Abstract
Until recently, diazotrophy was known in only one of the 30 formally described species of Burkholderia. Novel N2-fixing plant-associated Burkholderia species such as B. unamae, B. tropica, and B. xenovorans have been described, but their environmental distribution is scarcely known. In the present study, the occurrence of N2-fixing Burkholderia species associated with different varieties of sugarcane and maize growing in regions of Mexico and Brazil was analyzed. Only 111 out of more than 900 isolates recovered had N2-fixing ability as demonstrated by the acetylene reduction assay. All 111 isolates also yielded a PCR product with primers targeting the nifH gene, which encodes a key enzyme in the process of nitrogen fixation. These 111 isolates were confirmed as belonging to the genus Burkholderia by using a new 16S rRNA-specific primer pair for diazotrophic species (except B. vietnamiensis) and closely related nondiazotrophic Burkholderia. In Mexico, many isolates of B. unamae (predominantly associated with sugarcane) and B. tropica (more often associated with maize) were recovered. However, in Brazil B. tropica was not identified among the isolates analyzed, and only a few B. unamae isolates were recovered from one sugarcane variety. Most Brazilian diazotrophic Burkholderia isolates (associated with both sugarcane and maize plants) belonged to a novel species, as revealed by amplified 16S rRNA gene restriction profiles, 16S rRNA gene sequencing, and protein electrophoresis. In addition, transmissibility factors such as the cblA and esmR genes, identified among clinical and environmental isolates of opportunistic pathogens of B. cenocepacia and other species of the B. cepacia complex, were not detected in any of the plant-associated diazotrophic Burkholderia isolates analyzed.
Until very recently, the genus Burkholderia included 30 properly described species (13), but the number of novel Burkholderia species has continuously increased. Unfortunately, species such as B. kururiensis (50), B. sacchari (7), B. phenoliruptrix (12), B. xenovorans (21), and B. tuberum and B. phymatum (45) have been proposed on the basis of a very limited set of isolates (one to three) analyzed, and consequently, their environmental distribution and ecology are poorly understood. It is well known that bacteria of the B. cepacia complex (Bcc), which includes nine species or genomovars (26), may be found in soils (including polluted soils), the rhizospheres of crop plants, and water, as well as in various animal species, in humans, and in the hospital environment (13). Studies published in Europe have shown that Bcc bacteria are good colonizers of the maize rhizosphere, and they may represent one of the predominant bacterial groups in maize grown in Italy (10, 17, 19). In addition, the species B. graminis (48) and B. cepacia genomovar III (2), presently formally classified as B. cenocepacia (46), were shown to be abundant in maize fields analyzed in France. Although the nine species of the Bcc have been isolated from cystic fibrosis patients, B. cenocepacia has been the predominant species recovered (13, 32). B. cenocepacia cystic fibrosis strains have been shown to possess transmissibility factors such as the cblA gene, encoding giant cable pili (25, 37), and the epidemic strain marker regulator or esmR gene, designated B. cepacia epidemic strain marker (BCESM) (25), which is part of a genomic island (3). cblA and esmR genes have been found in a few environmental isolates (25, 34), but only a limited number of these have been analyzed.
For a long time, N2-fixing ability in bacteria of the genus Burkholderia was recognized only in the species B. vietnamiensis (20), a member of the Bcc (13). This species has been found in the rhizospheres and in the rhizoplanes of maize, coffee, and sorghum plants cultivated in different regions of Mexico, as well as inside maize roots in the state of Morelos, Mexico (18). Recently, two nodulating N2-fixing strains recovered from legume plants were assigned to the genus Burkholderia according to their 16S rRNA sequences (27), and they were formally classified as B. phymatum and B. tuberum (45). B. kururiensis, a trichloroethylene-degrading bacterium (50), has been identified as a diazotrophic species (18). Very recently, many diazotrophic isolates found in rhizospheric and/or endophytic association with maize and coffee plants (18) were identified as novel Burkholderia species, i.e., B. unamae (9), B. tropica (33), and B. xenovorans (21). In addition, B. unamae isolates have been recovered from a few field-grown sugarcane plants in Mexico (9), and a few B. tropica isolates have been recovered from within sugarcane stems cultivated in Brazil and from the rhizosphere of sugarcane in South Africa (33).
