Abstract
The PCR-single-strand conformation polymorphism (SSCP) technique was used to assess the diversity and distribution of Rieske nonheme iron oxygenases of the toluene/biphenyl subfamily in soil DNA and bacterial isolates recovered from sites contaminated with benzene, toluene, ethylbenzene, and xylenes (BTEX). The central cores of genes encoding the catalytic α subunits were targeted, since they are responsible for the substrate specificities of these enzymes. SSCP functional genotype fingerprinting revealed a substantial diversity of oxygenase genes in three differently BTEX-contaminated soil samples, and sequence analysis indicated that in both the soil DNA and the bacterial isolates, genes for oxygenases related to the isopropylbenzene (cumene) dioxygenase branch of the toluene/biphenyl oxygenase subfamily were predominant among the detectable genotypes. The peptide sequences of the two most abundant α subunit sequence types differed by only five amino acids (residues 258, 286, 288, 289, and 321 according to numbering in cumene dioxygenase α subunit CumA1 of Pseudomonas fluorescens IP01). However, a strong correlation between sequence type and substrate utilization pattern was observed in isolates harboring these genes. Two of these residues were located at positions contributing, according to the resolved crystal structure of cumene dioxygenase from Pseudomonas fluorescens IP01, to the inner surface of the substrate-binding pocket. Isolates containing an α subunit with isoleucine and leucine at positions 288 and 321, respectively, were capable of degrading benzene and toluene, whereas isolates containing two methionine substitutions were found to be incapable of degrading toluene, indicating that the more bulky methionine residues significantly narrowed the available space within the substrate-binding pocket.
Aromatic hydrocarbons such as benzene, toluene, and ethylbenzene are common contaminants of soil and groundwater (41) and are listed as priority pollutants by the U.S. Environmental Protection Agency (http://www.epa.gov/waterscience/criteria/wqcriteria.html). One of the most attractive means to remove these compounds from polluted environments is through bioremediation, which can be achieved by natural attenuation, by stimulation of the local microbial activity through the addition of nutrients and/or electron acceptors, or by bioaugmentation (65).
The aerobic degradation of aromatic compounds is frequently initiated by Rieske nonheme iron oxygenases, which catalyze the incorporation of two oxygen atoms into the aromatic ring to form arene cis-diols (30), followed by a dehydrogenation reaction catalyzed by a cis-dihydrodiol dehydrogenase to give catechol or substituted catechols which serve as substrates for oxygenolytic aromatic ring cleavage. Rieske nonheme iron oxygenases are multicomponent enzyme complexes composed of a terminal oxygenase component (iron-sulfur protein [ISP]) and different electron transport proteins (a ferredoxin and a reductase or a combined ferredoxin-NADH-reductase) (13). The catalytic iron-sulfur proteins are heteromultimers, comprising a large (α) and a small (β) subunit, with the former containing a Rieske-type [2Fe-2S] cluster, a mononuclear nonheme iron oxygen activation center, and a substrate-binding site (13, 27) and being responsible for substrate specificity (30). Comparison of the amino acid sequences of the terminal oxygenase α subunits (ISPα) revealed that they form a family of diverse but evolutionarily related sequences, and four distinct major lineages have been identified (30). Although none of the enzymes is completely specific, a broad correlation between the grouping in toluene/biphenyl, naphthalene, benzoate, or phthalate subfamilies and the native substrates oxidized by the subfamily members can be observed. However, various enzyme-engineering studies on biphenyl, benzene, chlorobenzene, and naphthalene dioxygenases also showed that only slight sequence differences in the amino acid sequences of ISPα can be associated with dramatic changes in substrate specificity or regiospecificity (8, 24, 50, 69). Information on Rieske nonheme iron oxygenases has been obtained mainly from cultured strains. However, various culture-independent studies suggest that distinct and potentially numerically dominant sequence types of as-yet-unrecognized functional genes exist in natural and anthropogenically influenced environments and that the sequences from cultured strains are not likely to represent the microbial functional gene diversity in the environment (63, 64, 68).
Members of the toluene/biphenyl oxygenase subfamily have been reported to be involved in the degradation of benzene, chlorobenzenes, toluene, ethylbenzene, isopropylbenzene, and biphenyl, and phylogenetic analysis of their ISPα peptide sequences (47) indicated the presence of several distinct sequence clusters, such as benzene/toluene oxygenases, isopropylbenzene oxygenases, and biphenyl oxygenases. The amino acid residues reported to be responsible for the substrate specificity and regioselectivity of members of the subfamily are located within a central core of the α subunit (20, 27, 69). ISPα DNA sequence alignments had identified highly conserved regions for use as primer binding sites in PCR, including conserved regions flanking the central core, thus facilitating molecular studies on the diversity of these catabolic genes in the environment (38, 64, 68). Detection and differentiation of toluene/biphenyl oxygenases by using PCR-based genetic profiling techniques targeting regions responsible for substrate specificity could provide hints on the overall metabolic network acting cooperatively for in situ biodegradation. Monitoring the structure and diversity of predominant catabolic gene polymorphisms can give indications on functional adaptations, as optimized catabolic enzyme variants are likely to be selected for functioning under the existing environmental conditions.
PCR-single-strand conformation polymorphism (SSCP) analysis has been demonstrated to be a powerful technique for elucidating the structure of complex microbial communities as well as the diversity of catabolic genes (37, 52-54). Based on sequence-dependent folding of single-stranded DNA (ssDNA) into secondary-structure conformations, which exhibit different electrophoretic mobilities in nondenaturing polyacrylamide gels, DNA molecules of the same length but with different nucleotide sequences can be separated (54). Here we describe the application of the PCR-SSCP technique for the simultaneous detection and discrimination of ISPα gene segments retrieved from pure culture strains exhibiting different aromatic hydrocarbon-degradative capabilities and from DNA extracted from soil samples differently contaminated with benzene, toluene, ethylbenzene, and xylenes (BTEX). The translation products of ISPα gene segments recovered from SSCP fingerprints were analyzed on the basis of structural information of related dioxygenase enzymes in order to evaluate the functional significance of structurally important amino acid residues.
MATERIALS AND METHODS
Bacterial strains.
Reference strains used in this study are listed in Table 1 and comprised bacteria which are known to possess genes coding for Rieske nonheme iron oxygenase of the toluene/biphenyl subfamily and bacteria for which the presence of such genes has not been reported. The 16S rRNA gene sequence from the reference strain Pandoraea sp. strain JB1*, which we received as Burkholderia sp. strain JB1, was reevaluated, since an ISPα gene segment differing from that reported previously (GenBank accession no. AJ010057) was detected (see Results). The nearly complete 16S rRNA gene sequence (1,455 nucleotides) obtained showed 100% sequence identity with 16S rRNA genes from Pandoraea spp. but differed from the sequence previously reported for strain JB1 (X92188) by seven nucleotides. Additionally, 36 bacterial isolates obtained from BTEX-contaminated soil samples (37), the majority of which were capable of mineralizing benzene, were analyzed for the presence of ISPα genes of the toluene/biphenyl subfamily.
TABLE 1.
Reference strains used in this study and results of the PCR targeting ISPα genes of the toluene/biphenyl oxygenase subfamily
| Organism | Relevant aromatic growth substratea | Reference(s) | Reported ISPα gene(s) | GenBank accession no. | PCR primer setc
|
|
|---|---|---|---|---|---|---|
| BPDOXF/ BPDOXR | bphAf668-3/bphAf1153-2 | |||||
| Burkholderia xenovorans LB400 | Bphb | 10, 31 | bphA1 | M86348 | + | + |
| Cupriavidus necator H850 | Bph | 6 | bphA1 | AJ544525 | + | + |
| Ralstonia sp. strain PS12 | Tcb | 7 | tecA1 | U78099 | + | + |
| Rhodococcus globerulus P6 | Bph | 2 | bphA1 | X80041 | (+) | + |
| Pandoraea sp. strain JB1* | NS | + | + | |||
| Pseudomonas sp. strain Cam-1 | Bph | 45 | bphA1 | AY027651 | + | + |
| Pseudomonas sp. strain IC | Bph, Tt | 14 | bphX | AF077669 | + | + |
| Pseudomonas aeruginosa JI104 | B | 42 | bnzA | E04215 | + | + |
| Pseudomonas fluorescens IP01 | T, Eb, Ipb | 1, 16 | cumA1 | D37828 | + | + |
| Pseudomonas pseudoalcaligenes KF707 | Bph | 25 | bphA1 | M83673 | + | + |
| Pseudomonas stutzeri AN10 | Nah | 11 | nahAc | AF039533 | − | − |
| Pseudomonas stutzeri OM1 | Car | 49 | carAa | AB001723 | − | − |
| Pseudomonas stutzeri OX1 | B, T, Ph, o-X | 9 | − | − | ||
| Pseudomonas putida F1 | B, T, Eb, p-C | 21, 29 | todC1, cmtAb | J04996, U24215 | + | + |
| Pseudomonas putida G7 | Nah | 58 | nahAc | M83949 | − | − |
| Pseudomonas sp. strain CF600 | Ph | 56 | − | − | ||
| Pseudomonas putida mt-2 | T, m-X, p-X, Tt, Bt | 12 | xylX, benA | M64747, AAN68769 | − | − |
| Pseudomonas putida MT53 | T, m-X, p-X, Tt | 40 | xylX | − | − | |
| Pseudomonas putida HS1 | T, m-X, p-X | 43 | xylX | AF134348 | − | − |
| Pseudomonas putida 3,5X | Ph | 48 | − | − | ||
B, benzene; T, toluene; Eb, ethylbenzene; Ipb, isopropylbenzene; m-X, m-xylene; o-X, o-xylene; p-X, p-xylene; p-C, p-cumate; Tt, toluate; Bt, benzoate; Ph, phenol; Tcb, tetrachlorobenzene; Bph, biphenyl; Nah, naphthalene; Car, carbazole. NS, not specified.
