplankton assemblage composition in lakes of different hydraulic retention time. Limnol. Oceanogr. 49:125-136. [Google Scholar]
- 21.Lüdemann, H., and R. Conrad. 2000. Molecular retrieval of large 16S rRNA fragments from an Italian rice paddy soil affiliated with the class Actinobacteria. Syst. Appl. Microbiol. 23:582-584. [DOI] [PubMed] [Google Scholar]
- 22.Methé, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition—analyses of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 23.Methé, B. A., and J. P. Zehr. 1999. Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77-96. [Google Scholar]
- 24.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-23. In A. D. L. Akkermans, J. D. van Elsas, and F. J. Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 26.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pernthaler, J., T. Posch, K. Šimek, J. Vrba, A. Pernthaler, F. O. Glöckner, U. Nübel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of Actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Premazzi, G., and G. Chiaudani. 1992. Ecological quality of surface waters—quality assessment schemes for European Community lakes. JRC report EUR 14563 EN. Commission of the European Communities, Brussels, Belgium.
- 29.Rappé, M. S., D. A. Gordon, K. L. Vergin, and S. J. Giovannoni. 1999. Phylogeny of actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst. Appl. Microbiol. 22:106-112. [Google Scholar]
- 30.Rheims, H., A. Felske, S. Seufert, and E. Stackebrandt. 1999. Molecular monitoring of an uncultured group of the class Actinobacteria in two terrestrial environments. J. Microbiol. Methods 36:65-75. [DOI] [PubMed] [Google Scholar]
- 31.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 32.Sekar, R., A. Pernthaler, J. Pernthaler, F. Warnecke, T. Posch, and R. Amann. 2003. An improved protocol for the quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selje, N., T. Brinkhoff, and M. Simon. 2005. Detection of abundant bacteria in the Weser estuary using culture-dependent and culture-independent approaches. Aquat. Microb. Ecol. 39:17-34. [Google Scholar]
- 35.Semenova, E. A., and K. D. Kuznedelov. 1998. A study of the biodiversity of Baikal picoplankton by comparative analysis of 16S rRNA gene 5′-terminal regions. Mol. Biol. 32:754-760. [PubMed] [Google Scholar]
- 36.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 37.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 38.Strickland, J. D. H., and T. R. Parsons. 1978. A practical handbook of seawater analysis, 2nd ed. Fisheries Research Board of Canada, Ottawa, Canada.
- 39.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 40.Van der Gucht, K., T. Vandekerckhove, N. Vloemans, S. Cousin, K. Muylaert, K. Sabbe, M. Gillis, S. Declerk, L. De Meester, and W. Vyverman. 2005. Characterization of bacterial communities in four freshwater lakes differing in nutrient load and food web structure. FEMS Microbiol. Ecol. 53:205-220. [DOI] [PubMed] [Google Scholar]
- 41.Warnecke, F., R. Amann, and J. Pernthaler. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 42.Warnecke, F., R. Sommaruga, R. Sekar, J. S. Hofer, and J. Pernthaler. 2005. Abundance, identity, and growth state of Actinobacteria in mountain lakes of different UV transparency. Appl. Environ. Microbiol. 71:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, K., M. Teramoto, and S. Harayama. 1999. An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 65:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wetzel, R. G., and G. E. Likens. 1991. Limnological analyses, 2nd ed. Springer-Verlag, Inc., New York, N.Y.
- 45.Zwart, G., B. C. Crump, M. P. Agterveld, F. Hagen, and S.-K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]