Although B. cepacia and B. vietnamiensis are species found in the rhizospheres of crop plants and both are able to promote plant growth (6, 43), their taxonomical position as members of the Bcc has limited their potential use in agriculture. Understanding the taxonomy and ecological distribution of Burkholderia species associated with plants is an important step towards the possibility of using some of them for improving plant growth. Hitherto a detailed study on the occurrence of N2-fixing species of the genus Burkholderia associated with plants has not been described. The aim of this work was to isolate, identify, and compare the diazotrophic species of the genus Burkholderia associated with sugarcane and maize varieties planted in Brazil and Mexico.
MATERIALS AND METHODS
Plant samples.
In Brazil, the plants of sugarcane varieties SP 70-1143 and RB 72-454 and maize variety Aventis A2345 were collected at the Experimental Campus of Embrapa Agrobiology in Seropédica, Rio de Janeiro. In Mexico, the plants of sugarcane varieties MEX 69-290 and CP 72-2086 were collected in Tuxtepec, Oaxaca state, and domestic varieties of maize plants were collected in Tlayacapan, Morelos, and in Coatepec, Veracruz state (Table 1).
TABLE 1.
Nitrogen-fixing Burkholderia species isolated from maize and sugarcane
| Crop and locality | Variety | Species | No. of isolates | Sourcea |
|---|---|---|---|---|
| Maize | ||||
| Mexico | ||||
| Tlayacapan, Morelos | Criollo ancho | B. tropica | 7 | Rhizosphere |
| Rhizoplane | ||||
| 2 | Roots | |||
| Coatepec, Veracruz | Criollo | B. tropica | 12 | Rhizosphere |
| 1 | Rhizoplane | |||
| 23 | Roots | |||
| B. unamae | 2 | Roots | ||
| Brazil | ||||
| Seropédica, Rio Janeiro | Aventis A2345 | Burkholderia sp.b | 11 | Rhizosphere |
| 2 | Roots | |||
| Sugarcane | ||||
| Mexico | ||||
| Tuxtepec, Oaxaca | CP 72-2086 | B. unamae | 28 | Rh-Rp |
| B. tropica | 2 | Rh-Rp | ||
| MEX 69-290 | B. unamae | 3 | Rh-Rp | |
| 2 | Roots | |||
| B. tropica | 1 | Roots | ||
| Brazil | ||||
| Seropédica, Rio Janeiro | SP 70-1143 | B. unamae | 3 | Rhizosphere |
| RB 72-454 | Burkholderia sp.b | 5 | Leaves |
Roots and leaves were surface sterilized. Rh-Rp, rhizosphere and rhizoplane.
Corresponds to Burkholderia NAR group.
Isolation, culture conditions, and biochemical tests.
In Mexico, five field-grown maize plants (in flowering) from each region and 16 sugarcane plants of each variety field grown for 10 months were randomly collected, with a distance of 10 m between plants. Samples of rhizosphere and rhizoplane, as well as surface-sterilized roots and stems of maize, were analyzed for recovery of diazotrophic isolates. Rhizosphere soil and plant samples were treated as described previously (18). Briefly, the root was shaken gently to remove the loosely attached soil, and the adhering soil was rinsed in 10 mM MgSO4 · 7H2O (Mgsol). The resulting rinse solution was considered to contain the rhizosphere bacteria; this solution was 10-fold serially diluted and used for isolation of the rhizosphere bacteria. The root was subsequently washed with Mgsol containing 0.01% (vol/vol) Tween 20, followed by two rinses with Mgsol, and then immersed in Mgsol and vortexed for 3 min. The resulting suspension was considered to contain bacteria from the rhizoplane, and the solution was serially diluted and used for the bacterial isolation. N-free semisolid BAz medium was used as an enrichment culture for N2-fixing Burkholderia, and BAc agar plates were used for isolation and pure cultures (18). Sugarcane roots were excised from 16 plants, and loosely adhering soil was removed; roots were then grouped into four samples, each comprising four root systems. Afterwards, a portion from the roots was cut into small pieces (1 cm), and 5 grams of each root sample was vortexed at 3,000 rpm for 3 min in Mgsol. This suspension was considered to contain bacteria from the rhizosphere-rhizoplane. Other root portions were washed with tap water and subsequently immersed for 5 min with agitation in full-strength bleach solution containing Tween 20 and rinsed three times in sterile water. Stems were cut in 10- to 15-cm pieces, immersed in 96% alcohol, and flamed for 30 to 45 seconds; thereafter, stems were surface sterilized as mentioned above for the roots. The rind was discarded under sterile conditions, and 5 grams of each stem (group of four stems) was macerated in a blender to give a 10−1 dilution. All macerated sugarcane samples were serially diluted, and 100-μl aliquots were placed in vials containing 5 ml of N-free semisolid BAz medium and incubated at 29°C for 4 to 5 days. Vials showing a fine subsurface pellicle were transferred to fresh semisolid BAz medium and new growth streaked out on BAc agar plates. Colonies were purified and assayed for nitrogenase activity by the acetylene reduction activity (ARA) method (8). ARA-positive colonies were maintained in 20% glycerol at −80°C prior to characterization.