Boldface type indicates that initial attack proceeds via a Rieske nonheme iron oxygenase of the toluene/biphenyl subfamily (30).
+, PCR signal; (+), weak PCR signal; −, no PCR signal.
Soil samples.
Soil samples were collected from the unsaturated zone (X, zone above the water table in which the pore spaces are only partially filled with water) and the capillary fringe zone (Y, zone above the water table within which the soil is saturated by water under less than atmospheric pressure) at two sampling sites of a BTEX-contaminated aquifer located in the Czech Republic. The BTEX concentrations in the groundwater (determined by Aquatest a.s., Prague) ranged from 320, 97, 6, and 13 mg/liter of benzene, toluene, ethylbenzene, and xylenes, respectively (site 1), to 55, 2, 0.007, and 0.012 mg/liter of benzene, toluene, ethylbenzene, and xylenes, respectively (site 3). Further characteristics of the soil samples were described previously (34, 37).
Chemicals.
Chemicals were purchased from Aldrich Chemie (Steinheim, Germany), Fluka AG (Buchs, Switzerland), or Merck AG (Darmstadt, Germany). Benzene-cis-dihydrodiol (cis-3,5-cyclohexadiene-1,2-diol) was obtained from Sigma-Aldrich Chemie (Deisenhofen, Germany).
Oxygen uptake measurements with whole cells.
Transformation of benzene, toluene, phenol, and benzene-cis-dihydrodiol was determined polarographically by measuring O2 consumption with a Clark-type oxygen electrode (Bachhofer, Reutlingen, Germany). Benzene-grown cells were harvested during late-exponential-phase growth, washed, and resuspended to a turbidity (A546) of 30 in 50 mM phosphate buffer (pH 7.5). Of this suspension, 0.1 ml was added to 2.9 ml phosphate buffer (50 mM, pH 7.5) saturated with air. After 5 min of constant endogenous oxygen uptake, the reaction was started by adding the substrate (100 mM aqueous stock solutions or solutions in dimethyl sulfoxide in the cases of benzene and toluene) to a final concentration of 0.2 mM. Uptake rates were determined at 25°C and corrected for endogenous O2 uptake. In the case of benzene-cis-dihydrodiol transformation, metabolism was confirmed by high-performance liquid chromatography analysis. Samples were taken during oxygen uptake measurements, and 10 μl of cell-free supernatants was analyzed with a Shimadzu high-performance liquid chromatography system (LC-10AD liquid chromatograph, DGU-3A degasser, SPD-M10A diode array detector, and FCV-10AL solvent mixer) equipped with an SC125/LiChrospher column (Bischoff, Leonberg, Germany) by use of an aqueous solvent system (flow rate, 1 ml/min) containing 0.01% (vol/vol) H3PO4 (87%) and 30% (vol/vol) methanol.
DNA extraction.
DNA from eight reference strains (Burkholderia xenovorans LB400, Cupriavidus necator H850, Pseudomonas sp. strain Cam-1, Pseudomonas pseudoalcaligenes KF707, Pandoraea sp. strain JB1*, Pseudomonas putida F1, Ralstonia sp. strain PS12, and Rhodococcus globerulus P6) was extracted using the QIAGEN Genomic DNA Tip 100/G system (QIAGEN, Hilden, Germany). DNA concentrations in the extracts were quantified using a PicoGreen double-stranded DNA (dsDNA) quantitation kit (Molecular Probes, Leiden, The Netherlands) and adjusted to a final concentration of 1 ng/μl with Tris-HCl buffer (10 mM, pH 8.0). To extract DNA from other reference strains and from the BTEX isolates, single colonies were boiled and centrifuged (39) and the supernatants were used either 10-fold or 100-fold diluted in Tris-HCl buffer (10 mM, pH 8.0) as template DNA in PCR. Total DNA from soil samples (wet weight, 500 mg) was extracted using a FastDNA spin kit for soil (Bio101 Systems, Q-BIOgene, Heidelberg, Germany) and DNA concentrations in extracts were determined as described above. The soil extracts, containing approximately 2 ng/μl DNA, were used either directly or 10-fold diluted in Tris-HCl buffer (10 mM, pH 8.0) as template DNA in PCR.
Evaluation of primer binding sites suitable for amplification of ISPα gene segments.
DNA and amino acid sequences of the α subunits of Rieske nonheme iron oxygenases (ISPα) were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/) and aligned with ClustalW (http://www.ebi.ac.uk/clustalw/). A subset of 31 ISPα genes (GenBank accession numbers AF006691, AJ293587, U53507, D37828, AF049851, AJ010057, U47637, AB086835, M86348, U95054, M83673, AY027651, E04215, D88020, X80041, U27591, D32142, U24277, AF148496, AJ006307, J04996, U15298, U78099, D17319, U51165, AF452376, AB121977, AY554272, AJ536756, AY831463, and AB193045), comprising the majority of currently known members of the toluene/biphenyl subfamily (30), was examined for conserved regions for use as PCR primer binding sites. Amino acid residues which were reported to have an influence on substrate specificity of oxygenases of the toluene/biphenyl subfamily are encoded by ISPα gene segments flanked by conserved sites at approximately positions 670 and 1170 of the bphA1 gene from B. xenovorans LB400 (8, 26, 46, 60, 69). Two degenerate primer sets targeting these conserved regions were used to amplify 525- to 531-bp segments of ISPα genes. The primer set BPDOXF/BPDOXR was designed by Yeates et al. (68); however, DNA sequence alignments revealed a critical Y-A mismatch between reverse primer BPDOXR and the ISPα genes bphA1 from R. globerulus P6 and Rhodococcus erythropolis TA421 and bpdC1 from Rhodococcus sp. M5 (GenBank accession numbers X80041, D88020, and U27591, respectively) at position 3 from the primer 3′ end. To increase their specificity for amplification of ISPα genes of the toluene/biphenyl subfamily, these primers were modified. The modified forward and reverse primers (bphAf668-3 [5′-GT TCC GTG TAA CTG GAA RTW YGC-3′] and bphAr1153-2 [5′-CCA GTT CTC GCC RTC RTC YTG HTC-3′], respectively) comprised a pool of degenerate primers in which a minimum of 14 bases at the 3′ end corresponded to the aligned toluene/biphenyl ISPα gene sequences. For PCR-SSCP analysis, ISPα gene segments were amplified with the modified primer set by using a 5′-end-phosphorylated reverse primer (54). The forward primer, bphAf371B (5′-GGC TTT CAC CTG CAS YTA YCA YGG-3′) (targeting positions 348 to 371 in bphA1 of LB400) was used together with bphAr1153-2 to amplify 799- to 826-bp ISPα segments, which comprised the primer binding site for bphAf668-3.
PCR amplification of ISPα gene segments.
PCR amplification was performed in a final volume of 50 μl containing 1× PCR buffer (QIAGEN, Hilden, Germany) supplemented with 1.5 mM MgCl2, 200 μM of each dNTP (Bioline, Luckenwalde, Germany), 0.5 μM of each primer (MWG-Biotech, Ebersberg, Germany), 2.5 U of HotStarTaq polymerase (QIAGEN), and 2 μl of template DNA. The PCR program comprised an initial denaturation for 15 min at 95°C followed by 32 cycles of 40 s at 94°C, 40 s at 58°C, and 60 s at 72°C and a final elongation for 10 min at 72°C. For amplification of soil DNA, the cycle number was increased to 35 cycles (sample 1Y or 3Y) or 40 cycles (sample 1X). The PCR conditions for amplification with BPDOXF and BPDOXR were as described previously (68). PCR products (2 μl of the PCR mixture) were separated by agarose gel electrophoresis (1.5% agarose, 1× Tris-acetate-EDTA buffer; 1 h at 90 V) and visualized by ethidium bromide staining. PCR yield estimates were obtained by quantitative image analysis of digitized agarose gel images using Scion Image Beta version 4.0.2 (Scion Corporation, Frederick, MD).
PCR-SSCP analysis of ISPα gene segments.
PCR products were purified with a MinElute PCR purification kit (QIAGEN) as recommended by the manufacturer and eluted with 20 μl Tris-HCl buffer (10 mM, pH 8.0). The phosphorylated strands were removed by digestion with 12.5 U lambda exonuclease (New England Biolabs, Beverly, MA) in a final volume of 25 μl for 1 h at 37°C (54). The digestion was stopped by the addition of 5 μl sodium acetate (3 M, pH 5.5), and the ssDNA was purified with a MinElute PCR purification kit and eluted with 20 μl sterile water. The ssDNA concentration was assessed by comparing the fluorescence intensities of ssDNA sample bands to the fluorescence intensities of dsDNA standard bands (Low DNA Mass Ladder; Invitrogen) after agarose gel electrophoresis (1.5% agarose, 1× Tris-acetate-EDTA buffer; 1 h at 90 V) and ethidium bromide staining. Prior to the SSCP procedure, samples were diluted with sterile water to yield an ssDNA concentration equivalent to 1.7 ng/μl dsDNA per sample. PCR-negative controls were used without further dilution. Aliquots of 17 μl were evaporated for 15 min at 30°C in a vacuum concentrator to yield a final volume of approximately 4 μl. The samples were mixed with 4 μl SSCP loading buffer (95% [vol/vol] formamide, 10 mM NaOH, 0.025% [wt/vol] bromophenol blue, and 0.025% [wt/vol] xylene cyanol), incubated for 2 min at 95°C in a water bath, and placed on ice water for 5 min. The influence of gel temperature and MDE gel solution (Cambrex Bioscience, Rockland, ME) concentration on the resolution performance of the SSCP method was tested with ssDNA ISPα segments generated from six reference strains (LB400, H850, Cam-1, KF707, JB1*, F1, PS12, and P6) in individual runs using the following conditions (gel temperature/MDE gel solution concentration): (i) 20°C/0.6×, (ii) 24°C/0.6×, (iii) 28°C/0.6×, (iv) 28°C/0.7×, and (v) 30°C/0.7×. Electrophoresis under optimized conditions was performed on 21- by 21-cm 0.7× MDE gels in 1× Tris-borate-EDTA as the running buffer at 400 V and 10 mA for 20 h at 30°C in a Pharmacia Macrophor electrophoresis unit 2010-001 connected to a circulating water bath (Lauda Ecoline RE104 thermostat). The gels were silver stained according to the procedure of Bassam et al. (5) and dried at room temperature. ISPα SSCP fingerprints from pure cultures were reproduced in at least two independent SSCP analyses. Silver-stained gels were scanned on a flatbed scanner (Epson Expression 1600 PRO) to create 8-bit TIFF images, which were then analyzed with GelCompar II software (Applied Maths, Kortrijk, Belgium).
Isolation and reamplification of single-strand products from SSCP profiles.
Selected single-strand products were excised from silver-stained polyacrylamide gels and transferred to microtubes containing 50 μl of elution buffer (10 mM Tris-HCl [pH 8.8], 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100). The tubes were incubated for 20 min at 95°C, and 3 μl of eluted ssDNA was subjected to PCR reamplification using the same primers and conditions used to generate the original dsDNA segments.
Cloning of PCR-amplified ISPα segments.
PCR-reamplified products from selected SSCP bands and ISPα gene segments amplified with the primer set bphAf371B/bphAr1153-2 were purified with a QIAquick PCR purification kit (QIAGEN), ligated into pGEM-T-vector system (Promega), and transformed into Escherichia coli JM109 competent cells (Promega) as specified by the manufacturers. Plasmid inserts were amplified by PCR with vector-specific primers M13 forward and M13 reverse (Promega), annealing in the flanking cloning region.
DNA sequencing and sequence analyses.
PCR products were purified with a QIAquick PCR purification kit and sequenced using an ABI PRISM BigDye Terminator v1.1 Ready Reaction cycle sequencing kit (Applied Biosystems, Darmstadt, Germany). Primers used for sequencing reactions were the same as those used in the original PCR, and sequencing was performed on an ABI PRISM 3100 genetic analyzer (Applied Biosystems). Raw sequence data from both strands were assembled with Sequencher software version 4.0.5 (Gene Codes Corporation, Ann Arbor, MI), and consensus sequences were translated in frame (omitting the primer sequences) with BioEdit software version 5.0.9 (http://www.microbiology.ncsu.edu/resources/index.html). For cloned PCR products, only consensus sequences deduced from at least three identical or nearly identical clones (difference of up to four bases) were included in the sequence analysis in order to reduce potential sequence diversity biases due to PCR and/or sequencing errors. DNA similarity searches were performed with the BLASTN algorithm and public nucleotide databases (http://www.ncbi.nlm.nih.gov/). DNA and protein alignments were conducted with ClustalW. Phylogenetic analyses were done with MEGA software version 3.0 (http://www.megasoftware.net/), by using the neighbor-joining method with the p-distance model and pairwise deletion of gaps/missing data. A consensus tree was inferred from a total of 1,000 bootstrap trees generated for each data set.
AFDRA and ARDRA.
The presence of a catechol 2,3-dioxygenase (C23O) gene was determined by using PCR amplification with primers C23O-ORF-F and C23O-ORF-R, and C23O types were identified by amplified functional DNA restriction (AFDRA) analysis as reported previously (36). Amplified ribosomal DNA restriction analysis (ARDRA) was performed by using protocols described previously (37).
Nucleotide sequence accession numbers.
The ISPα sequences and the 16S rRNA gene sequence of Pandoraea sp. strain JB1* are available under GenBank accession numbers DQ166964 to DQ167022 and DQ336933 to DQ336947.
RESULTS
Primer evaluation.
Twenty reference strains were chosen to assess the specificity of the primer set BPDOXF/BPDOXR (68), which had been designed for use in studies on the diversity of Rieske nonheme ISPα genes. Using this primer set, single products of the expected size were amplified from 11 reference strains known to possess an ISPα gene of the toluene/biphenyl oxygenase subfamily (Table 1). No products were amplified from reference strains containing α subunits of the naphthalene (P. putida G7 [58] and P. stutzeri AN10 [11]) or benzoate/toluate (P. putida mt-2 [67] and MT53 [40]) oxygenase subfamilies and other reference strains obviously devoid of genes encoding Rieske nonheme iron oxygenase α subunits of the toluene/biphenyl subfamily (Table 1). However, compared to the other positively tested target genes, amplification of bphA1 from R. globerulus P6 resulted in a considerably lower product yield, as judged from quantitative image analysis of ethidium bromide-stained agarose gels (data not shown). To increase the specificity of the primers for amplification of ISPα genes of the toluene/biphenyl subfamily, the primers were modified to match with an updated DNA alignment comprising 31 reference sequences. PCR performed with the modified primers resulted in similar product yields for all reference strains containing target genes.
SSCP analysis of ISPα gene segments.
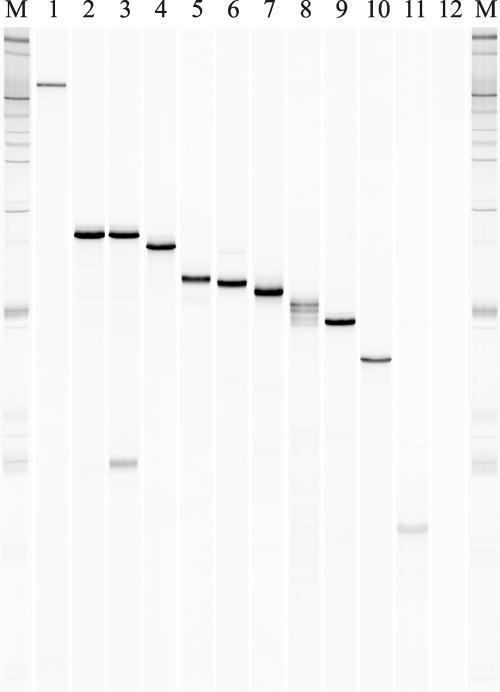
A PCR-SSCP method was established for the detection and discrimination of ISPα genes of the toluene/biphenyl oxygenase subfamily using α subunit gene segments amplified from reference strains with the primer set bphAf668-3/bphAr1153-2. Five different combinations of gel temperature and MDE gel solution concentration were tested to determine the optimum conditions for differentiation of the approximately 530-base single-strand ISPα segments, and best resolution and selectivity of SSCP profiles were achieved using an MDE gel concentration of 0.7× and a gel temperature of 30°C, allowing the discrimination of the predominant bands of 11 reference ssDNA products (Fig. 1). Single-strand ISPα gene segments of identical mobilities were obtained from B. xenovorans LB400 and C. necator H850, which was expected, as both strains were reported to contain ISPα genes identical in sequence in the fragment analyzed here (38). The developed PCR-SSCP method was able to separate even highly homologous ISPα gene segments, as indicated by the different migration patterns of the single strands originating from Pseudomonas sp. strain Cam-1 and P. pseudoalcaligenes KF707, which differed by only one nucleotide. Sequences retrieved from predominant bands were identical to those previously reported, except that of the Pandoraea sp. strain JB1* bphA1 sequence, which differed in seven nucleotides from the sequence reported for Burkholderia sp. strain JB1 (AJ010057). The ISPα gene fragment recovered from the PCR-SSCP profile of the biphenyl degrader Pseudomonas sp. strain IC (14) was found to be identical to bphA1 from Pseudomonas sp. strain B4 (U95054). SSCP analysis of the majority of reference strains resulted in the identification of one ISPα gene only; however, a second ISPα gene could be observed with C. necator H850 (Fig. 1, lane 3, lower band), the deduced amino acid sequence of which exhibited the highest levels of similarity to putative ISPα proteins from Bordetella bronchiseptica strain RB50 (BX640439) and Variovorax paradoxus strain E4C (AF209470) (84% to 86% amino acid sequence identity).