In Brazil, roots were excised from four maize plants (two plants that were 2 months old [vegetative state] and two plants that were 3 months old [reproductive state]) as well as from five sugarcane plants of each variety growing for 10 months (second harvest), and loose soil was separated by agitation. From each plant, a portion of root with adhering soil was rinsed in sterile distilled water; the resulting rinse solution was 10-fold serially diluted and used for isolation of the rhizosphere bacteria. Other root portions were washed with tap water and subsequently surface sterilized with 1% chloramine T for 15 min and rinsed three times in sterile distilled water. Stems of maize and sugarcane stems or leaves were similarly disinfected with chloramine T. Surface-sterilized roots as well as stems and leaves were macerated in a blender at high speed for 3 min to give a 10−1 dilution. All macerated maize or sugarcane samples were serially diluted, and 100-μl aliquots were inoculated into 5 ml of N-free semisolid BAz and JMV media (33). Vials containing BAz medium were autoclaved at 121°C for 20 min, and filter-sterilized cycloheximide (200 μg/tube) was then added. After incubation for 4 to 5 days at 30°C, a fine subsurface pellicle was formed. Vials showing pellicles were transferred to fresh semisolid BAz or JMV medium and new growth streaked out on BAc agar plates. Colonies formed were inoculated into new semisolid JMV medium. All colonies obtained were transferred to LB medium and conserved for further observations. Colonies were assayed for ARA, and the isolates that exhibited activity after 13 h of incubation were used for further characterization. These isolates were maintained in 20% glycerol at −80°C prior to analysis.
Acetylene-reducing isolates were analyzed with the API 20NE system. The inoculum and the assay conditions were according to recommendations of the manufacturer (bioMérieux). The results were interpreted using the API analytical profile index, which provided a percent probability of identification.
Total DNA isolation and 16S rRNA-specific primers.
Genomic DNA was isolated from bacterial cells by using published protocols (1). ARA-positive isolates were presumptively assigned to the genera Burkholderia-Ralstonia by amplifying the 16S rRNA gene with the specific primers BuRa-16-1 and BuRa-16-2, using the PCR conditions described previously (4). ARA-positive isolates were confirmed to belong to the genus Burkholderia by amplifying the 16S rRNA gene with new specific primers (GB-F and GBN2-R). Burkholderia and Ralstonia 16S rRNA sequences available in the NCBI database were aligned to identify conserved regions in order to design and test specific PCR primers which would give an amplicon only with N2-fixing Burkholderia species (but excluding B. vietnamiensis) and closely related nondiazotrophic Burkholderia species with greater than 97% identity between their 16S rRNA gene sequences. In the 16S rRNA gene sequences a region corresponding to positions 85 to 104 of B. unamae (GenBank accession no. AY221956) was identified as specific for the genus Burkholderia. This region was chosen to define the 20-mer forward primer GB-F (5′-AGTAATACATCGGAACRTGT-3′). Another 16S rRNA gene region, at positions 1091 to 1110 of B. unamae, was identified as being specific for N2-fixing Burkholderia (except B. vietnamiensis) and closely related (97% identity or higher) nondiazotrophic Burkholderia species. The sequence allowed the design of the 19-mer reverse primer GBN2-R (5′-GCTCTTGCGTAGCAACTAG-3′). The specificity of the primer pair was tested with most of the well-known Burkholderia species. The PCR mix was comprised of 20 ng of genomic DNA, 1.5 mM MgCl2, 250 μM deoxynucleoside triphosphates, 5 pmol of each primer, and 1.0 U of Taq polymerase. PCR conditions were as follows: initial denaturation for 5 min at 94°C; followed by 30 cycles of 30 s of denaturation at 94°C, 45 s of annealing at 60°C, and 1 min of elongation at 72°C; followed by a final 5-min elongation at 72°C. The reaction amplified a 1,025-bp fragment.