FIG. 1.
PCR-SSCP analysis of ISPα gene segments obtained from reference strains. The following strains were analyzed: P. putida F1 (todC1, lane 1); B. xenovorans LB400 (bphA1, lane 2); C. necator H850 (bphA1, lane 3); Pseudomonas sp. strain IC (bphA1, lane 4); R. globerulus P6 (bphA1, lane 5); P. pseudoalcaligenes KF707 (bphA1, lane 6); P. aeruginosa JI104 (bnzA, lane 7); Ralstonia sp. strain PS12 (tecA1, lane 8); Pseudomonas sp. strain Cam-1 (bphA1, lane 9); Pandoraea sp. strain JB1* (bphA1, lane 10); and P. fluorescens IP01 (cumA1, lane 11). Lane 12 shows the PCR-negative control. Molecular weight marker III (M) (250 ng per lane; Roche) was used to normalize the band patterns.
Diversity of ISPα genes in isolates from BTEX-contaminated soil samples.
Recently, a large set of bacterial strains was isolated from BTEX-contaminated soil samples based on their ability to convert catechol by meta-cleavage to the yellow product 2-hydroxymuconic semialdehyde (37). A high proportion of these isolates was found to be capable of mineralizing benzene and/or toluene and harbored C23O subfamily I.2.A variants, which were also observed to be abundant in the study sites (37). In this study, a subset (36 isolates) of these bacterial strains was screened for the presence of ISPα genes of the toluene/biphenyl oxygenase subfamily using primers bphAf668-3 and bphAr1153-2, and products of the expected size could be amplified with DNA extracted from 33 out of the 36 bacterial isolates (Table 2). ISPα gene segments could not be amplified from the isolates IA1YICDA and IA1YICDB, which were capable of mineralizing toluene, as well as m- and p-xylene, but not benzene (37). This phenotype is similar to that of strains harboring the well-studied toluene degradation pathway encoded by TOL plasmids (12, 67), where degradation is initiated by oxidation of the methyl group. Additionally, isolate 3YC1b failed to give a PCR product. Its capability to grow on benzene, however, suggests that it either harbors an ISPα gene exhibiting low gene sequence homology with the primers used here or possibly initiates the degradation by means of two successive monooxygenation reactions.
TABLE 2.
Substrate utilization profiles and genotypic characteristics of BTEX isolatesa
| Isolate name | Growth substrateb
|
C23O of subfamily I.2.A | C23O typec | ISPα typed | ISPα/C23O combinationf | ARDRA typeg | ||
|---|---|---|---|---|---|---|---|---|
| Benzene | Toluene | Ethylbenzene | ||||||
| IA1YICDA | − | + | − | + | D | − | PS2 | |
| IA1YICDB | − | + | − | + | D | − | PS2 | |
| 3YC1b | + | + | − | − | − | ND | ||
| 3YC2 | + | + | + | − | Ae | I | PS2• | |
| 3YC4 | + | + | + | − | A | I | PS2• | |
| 3YBTEX2b | + | + | + | − | A | I | PS2 | |
| 1XXyl1 | + | + | + | − | A | I | ND | |
| 1YC1 | + | + | + | + | ND | Ae | PS1• | |
| 1XC2 | + | + | − | + | ND | Ae | PS2 | |
| 1YXyl2 | + | + | + | + | A• | A | II | PS1 |
| 1YXyl3 | + | + | + | + | A• | A | II | PS1 |
| 1XBTEX3 | + | + | + | +• | A• | A | II | PS1 |
| 1XBTEX4 | + | + | + | + | A• | A | II | PS1 |
| 1YXyl1 | + | + | + | + | A2 | Ae | II | PS1 |
| 1YdBTEX2 | + | + | + | + | A2 | A | II | PS1 |
| 1XBTEX1 | + | + | + | + | B | A | III | PS1 |
| 1XB2 | + | + | + | + | B | A | III | PS1 |
| 1XC1 | + | + | + | +• | C• | Ae | IV | PS1 |
| 3YXyl2 | + | + | − | +• | C• | Ae | IV | PS2 |
| 3YC5 | + | + | − | + | C• | Ae | IV | PS2 |
| 3YdBTEX2 | + | + | + | + | C | Ee | V | PS2• |
| 1YB1 | + | − | − | + | A• | C | VI | PS1 |
| 1YB2 | + | − | − | + | A1 | Ce | VI | PS1 |
| 1YB3 | + | − | − | + | A1 | C | VI | PS1 |
| 1YC2 | + | − | − | + | A1 | Ce | VI | PS1 |
| 1YC3 | + | − | − | + | A1 | C | VI | PS1 |
| 1YBTEX1 | + | − | − | + | A1 | C | VI | PS1 |
| 1YBTEX3 | + | − | − | + | A1 | C | VI | PS1 |
| 1YdBTEX3 | + | − | − | + | A1 | C | VI | PS1 |
| 3YdBTEX1b | + | − | − | + | A1 | C | VI | PS2 |
| 3YXyl2b | + | − | − | + | A1 | Ce | VI | PS2 |
| 3YXyl3 | + | − | − | + | A1 | C | VI | PS2 |
| 3YXyl1 | + | + | − | + | A1 | Ce | VI | PS2 |
| 1XB1 | + | − | − | + | A• | C**e | VII | PS3 |
| 3YC3 | + | − | − | − | Be | VIII | AR• | |
| 1XXyl1b | + | − | − | − | De | IX | SP• | |
Data on substrate utilization, presence/type of C23O gene, and ARDRA type were reported previously (37); data indicated by • were obtained in this study by using previously reported procedures (36, 37).
+, growth on substrate vapor; −, no growth on substrate vapor.
C23O types marked by • were identified by AFDRA as reported previously (36), whereas all other C23O types were previously identified by using both AFDRA and DNA sequencing (37). ND, not determined.
ISPα type was determined by sequencing of reamplified SSCP products; −, no PCR product.
ISPα sequence was also determined by sequencing of cloned PCR products amplified with primers bphAf371B and bphAr1153-2.
Roman numerals are used to distinguish between different combinations of C230 and ISPα genotypes observed in a given isolate.
The major portion of isolates from the highly contaminated site 1 was closely related to Pseudomonas veronii CIP 104663 (AF064460, ARDRA profile PS1), whereas isolates from the slightly contaminated site 3 were predominantly related to Pseudomonas mandelii CIP 105273 (AF058286, ARDRA profile PS2). Strain 3YC3 exhibited an ARDRA fingerprint identical to those of previously analyzed Arthrobacter isolates (e.g., AY512644) and was thus considered to be a member of the genus Arthrobacter. Strain 1XXyl1b had previously been identified as a member of the genus Sphingomonas (AY512601) (34).
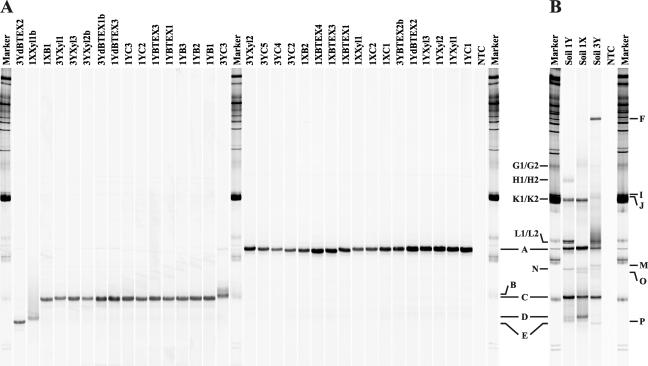
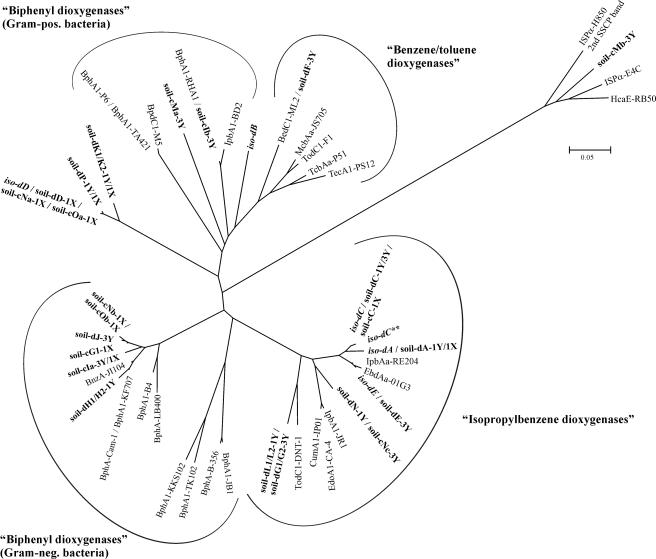
PCR-SSCP analysis of the ISPα gene segments revealed the presence of only one predominant single-strand product per isolate and allowed five different electrophoretic mobilities to be distinguished (Fig. 2A, band positions A, B, C, D, and E). The nucleotide sequence of the most frequently occurring band, designated ISPα type A, was, as verified by sequencing, identical in 17 out of the 33 positively tested isolates, and the derived translation product showed highest identity to the ISPα peptide (IpbAa) of P. putida RE204 isopropylbenzene dioxygenase (22) (Fig. 3). The second predominant ISPα type, referred to as ISPα type C, was observed in 13 isolates. All but one of these isolates contained an ISPα gene sequence that differed from sequence type A by only six nucleotides, corresponding to a difference of five amino acids in the deduced peptide sequence. The ISPα sequence retrieved from isolate 1XB1, designated ISPα type C**, differed from sequence type C by a single nucleotide, resulting in one amino acid difference. Three further ISPα sequence types, detectable in one isolate each, were found to be either highly homologous to ISPα types A and C (sequence type E) or more closely related to the ISPα sequence cluster encoding biphenyl dioxygenases of gram-positive strains and (chloro-)benzene/toluene dioxygenases (ISPα types B and D) (Fig. 3). ISPα PCR-SSCP analysis performed on artificial mixtures of genomic DNA extracted from three strains harboring different ISPα types showed that each component was amplified by PCR, and product yields reflected the abundance of each ISPα gene in the template mixture (see Fig. S1 in the supplemental material).
FIG. 2.
SSCP profiles of ISPα gene segments obtained from BTEX-degrading isolates (A) and BTEX-contaminated soil samples (B). Bands selected for reamplification (by PCR), cloning, and sequencing are indicated by letters. Molecular weight marker III (Roche) (250 ng per lane) was used to normalize the band patterns. ISPα types were named according to their band positions as types A through E for the isolate profiles and types A through P for the soil profiles. NTC, PCR nontemplate control.
FIG. 3.
Phylogenetic analysis of deduced peptide sequences from ISPα gene segments recovered from SSCP profiles (comprising 158 to 161 amino acids, corresponding to positions 224 to 384 of BphA1 from B. xenovorans LB400). Characterized ISPα peptides of the toluene/biphenyl oxygenase subfamily (IpbAa, Pseudomonas putida RE204 [AF006691]; EbdAa, Pseudomonas putida 01G3 [AJ293587]; IpbA1, Pseudomonas sp. strain JR1 [U53507]; CumA1, Pseudomonas fluorescens IP01 [D37828]; EdoA1, Pseudomonas fluorescens CA-4 [AF049851]; BphA1, Burkholderia sp. strain JB1 [AJ010057]; BphA1, Comamonas testosteroni B-356 [U47637]; BphA1, Comamonas testosteroni TK102 [AB086835]; BphA1, Pseudomonas sp. strain KKS102 [D17319]; BphA1, Burkholderia xenovorans LB400 [M86348]; BphA1, Pseudomonas sp. strain B4 [U95054]; BphA1, Pseudomonas pseudoalcaligenes KF707 [M83673]; BphA, Pseudomonas sp. strain Cam-1 [AY027651]; BnzA, Pseudomonas aeruginosa JI104 [E04215]; BphA1, Rhodococcus erythropolis TA421 [D88020]; BphA1, Rhodococcus globerulus P6 [X80041]; BpdC1, Rhodococcus sp. strain M5 [U27591]; BphA1, Rhodococcus sp. strain RHA1 [D32142]; IpbA1, Rhodococcus erythropolis BD2 [U24277]; BedC1, Pseudomonas putida ML2 [AF148496]; McbAa, Ralstonia sp. strain JS705 [AJ006307]; TodC1, Pseudomonas putida F1 [J04996]; TcbAa, Pseudomonas sp. strain P51 [U15298]; TecA1, Ralstonia sp. strain PS12 [U78099]; ISPα, Variovorax paradoxus E4C [AF209469]; HcaE, Bordetella bronchiseptica RB50 [NC_002927]; ISPα, Cupriavidus necator H850 [2nd SSCP band]) and ISPα peptides from bacterial isolates (bold italics) and soil samples (bold) are shown. The ISPα peptides obtained from soil (soil-) were designated according to the procedure of sequence recovery, i.e., direct sequencing of reamplified SSCP products (d) or sequencing of cloned PCR-amplified products (c), followed by band position (A through P) and sample origin (1X, 1Y, or 3Y). In cases where more than one consensus sequence was retrieved by cloning, the different sequence types are indicated by lowercase letters (a, b, or c) after the band position designation. The ISPα peptide sequences from isolates (iso-) were deduced from directly sequenced reamplification products. The tree was constructed by using the neighbor-joining method in MEGA with the p-distance model and pairwise deletion of gaps/missing data. The scale bar indicates the percent divergence. The names of ISPα peptide clusters were assigned according to the native substrates used for isolation of their hosts and characterization of their activity. pos., positive; neg., negative.
There was a strong correlation between the ISPα type and the substrate utilization profile observed with the respective isolates (Table 2), as all isolates harboring ISPα type A could grow on benzene and toluene as a sole source of carbon and energy, while all but 1 of the 13 isolates containing ISPα type C were able to grow on benzene but not on toluene. To provide evidence that degradation of benzene by the isolates is initiated by dioxygenation rather than two successive monooxygenations, oxygen uptake rates by two representative isolates containing ISPα type A and two isolates containing ISPα type C as well as that by the isolate containing ISPα type B were assessed (Table 3). All isolates exhibited a significant oxygen uptake with benzene cis-dihydrodiol, indicating that this compound is dehydrogenated and subsequently subject to ring cleavage, whereas oxygen uptake with phenol was absent. While all tested isolates showed significant oxygen uptake with benzene, which had been used as growth substrate, only the two isolates harboring ISPα type A were active with toluene.
TABLE 3.
Oxygen uptake rates of selected benzene-degrading isolates
| Isolate name | ARDRA type | Phenotypea | Genotype
|
Oxygen uptake rate (μmol of O2 min−1) withb:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C23O subfamily I.2.A | ISPα | xylMc | tbmDc | tmoAc | Benzene | Toluene | Phenol | DHDd | |||
| 1YdBTEX2 | PS1 | BT | + | A | + | + | − | 5 ± 1 | 14 ± 4 | <1 | 90 ± 15 |
| 1XB2 | PS1 | BT | + | A | + | − | − | 4 ± 1 | 8 ± 2 | <1 | 65 ± 10 |
| 3YC3 | AR | B | − | B | ND | ND | ND | 15 ± 3 | <1 | <1 | 35 ± 5 |
| 1YB2 | PS1 | B | + | C | + | + | + | 4 ± 1 | <1 | <1 | 35 ± 5 |
| 3YdBTEX1b | PS2 | B | + | C | + | − | − | 4 ± 1 | <1 | <1 | 50 ± 10 |
BT, benzene and toluene degradation; B, benzene degradation.
Oxygen uptake rates were determined with whole cells grown on benzene and are expressed as the rate of O2 uptake measured polarographically using cell densities (A546) of 1. Rates given are means of three independent determinations.
PCR result was reported by Hendrickx et al. (34). ND, not determined.
DHD, benzene cis-dihydrodiol.
To obtain additional sequence information in an approximately 300-bp stretch of the ISPα genes upstream of the region used for PCR-SSCP, the isolates were subjected to a second PCR analysis using the primer set bphAf371B/bphAr1153-2. PCR amplification of ISPα gene segments was observed with the same 33 isolates described above (data not shown), and sequencing of the cloned products from seven strains harboring ISPα type A, four strains harboring ISPα type C, and the one strain harboring ISPα type C** (Table 2) revealed that they were identical in the 300-bp DNA stretch upstream of the sequence derived from SSCP analysis, indicating that sequence differences were confined to a region of the carboxy-terminal portion of the α subunits that was previously identified to be of crucial importance for determining substrate specificity (8, 24, 26, 60, 69).
ISPα gene diversity in BTEX-contaminated soil samples.