ARDRA.
Primers fD1 and rD1 were used for the amplification of the 16S rRNA gene (49), using the PCR conditions described previously (18). The PCR-amplified 16S rRNA genes (ca. 1.5 kb) were restricted with AluI, DdeI, HaeIII, HhaI, HinfI, MspI, and RsaI. The restriction fragments were separated by electrophoresis in 3% agarose gels, and the restriction patterns were compared. Each isolate was assigned to an amplified 16S rRNA gene restriction analysis (ARDRA) genotype, defined by the combination of the restriction patterns obtained with the seven restriction endonucleases (18). Similarities among the 16S rRNA gene sequences were estimated from the proportion of shared restriction fragments by using the method of Nei and Li (29). A dendrogram was constructed from the resulting distance matrix by using the unweighted pair group method with averages (40).
16S rRNA gene sequencing and phylogenetic analysis.
Strains SRMrh-20, SMrh-85, and SRCL-318, which corresponded to a predominant unknown ARDRA genotype recovered from plants that were field grown in Brazil, were chosen for 16S rRNA gene sequencing. In order to obtain the 16S rRNA sequence, PCR products were cloned into the pCR2.1 vector according to the manufacturer's instructions (Invitrogen). 16S rRNA genes were restricted in fragments from 850 and 650 bp using the enzyme EcoRI and subcloned into vector pUC18. 16S rRNA gene sequencing of strains SRMrh-20 and SRCL-318 was performed by the Biotechnology Institute, UNAM (Mexico), and strain SRMrh-85 was sequenced by Embrapa Agrobiology (Brazil). These sequences were compared with previously published 16S rRNA gene sequences from Burkholderia species and related bacteria such as Pandorea and Ralstonia. The multiple alignments of the sequences were performed with CLUSTAL W software (42). The tree topology was inferred by the neighbor-joining method (36), based on 1,299 DNA sites, and distance matrixes were determined according to the method of Jukes and Cantor (23) using the program MEGA version 2.1 (24).
SDS-PAGE of whole-cell proteins.
Preparation of whole-cell proteins from diazotrophic isolates and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) assays were performed as described by Estrada-de los Santos et al. (18).
PCR amplification of nifH genes.
Primers IGK (30) and NDR-1 (44) were used for the amplification of the nifH genes. The PCR mix was comprised of 20 ng of genomic DNA, 1.5 mM MgCl2, 100 μM deoxynucleoside triphosphates, 0.005 μmol of each primer, and 1.0 U of Taq polymerase. The amplification was done with an initial denaturation step at 94°C for 5 min; followed by 30 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 1 min; with a final renaturation step for 10 min. The reaction amplified a 1.2-kb fragment comprising the entire nifH gene, the intergenic spacer region, and the 5′ end of the nifD gene (44).
Detection of transmissibility marker genes.
cblA and esmR genes in acetylene-reducing isolates were investigated with the specific primers CBL1/CBL2 and BCESM1/BCESM2, respectively, using PCR conditions described previously (25, 37). In addition, total DNA from acetylene-reducing Burkholderia isolates was digested with EcoRI, and restriction fragments were electrophoresed and transferred from gels to nylon filters by the Southern procedure and hybridized as described previously (18). PCR-amplified cblA and esmR genes from B. cenocepacia strain J2315T were used as 32P-labeled probes.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of strains SRMrh-20, SMrh-85, and SRCL-318 have been deposited in the EMBL/GenBank database under accession numbers AY96520, AY965242, and AY965241, respectively.
RESULTS
Isolation and biochemical tests.