The developed PCR-SSCP technique was used to assess the diversity and distribution of Rieske nonheme iron oxygenases of the toluene/biphenyl subfamily in differently contaminated soil samples from a BTEX-contaminated site. Comparison of the SSCP profiles revealed significant differences in the catabolic gene structure between the three soil samples (Fig. 2B). Bands that were detectable against a low surrounding background could be sequenced directly from reamplified ssDNA, resulting in unambiguous identification of the single-strand products. The majority of bands with high relative intensities and identical mobilities, like bands A and C, were found to contain identical single-stranded DNA species. Several of the closely migrating single-strand products represented identical ssDNA sequence types (bands G1/G2, H1/H2, K1/K2, and L1/L2), suggesting that the same single strand formed two conformers migrating with slightly different mobilities. In the case of faint bands, direct sequencing of reamplified single-strand products did not result in conclusive sequences. To reveal the identity of the ISPα types contained in these bands, the reamplification products were cloned and sequenced. For each band, one (or two distinct) consensus sequence(s) could be inferred from at least three clones containing nearly or completely identical sequence types.
Phylogenetic analysis of the peptide sequences deduced from directly sequenced PCR-amplified SSCP products revealed the majority of the soil peptides to be related to ISPα sequences of ethylbenzene/isopropylbenzene dioxygenases (Fig. 3), with the nucleotide sequences retrieved from bands A, C, and E being identical to the corresponding ISPα segments retrieved from isolates. The ISPα gene sequences retrieved from bands G1/G2 and L1/L2, respectively, differed by nine nucleotides; however, none of them caused a difference in the deduced peptide sequences, which were most closely related to TodC1 from the toluene-degrading bacterium Thauera sp. strain DNT-1 (57). The ISPα sequence types soil-dK, soil-dP, and soil-dD, which were, among the soil samples tested, detectable only in the highly contaminated soil samples 1X and 1Y, grouped together in a divergent branch of the toluene/biphenyl oxygenase subfamily and exhibited less than 75% identity to previously described ISPα peptide sequences. The peptide sequence inferred from an intense band, which was detected only in the SSCP profile of soil 3Y, was found to be identical to benzene dioxygenase ISPα from P. putida ML2 (62). ISPα sequences closely related to P. pseudoalcaligenes KF707 (61) and Pseudomonas sp. strain Cam-1 (45) biphenyl dioxygenase ISPα peptides were inferred from bands H1/H2 and J.
DISCUSSION
In this study, the PCR-SSCP technique was used for detection and discrimination of ISPα gene homologues amplified from bacterial isolates and environmental samples. Several primer sets suitable for amplification of Rieske nonheme iron oxygenase ISPα gene segments have been reported previously (4, 34, 38, 64, 68). Since it has been noted that amino acid residues important for the substrate specificity of Rieske nonheme iron oxygenases of the toluene/biphenyl subfamily are concentrated in the C-terminal portion of the α subunits (26, 51, 69), primer sets enabling amplification of this region are of special interest. SSCP analysis has been shown to be a sensitive method for the discrimination of gene homologues that is able to distinguish sequence differences as small as one base substitution in several hundred bases (33, 37, 54). However, it has also been demonstrated that the sensitivity of PCR-SSCP in the detection of single base substitutions decreased with segments larger than 300 bases (33, 55). Thus, the primer binding sites previously selected by Yeates et al. (68) were chosen, as these produced the shortest possible gene segment encoding the major portion of amino acids crucial for substrate specificity. Adjusting the reported primer sequences to match the toluene/biphenyl dioxygenase subfamily members together with careful optimization of the PCR-SSCP conditions allowed detection and differentiation of all reference ISPα gene segments down to single base differences.
PCR-SSCP analysis of ISPα gene segments obtained from the three differently contaminated soil samples showed that there was a high degree of catabolic genotype diversity. Sequence analysis indicated that the detectable polymorphisms were dominated by ISPα sequence types related to the ethylbenzene/isopropylbenzene oxygenase branch rather than by sequences related to the expected toluene/benzene oxygenase branch (Fig. 3). In particular, ISPα types A and C not only were observed in all three soil samples, suggesting that the environmental conditions were selective for strains possessing these ISPα types, but also were predominant in benzene- and/or toluene-degrading bacterial isolates previously isolated from these soil samples (Fig. 2A and B).
This finding was unexpected as, to our knowledge, Rieske nonheme iron oxygenases closely related to the ethylbenzene/isopropylbenzene oxygenase branch were previously not observed to be prevalent in BTEX-contaminated environments. Previous investigations, however, have concentrated on the presence and abundance of ISPα genes more closely related to the toluene/benzene and biphenyl dioxygenase branches by using DNA:DNA hybridization (59) or PCR-based assays (15, 44). The use of specific DNA probes and oligonucleotide primers may have failed to detect genes exhibiting lower levels of homology with the targeted genes. More-detailed studies on the diversity of Rieske nonheme iron oxygenases in pristine soil and soil with a history of aromatic hydrocarbon pollution (68) as well as in soil and groundwater at an aromatic hydrocarbon-contaminated site (63, 64) have been performed by analyzing clone libraries of PCR products which were amplified with degenerate primers targeting the same primer binding sites as used here (68) or primer binding sites flanking the ISPα gene region which encodes the Rieske-type [2Fe-2S] cluster (63, 64). In these studies, amplification of sequence types showing no clear affiliation to previously described ISPα proteins (68) or of a significant portion of gene segments which did not code for Rieske nonheme iron oxygenase ISPα proteins (63) was reported. In contrast to these findings, the vast majority of the peptide sequences retrieved from the BTEX-contaminated soil samples analyzed here showed a clear membership to the toluene/biphenyl oxygenase subfamily, with only a small fraction of the clones (less than 10%) prepared from faint SSCP bands exhibiting no clear pattern of relationship to previously described Rieske nonheme iron oxygenase ISPα proteins. A comparison of the ISPα peptide sequences obtained in this study with those reported by Taylor et al. (63, 64) suggests that the structures of dioxygenases putatively involved in pollutant degradation are clearly different in the organic waste landfill soil and groundwater samples and the industrial site analyzed here. Common to both environments seems to be the low abundance of genes closely related to the toluene/benzene dioxygenase branch of the toluene/biphenyl subfamily, suggesting that the previously characterized dioxygenases may not play significant roles in the sites under investigation.
As indicated above, the ISPα sequence types A and C were not only abundant in all three contaminated soil samples analyzed in this study but also predominant among the benzene-degrading isolates recovered from these soil samples. Overall, ISPα gene segments of the toluene/biphenyl subfamily were amplifiable from 33 out of 34 isolates that were capable of growth on benzene, and biochemical analysis supported the notion that, at least in the five representative strains analyzed, benzene metabolism is initiated by dioxygenation, yielding benzene dihydrodiol as an intermediate, rather than monooxygenation, which would result in phenol as intermediate (Table 3). In a recent study, the presence of alternative modes of benzene/toluene activation has been suggested for a subset of the isolates analyzed here, whereas genes encoding Rieske nonheme iron oxygenases were not detected (34). Evidently, the primers designed by Hendrickx et al. (34) for detection of toluene/biphenyl ISPα genes failed to amplify the genes reported here. Moreover, there was no correlation between the growth phenotype and the catabolic genotypes detected by Hendrickx et al. (34) (Table 3). As benzene lacks a side chain, this substrate cannot be transformed by xylene monooxygenases (genotype xylM). Furthermore, neither the strains harboring a gene coding for a phenol hydroxylase α subunit (genotype tbmD) nor strain 1YB2, which contained a gene fragment probably encoding an α subunit of aromatic monooxygenases (genotype tmoA), showed significant oxygen uptake with phenol. Therefore, degradation of benzene via phenol as the intermediate seems rather unlikely.
The fact that all isolates harboring genes similar to those encoding isopropylbenzene dioxygenases were capable of mineralizing benzene but only a subset was able to grow on toluene was on the first view astonishing, as all previously described isolates harboring ethylbenzene/isopropylbenzene dioxygenase genes (P. putida RE204, P. putida 01G3, Pseudomonas sp. strain JR1, Pseudomonas fluorescens IP01, P. fluorescens CA-4, and Thauera sp. strain DNT-1) were reported to mineralize toluene (1, 16-19, 22, 57). However, there was a strong correlation between the growth phenotype and the ISPα type detected (Table 2), with only isolates containing ISPα type C, B, or D unable to grow on toluene, suggesting that the differences in substrate specificity are related to structural differences in the α subunits of the terminal oxygenase components. Alignments of ISPα peptides retrieved from BTEX-contaminated soil and isolates to those of related proteins (see Fig. S2 in the supplemental material) revealed that the residues His234 and His240 (numbered according to P. fluorescens IP01 cumene dioxygenase [IP01-CumDO]), which are involved in the coordination of the mononuclear iron in the active site of IP01-CumDO and RHA1-BphDO (20, 27), as well as Asp231, which was postulated to play the role of an electron transfer bridge (20, 27), were completely conserved among the ISPα peptides retrieved from bacterial isolates and environmental samples. Among the 14 residues proposed to define the inner surface of the substrate-binding pocket in IP01-CumDO (20), 8 were found to be strictly conserved among the ISPα sequences retrieved in this study (Gln227, Phe228, Met232, Ala235, His323, Leu333, and Phe378, in addition to the above-mentioned His240). The isopropylbenzene dioxygenase-like ISPα sequences derived from the soil samples and isolates in this study differed from the IP01-CumDO sequence at up to three residues defining the inner surface of the substrate-binding pocket (see Fig. S2 in the supplemental material). Strikingly, isolates harboring an A321L ISPα variant (ISPα types A and E) were capable of mineralizing both toluene and benzene, whereas those harboring I288M and A321M variants (ISPα type C and C**) were capable of mineralizing benzene only. Modeling performed on the basis of the IP01-CumDO crystal structure (pdb-id 1WQL [20]), using “O” (35) for manual replacement of side chains and choice of the best-fitting rotamer, revealed that replacing Ile288 and Ala321 in IP01-CumDO with bulky methionine significantly narrowed the available space in the substrate-binding pocket (data not shown). This gives a possible explanation for the different phenotypes and the negligible oxygen uptake of isolates 1YB2 and 3YdBTEX1b with toluene as the substrate. Also, ISPα types B and D as well as K1/K2 and P contained bulky residues (methionine and phenylalanine, respectively) in positions 288 and 321, and the two single isolates containing ISPα type B and D were also capable of mineralizing benzene only. Taken together, these results indicate a significant contribution of the amino acid side chains in positions 288 and 321 to the shape and size of the substrate-binding pocket. Bagnéris et al. (3) also had suggested that less bulky amino acids in position 321 should facilitate access to toluene, which, as a substrate, is larger than benzene.