The bacterial growth in N-free semisolid BAz and JMV media formed very thin and fine pellicles at a depth of 4 mm below the surface at 24 to 48 h; after 72 h the pellicles became yellowish, diffuse, and thick and moved up to the surface without pH changes in the medium. More than 900 pure isolates were recovered in BAc agar plates from the rhizosphere as well as from washed and surface-sterilized roots of maize and sugarcane plants (20 to 25 isolates from each rhizosphere and plant analyzed). However, only 111 isolates (Table 1), commonly recovered from vials inoculated with samples (100-μl aliquots) from 10−1 or 10−2 dilutions (data not shown), showed nitrogenase activity as measured by the acetylene reduction method. Although isolates were recovered from aerial parts of maize and sugarcane, very few of these, recovered from leaves but not from stems, showed acetylene reduction activity. All of the acetylene-reducing isolates recovered were gram negative and motile in N-free semisolid media at 29°C. API 20NE biochemical tests identified the isolates as B. cepacia (98.9 to 99.6% confidence limits based on the API analytical profile index).
16S rRNA-specific primers.
ARA-positive isolates gave a PCR-amplified product of the correct size (409 bp) with primers BuRa-16-1 and BuRa-16-2, confirming their taxonomic status as members of the genera Burkholderia-Ralstonia. Although primers (Burk3 and BurkR) specific for the genus Burkholderia have been reported (38), they fail to amplify 16S rRNA genes in recently described novel N2-fixing Burkholderia species such as B. unamae and B. tropica as well as in other well-known species such as the diazotrophic B. kururiensis and the nondiazotrophic B. sacchari. All of these N2-fixing Burkholderia species, the diazotroph B. xenovorans, and the legume-nodulating, N2-fixing B. tuberum and B. phymatum, as well as those nondiazotrophic closely related Burkholderia species with higher than 97% identity between their 16S rRNA gene sequences (e.g., B. sacchari, B. caribensis, B. graminis, B. phenoliruptrix, and B. phytofirmans), gave a PCR-amplified product of the expected size (1 kb) with the specific primer pair GB-F and GBN2-R developed in the present study (Fig. 1). However, the PCR product from B. kururiensis was a less intense band than those amplified from other diazotrophs (data not shown). All of the nine species of the Bcc, including the diazotroph B. vietnamiensis and the plant-pathogenic species (e.g., B. caryophylli, B. plantarii, and B. gladioli) closely related to the Bcc, as well as Ralstonia pickettii and Ralstonia solanacearum, did not give a PCR-amplified product with this primer pair (Fig. 1). The primers GB-F and GBN2-R allowed the confirmation of all the acetylene-reducing isolates recovered from maize and sugarcane as members of the genus Burkholderia (Fig. 1). In addition, a variable number of ARA-negative isolates (total of around 100) recovered from the rhizospheres, rhizoplanes, and plant tissues of maize and sugarcane were analyzed by PCR with primers BuRa-16-1 and BuRa-16-2. The assays revealed that over 95% of the isolates belong to the genera Burkholderia-Ralstonia (data not shown).
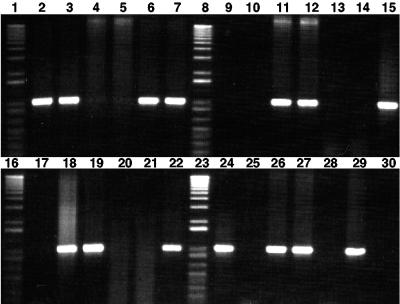
FIG. 1.
PCR-amplified product of 1,025 bp obtained with the 16S rRNA-specific primer pair GB-F and GBN2-R. One-kilobase plus DNA ladder: lanes 1, 8, 16, and 23. No-DNA control, lane 30. PCR amplification in diazotrophic Burkholderia species (except B. vietnamiensis) and closely related organisms; B. unamae: lane 2, MTl-641T, and lane 3, SCTx-181 isolated from rhizosphere of sugarcane (Mexico); B. tropica: lane 6, Ppe8T, and lane 7, SCTx-269 isolated from surface-sterilized roots of sugarcane; type NAR isolates: lanes 11 and 12, SRMrh-20 and SRCL-318 isolated from rhizosphere of maize and surface-sterilized leaves of sugarcane, respectively; lane 15, B. graminis CRD1MT; lane 18, B. caribensis MWAPT; lane 19, B. caledonica LMG 19076T; line 22, B. tuberum STM-678T; line 24, B. phenazinium LMG 2247T; lane 26, B. sacchari IPT101T; lane 27, B. phymatum STM815T; lane 29, B. xenovorans LB400T. PCR-negative amplification in B. cepacia complex species and closely related species: lane 4, B. vietnamiensis TVV75T; lane 5, B. multivorans LMG 13010T; lane 9, B. gladioli ATCC10248T; lane 10, B. cepacia ATCC25416T; lane 13, B. cenocepacia J2315T; lane 14, B. glumae LMG 2196T; lane 17, B. pyrrocinia ATCC15958T; lane 20, B. ambifaria LMG 11351; lane 21, B. caryophylli LMG 2155T; lane 25, Ralstonia pickettii LMG 5942T; lane 28, Ralstonia solanacearum LMG 2299T. Other Burkholderia species tested (data not shown): PCR-positive amplification, B. hospita LMG20598T, B. terricola LMG20594T, B. phytofirmans PsJNT, and B. fungorum LMG 16225T; PCR-negative amplification, B. stabilis LMG 6997.
ARDRA analysis.
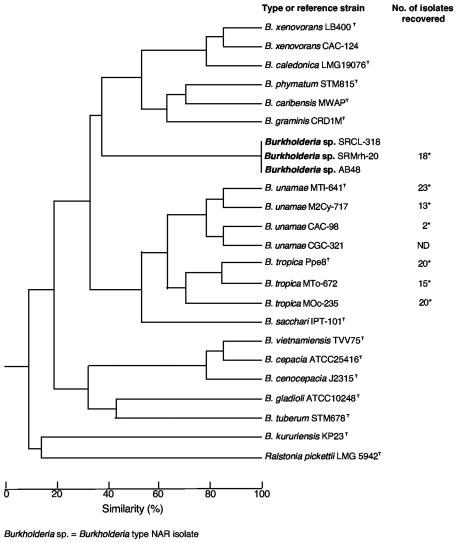
A total of eight ARDRA profiles, clearly different from those for Ralstonia species, were identified from among the ARA-positive isolates recovered from field-grown maize and sugarcane in Mexico and Brazil (data not shown). Five ARDRA profiles were identified among 54 acetylene-reducing isolates recovered from the rhizosphere-rhizoplane and from surface-sterilized roots of maize growing in Mexico (Table 1; Fig. 2). Three ARDRA genotypes corresponding to the species B. tropica (52 isolates) and one genotype belonging to B. unamae (2 isolates) were identified (Table 1). In contrast, only one ARDRA profile was identified among 13 acetylene-reducing isolates associated with maize grown in Brazil, which was different from those ARDRA profiles observed in other well-known N2-fixing Burkholderia species. The 13 Burkholderia isolates identified with such an ARDRA profile are hereafter referred to as the new acetylene-reducing (NAR) group or type NAR isolates. From among 36 isolates associated with sugarcane varieties grown in Mexico, three ARDRA genotypes were identified, corresponding to two genotypes of the species B. unamae (33 isolates) and one of B. tropica (3 isolates) (Table 1; Fig. 2). Associated with sugarcane varieties grown in Brazil were two identified ARDRA genotypes; one of them (three isolates recovered from variety SP 70-1143) showed a profile identical to that of B. unamae, and the other one (five isolates from variety RB 72-454) showed a profile identical to that of the Burkholderia NAR group (Table 1; Fig. 2). From among 90 ARA-negative isolates recovered from sugarcane and maize varieties cultivated in Mexico, one ARDRA genotype was predominant (data not shown). Associated with maize and sugarcane, this ARDRA genotype could correspond to the species B. cepacia, B. cenocepacia, and/or B. ambifaria, all of which are nondiazotrophic members of the B. cepacia complex. However, additional molecular tests are required for their definitive identification at the species level.
FIG. 2.
Dendrogram of genetic relatedness among diazotrophic Burkholderia isolates recovered from maize and sugarcane plants cultivated in Mexico and Brazil and related Burkholderia species, based on ARDRA. The total number of isolates recovered from each diazotrophic Burkholderia species is indicated. *, ARDRA genotypes corresponding to B. unamae and B. tropica strains used as reference have been described previously (9, 33). ND, ARDRA genotype not detected.
Phylogenetic analysis of 16S rRNA gene sequences.