In the well-characterized isopropylbenzene-degradative pathways of P. putida RE204 and P. fluorescens IP01 (1, 22), the genes coding for the oxygenase subunits are organized in operons together with genes encoding a cis-dihydrodiol dehydrogenase and an extradiol dioxygenase (C23O) of the subfamily I.3.A (23). In the isolates 1YB2 and 1YdBTEX2, which had been analyzed in more detail and obviously do not express any 3-isopropylcatechol 2,3-dioxygenase (subfamily I.3.A) during growth on benzene (37), benzene mineralization seems to involve an isopropylbenzene dioxygenase-like Rieske nonheme oxygenase for initial activation and a subfamily I.2.A C23O (36, 37) for catechol cleavage. As shown in Table 2, in addition to strain 1YB2, 12 further independently selected isolates (affiliated to three distinct Pseudomonas lineages) harbored the same combination of a type C (or C**) ISPα gene and a type A C23O gene. While the isolates harboring ISPα type C contained the C23O type A exclusively, the strains harboring ISPα type A were found to contain different C23O subfamily I.2.A genes or failed to yield a PCR product with primers targeting C23O subfamily I.2.A genes; the latter result indicated that these strains obviously contained a C23O of other subfamilies. The high degree of sequence homology between ISPα type A and C on the one hand and C23O type A2 and A1 on the other suggests a close evolutionary relationship between the two gene variants. The flexible combination of ISPα type A genes with different types of C23O genes indicates that the pathways were assembled in a modular fashion, in which the C23O genes were retrieved from different sources, possibly driven by the selective conditions of the environment, and the occurrence of identical or nearly identical genes and pathway module combinations in different hosts indicates that both the ethylbenzene/isopropylbenzene dioxygenase and the catechol 2,3-dioxygenase are localized on mobile genetic elements and have been spread through horizontal gene transfer.
C23Os of subfamily I.2.A have previously not been reported to be involved in the degradation of benzene or toluene via a dioxygenolytic route. They have, however, been commonly described as involved in phenol (56), toluate (12, 67), and salicylate (28) degradation. The respective catabolic operons comprising the C23O I.2.A gene have a rather stable structure and comprise genes coding for both the hydrolytic branch and the oxalocrotonate branch of the meta-cleavage pathway (66). Recruitment of both branches of the meta-cleavage pathway can be assumed to be advantageous to the presence of the hydrolytic branch only (as present in strains like RE204), particularly for benzene degradation (32). Both branches were expressed in isolates 1YB2 and 1YdBTEX2 (37).
In summary, bacteria from the examined BTEX-contaminated soil samples seem to possess a genotypic flexibility, which is important for their adaptation and evolution while facing challenging and continuously changing conditions in these ecosystems. Screening methods targeting genes encoding key enzymes in pollutant degradation as molecular markers, such as PCR-SSCP analyses, can be utilized to determine both the biodegradative potential and predominant catabolic pathways in an environment. Further studies on catabolic gene diversity and genetic flexibility will certainly be important elements in our understanding of natural bioremediation processes.
Supplementary Material
Acknowledgments
We thank W. Mohn for providing Pseudomonas sp. strain Cam-1 and J. Parsons for providing Pandoraea sp. strain JB1*. We also thank Julia Bötel, Iris Plumeier, and Theresa Namuth for excellent technical assistance.
This research was supported by project BIOTOOL (GOCE-003998) of the European Commission and PI 312/2-1 of the Deutsche Forschungsgemeinschaft.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aoki, H., T. Kimura, H. Habe, H. Yamane, T. Kodama, and T. Omori. 1996. Cloning, nucleotide sequence, and characterization of the genes encoding enzymes involved in the degradation of cumene to 2-hydroxy-6-oxo-7-methylocta-2,4-dienoic acid in Pseudomonas fluorescens IP01. J. Ferment. Bioeng. 81:187-196. [Google Scholar]
- 2.Asturias, J. A., E. Diaz, and K. N. Timmis. 1995. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from gram-negative bacteria. Gene 156:11-18. [DOI] [PubMed] [Google Scholar]
- 3.Bagnéris, C., R. Cammack, and J. R. Mason. 2005. Subtle difference between benzene and toluene dioxygenases of Pseudomonas putida. Appl. Environ. Microbiol. 71:1570-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, B. R., C. H. Nakatsu, and L. Nies. 2003. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassam, B. J., G. Caetano-Anolles, and P. M. Gresshoff. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196:80-83. [DOI] [PubMed] [Google Scholar]
- 6.Bedard, D. L., R. E. Wagner, M. J. Brennan, M. L. Haberl, and J. F. Brown, Jr. 1987. Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl. Environ. Microbiol. 53:1094-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beil, S., B. Happe, K. N. Timmis, and D. H. Pieper. 1997. Genetic and biochemical characterization of the broad spectrum chlorobenzene dioxygenase from Burkholderia sp. strain PS12—dechlorination of 1,2,4,5-tetrachlorobenzene. Eur. J. Biochem. 247:190-199. [DOI] [PubMed] [Google Scholar]
- 8.Beil, S., J. R. Mason, K. N. Timmis, and D. H. Pieper. 1998. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J. Bacteriol. 180:5520-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertoni, G., M. Martino, E. Galli, and P. Barbieri. 1998. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bopp, L. H. 1986. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J. Ind. Microbiol. 1:23-29. [Google Scholar]
- 11.Bosch, R., E. Garcia-Valdes, and E. R. Moore. 1999. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236:149-157. [DOI] [PubMed] [Google Scholar]
- 12.Burlage, R. S., S. W. Hooper, and G. S. Sayler. 1989. The TOL (pWW0) catabolic plasmid. Appl. Environ. Microbiol. 55:1323-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 14.Carrington, B., A. Lowe, L. E. Shaw, and P. A. Williams. 1994. The lower pathway operon for benzoate catabolism in biphenyl-utilizing Pseudomonas sp. strain IC and the nucleotide sequence of the bphE gene for catechol 2,3-dioxygenase. Microbiology 140:499-508. [DOI] [PubMed] [Google Scholar]
- 15.Cavalca, L., E. Dell'Amico, and V. Andreoni. 2004. Intrinsic bioremediability of an aromatic hydrocarbon-polluted groundwater: diversity of bacterial population and toluene monoxygenase genes. Appl. Microbiol. Biotechnol. 64:576-587. [DOI] [PubMed] [Google Scholar]
- 16.Chablain, P. A., G. Philippe, A. Groboillot, N. Truffaut, and J. F. Guespin-Michel. 1997. Isolation of a soil psychrotrophic toluene-degrading Pseudomonas strain: influence of temperature on the growth characteristics on different substrates. Res. Microbiol. 148:153-161. [DOI] [PubMed] [Google Scholar]
- 17.Chablain, P. A., A. L. Zgoda, C. O. Sarde, and N. Truffaut. 2001. Genetic and molecular organization of the alkylbenzene catabolism operon in the psychrotrophic strain Pseudomonas putida 01G3. Appl. Environ. Microbiol. 67:453-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corkery, D. M., and A. D. W. Dobson. 1998. Reverse transcription-PCR analysis of the regulation of ethylbenzene dioxygenase gene expression in Pseudomonas fluorescens CA-4. FEMS Microbiol. Lett. 166:171-176. [DOI] [PubMed] [Google Scholar]
- 19.Dabrock, B., J. Riedel, J. Bertram, and G. Gottschalk. 1992. Isopropylbenzene (cumene)—a new substrate for the isolation of trichloroethene-degrading bacteria. Arch. Microbiol. 158:9-13. [DOI] [PubMed] [Google Scholar]
- 20.Dong, X. S., S. Fushinobu, E. Fukuda, T. Terada, S. Nakamura, K. Shimizu, H. Nojiri, T. Omori, H. Shoun, and T. Wakagi. 2005. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J. Bacteriol. 187:2483-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eaton, R. W. 1996. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmt operon. J. Bacteriol. 178:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eaton, R. W., and K. N. Timmis. 1986. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putida RE204. J. Bacteriol. 168:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eltis, L. D., and J. T. Bolin. 1996. Evolutionary relationships among extradiol dioxygenases. J. Bacteriol. 178:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson, B. D., and F. J. Mondello. 1993. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl. Environ. Microbiol. 59:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa, K., and T. Miyazaki. 1986. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J. Bacteriol. 166:392-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furukawa, K., H. Suenaga, and M. Goto. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furusawa, Y., V. Nagarajan, M. Tanokura, E. Masai, M. Fukuda, and T. Senda. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342:1041-1052. [DOI] [PubMed] [Google Scholar]
- 28.Ghosal, D., I. S. You, and I. C. Gunsalus. 1987. Nucleotide sequence and expression of gene nahH of plasmid NAH7 and homology with gene xylE of TOL pWW0. Gene 55:19-28. [DOI] [PubMed] [Google Scholar]
- 29.Gibson, D. T., M. Hensley, H. Yoshioka, and T. J. Mabry. 1970. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry 9:1626-1630. [DOI] [PubMed] [Google Scholar]
- 30.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 31.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400(T) and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 32.Harayama, S., N. Mermod, M. Rekik, P. R. Lehrbach, and K. N. Timmis. 1987. Roles of the divergent branches of the meta-cleavage pathway in the degradation of benzoate and substituted benzoates. J. Bacteriol. 169:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi, K. 1991. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1:34-38. [DOI] [PubMed] [Google Scholar]
- 34.Hendrickx, B., H. Junca, J. Vosahlova, A. Lindner, I. Ruegg, M. Bucheli-Witschel, F. Faber, T. Egli, M. Mau, M. Schlomann, M. Brennerova, V. Brenner, D. H. Pieper, E. M. Top, W. Dejonghe, L. Bastiaens, and D. Springael. 2006. Alternative primer sets for PCR detection of genotypes involved in bacterial aerobic BTEX degradation: distribution of the genes in BTEX degrading isolates and in subsurface soils of a BTEX contaminated industrial site. J. Microbiol. Methods 64:250-265. [DOI] [PubMed] [Google Scholar]
- 35.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 36.Junca, H., and D. H. Pieper. 2003. Amplified functional DNA restriction analysis to determine catechol 2,3-dioxygenase gene diversity in soil bacteria. J. Microbiol. Methods 55:697-708. [DOI] [PubMed] [Google Scholar]
- 37.Junca, H., and D. H. Pieper. 2004. Functional gene diversity analysis in BTEX contaminated soils by means of PCR-SSCP DNA fingerprinting: comparative diversity assessment against bacterial isolates and PCR-DNA clone libraries. Environ. Microbiol. 6:95-110. [DOI] [PubMed] [Google Scholar]
- 38.Kahl, S., and B. Hofer. 2003. A genetic system for the rapid isolation of aromatic-ring-hydroxylating dioxygenase activities. Microbiology 149:1475-1481. [DOI] [PubMed] [Google Scholar]
- 39.Kanakaraj, R., D. L. Harris, J. G. Songer, and B. Bosworth. 1998. Multiplex PCR assay for detection of Clostridium perfringens in feces and intestinal contents of pigs and in swine feed. Vet. Microbiol. 63:29-38. [DOI] [PubMed] [Google Scholar]
- 40.Keil, H., S. Keil, R. W. Pickup, and P. A. Williams. 1985. Evolutionary conservation of genes coding for meta pathway enzymes within TOL plasmids pWW0 and pWW53. J. Bacteriol. 164:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keith, L. H., and W. A. Telliard. 1979. Priority pollutants. I. A perspective view. Environ. Sci. Technol. 13:416-423. [Google Scholar]
- 42.Kitayama, A., E. Suzuki, Y. Kawakami, and T. Nagamune. 1996. Gene organization and low regiospecificity in aromatic-ring hydroxylation of a benzene monooxygenase of Pseudomonas aeruginosa JI104. J. Ferment. Bioeng. 82:421-425. [Google Scholar]
- 43.Kunz, D. A., and P. J. Chapman. 1981. Isolation and characterization of spontaneously occurring TOL plasmid mutants of Pseudomonas putida HS1. J. Bacteriol. 146:952-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luz, A. P., V. H. Pellizari, L. G. Whyte, and C. W. Greer. 2004. A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can. J. Microbiol. 50:323-333. [DOI] [PubMed] [Google Scholar]
- 45.Master, E. R., and W. W. Mohn. 1998. Psychrotolerant bacteria isolated from Arctic soil that degrade polychlorinated biphenyls at low temperatures. Appl. Environ. Microbiol. 64:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nam, J. W., H. Nojiri, T. Yoshida, H. Habe, H. Yamane, and T. Omori. 2001. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 65:254-263. [DOI] [PubMed] [Google Scholar]
- 48.Ng, L. C., V. Shingler, C. C. Sze, and C. L. Poh. 1994. Cloning and sequences of the first eight genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene 151:29-36. [DOI] [PubMed] [Google Scholar]
- 49.Ouchiyama, N., S. Miyachi, and T. Omori. 1998. Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J. Gen. Appl. Microbiol. 44:57-63. [DOI] [PubMed] [Google Scholar]
- 50.Parales, R. E., S. M. Resnick, C. L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the α subunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pieper, D. H. 2005. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 67:170-191. [DOI] [PubMed] [Google Scholar]
- 52.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmalenberger, A., and C. C. Tebbe. 2003. Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol. Ecol. 12:251-261. [DOI] [PubMed] [Google Scholar]
- 54.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16s rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheffield, V. C., J. S. Beck, A. E. Kwitek, D. W. Sandstrom, and E. M. Stone. 1993. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics 16:325-332. [DOI] [PubMed] [Google Scholar]
- 56.Shingler, V., F. C. Franklin, M. Tsuda, D. Holroyd, and M. Bagdasarian. 1989. Molecular analysis of a plasmid-encoded phenol hydroxylase from Pseudomonas CF600. J. Gen. Microbiol. 135:1083-1092. [DOI] [PubMed] [Google Scholar]
- 57.Shinoda, Y., Y. Sakai, H. Uenishi, Y. Uchihashi, A. Hiraishi, H. Yukawa, H. Yurimoto, and N. Kato. 2004. Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl. Environ. Microbiol. 70:1385-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon, M. J., T. D. Osslund, R. Saunders, B. D. Ensley, S. Suggs, A. Harcourt, W. C. Suen, D. L. Cruden, D. T. Gibson, and G. J. Zylstra. 1993. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 127:31-37. [DOI] [PubMed] [Google Scholar]
- 59.Stapleton, R. D., and G. S. Sayler. 1998. Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microb. Ecol. 36:349-361. [DOI] [PubMed] [Google Scholar]
- 60.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taira, K., J. Hirose, S. Hayashida, and K. Furukawa. 1992. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 267:4844-4853. [PubMed] [Google Scholar]
- 62.Tan, H.-M., H. Y. Tang, C. L. Joannou, N. H. Abdel-Wahab, and J. R. Mason. 1993. The Pseudomonas putida ML2 plasmid-encoded genes for benzene dioxygenase are unusual in codon usage and low in G+C content. Gene 130:33-39. [DOI] [PubMed] [Google Scholar]
- 63.Taylor, P. M., and P. H. Janssen. 2005. Variations in the abundance and identity of class II aromatic ring-hydroxylating dioxygenase genes in groundwater at an aromatic hydrocarbon-contaminated site. Environ. Microbiol. 7:140-146. [DOI] [PubMed] [Google Scholar]
- 64.Taylor, P. M., J. M. Medd, L. Schoenborn, B. Hodgson, and P. H. Janssen. 2002. Detection of known and novel genes encoding aromatic ring-hydroxylating dioxygenases in soils and in aromatic hydrocarbon-degrading bacteria. FEMS Microbiol. Lett. 216:61-66. [DOI] [PubMed] [Google Scholar]
- 65.Timmis, K. N., and D. H. Pieper. 1999. Bacteria designed for bioremediation. Trends Biotechnol. 17:201-204. [DOI] [PubMed] [Google Scholar]
- 66.Williams, P. A., and J. R. Sayers. 1994. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5:195-217. [DOI] [PubMed] [Google Scholar]
- 67.Worsey, M. J., and P. A. Williams. 1975. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J. Bacteriol. 124:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeates, C., A. J. Holmes, and M. R. Gillings. 2000. Novel forms of ring-hydroxylating dioxygenases are widespread in pristine and contaminated soils. Environ. Microbiol. 2:644-653. [DOI] [PubMed] [Google Scholar]
- 69.Zielinski, M., S. Backhaus, and B. Hofer. 2002. The principal determinants for the structure of the substrate-binding pocket are located within a central core of a biphenyl dioxygenase alpha subunit. Microbiology 148:2439-2448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.