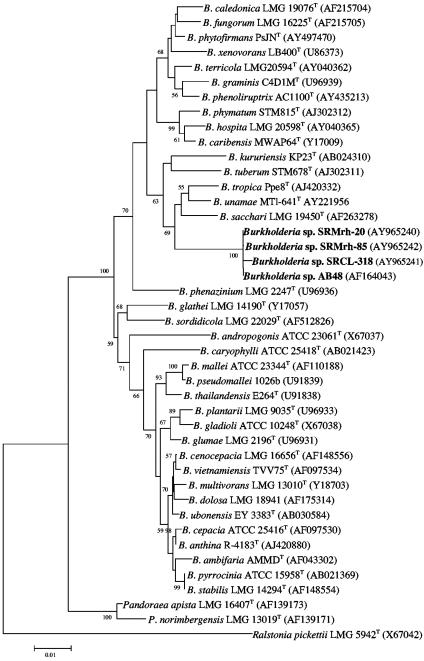
The sequences of Burkholderia strains SRMrh-20 and SRMrh-85, both isolated from the rhizosphere of maize, and of SRCL-318, isolated from surface-sterilized leaves of sugarcane, were compared with available 16S rRNA gene sequences from all of the Burkholderia species (Fig. 3). The strains SRMrh-20, SRMrh-85, and SRCL-318 were closely related, forming a cluster with a strain identified as Burkholderia sp. strain AB48 that was isolated from pineapple (Ananas comosus) in Brazil (14). Notably, strains SRMrh-20, SRMrh-85, and AB48 showed 100% identity between their 16S rRNA gene sequences and closely matched (99.77% identity) strain SRCL-318. Such an identity level strongly suggested that they belong to the same species. Comparison of the 16S rRNA gene sequences from all the well-known Burkholderia species showed that B. sacchari, a nondiazotrophic bacterium, was the closest species (97.1% identity) to the Burkholderia NAR group. This group, which apparently represents a novel Burkholderia species, clearly constituted a cluster largely distant (identity of <96%) from the cluster named the B. cepacia complex (Fig. 3), which includes the N2-fixing species B. vietnamiensis.
FIG. 3.
Phylogenetic tree based on 16S rRNA gene sequences, showing the relatedness among the Burkholderia NAR group, Burkholderia species, and related Betaproteobacteria. The bar represents 1 nucleotide substitution per 100 nucleotides. The GenBank accession number for each strain is shown in parentheses.
Protein electrophoregrams.
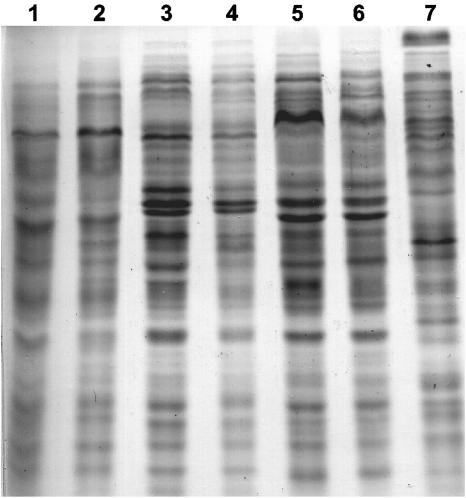
Acetylene-reducing isolates showing ARDRA patterns identical to ARDRA genotypes of B. unamae or B. tropica also showed SDS-PAGE protein profiles (evaluated by visual comparison) that were identical or almost identical to those from type strains of these species. Type NAR isolates showed protein patterns completely different from those of known N2-fixing Burkholderia species (Fig. 4).
FIG. 4.
Protein electrophoregrams of representative acetylene-reducing isolates recovered in the present study and type strains of known diazotrophic Burkholderia species. B. tropica: lane 1, MTl-632 isolated from rhizosphere of maize, and lane 2, Ppe8T; B. unamae: lane 3, MTl-641T; lane 4, SRCrh-274 isolated from rhizosphere of sugarcane (Brazil); type NAR isolates: lane 5, SRMrh-20, and lane 6, SRCL-318; lane 7, B. cepacia ATCC25416T.
PCR amplification of nifH genes.
All 111 of the acetylene-reducing isolates yielded a PCR product of the expected size of 1.2 kb (data not shown) with the nifH primers used. These results confirmed the diazotrophy of the Burkholderia isolates. In addition, PCR amplification of nifH genes was negative in 75 representative isolates that had no acetylene reduction activity (data not shown), which confirmed their lack of N2-fixing ability.
Transmissibility marker genes.
Two transmissibility factors associated with the highly transmissible epidemic strains of B. cenocepacia were analyzed by both PCR amplification and Southern blot assays. No cblA and esmR marker genes were detected among diazotrophic isolates by either PCR amplification or 32P hybridization assays (data not shown).
DISCUSSION
More than 900 pure isolates were recovered in BAc agar plates from the environment of maize and sugarcane plants cultivated in Mexico and Brazil. However, N2-fixing ability was demonstrated in only 111 isolates by means of ARA assays and confirmed with the detection of nifH genes amplified by PCR assays. Biochemical tests based on the API 20NE system identified the N2-fixing isolates as Burkholderia cepacia. However, it is known that this system is appropriate for the identification of the genus Burkholderia, but not at the species level (18, 39). In addition, it has been suggested that N2-fixing ability is used as a distinctive feature for the delineation of Burkholderia species, which is particularly useful for the differentiation of the type species B. cepacia and of the nondiazotrophic Bcc species (9). In fact, both the lack of acetylene reduction activity and the impossibility of detecting the nifH genes along with ARDRA profiles allowed the presumptive identification of most isolates associated with maize and sugarcane as belonging to the species B. cepacia, B. cenocepacia, and/or B. ambifaria. On this basis and considering the protein electrophoregrams as well as the molecular tests, the diazotrophic isolates associated with maize and sugarcane plants recovered in the present study were identified as belonging to the species B. unamae and B. tropica and one unknown Burkholderia species (NAR group).
In previous studies the diazotrophic species B. unamae and B. tropica were randomly recovered from the environments of both maize and sugarcane varieties cultivated in Mexico (9, 33). In the present study, predominant associations of B. tropica with maize and of B. unamae with sugarcane plants were clearly observed. This finding could be partially supported by the isolation, for the first time, of B. unamae from a sugarcane variety (SP 7011-43) grown in Brazil. In the present study B. tropica was not recovered from field-grown plants in Brazil; however, its association in low numbers with sugarcane cultivated in Brazil and South Africa has been documented (33). Though a low number of B. unamae isolates and no B. tropica isolates were recovered from field-grown plants in Brazil, these species appear to be widely distributed among geographical regions and plants. B. unamae has been isolated from the rhizosphere of sugarcane variety N16 cultivated in South Africa (NCBI GenBank database accession number AY391282) and from the rhizosphere of tomato plants in Mexico (unpublished results). Recently, based on the 16S rRNA sequence, one isolate recovered from within the dune grass Ammophila arenaria showed 99% identity with B. tropica strains (16). A possible explanation for the low number of B. unamae isolates or lack of isolation of B. tropica from plants in Brazil could be the dramatically uneven distribution of bacteria among individual plants, as has been observed in communities of B. cepacia complex species detected in the maize rhizosphere (31). A study by Tabacchioni et al. (41) showed that the genetic diversity levels of B. cepacia populations differ with the use of different isolation media. The lack of recovery of B. tropica and Burkholderia type NAR isolates from plants grown in Brazil and Mexico, respectively, could be attributed to the biases caused by using slightly different methodologies, but such a possibility should be discounted. The main difference in methodology was the use of N-free semisolid JMV medium, which was not used in Mexico, but several Burkholderia type NAR isolates were recovered from N-free semisolid BAz medium and BAc agar plates, both of which were used in Mexico. Other factors should account for the lack of recovery of these bacteria. Antagonism in agar plates is a well-known event (15), and recently, antagonistic activity between Gluconacetobacter diazotrophicus strains was demonstrated to occur in N-free semisolid LGI medium and was confirmed on agar plates as well as in endophytic association (28). On this basis, we speculate that antagonism could occur in N-free semisolid BAz or JMV medium during the enrichment subcultures of diazotrophic Burkholderia species as well as under natural conditions. However, such antagonistic activities between Burkholderia species should eventually be demonstrated to occur in further studies.
16S rRNA sequences and whole-cell protein profiles are strong evidence in the delineation of new bacterial species (47). In fact, over the last few years the descriptions of many novel bacterial species have been based on data generated from 16S rRNA sequencing studies, but unfortunately they have been based on the analysis of one or two isolates (35), consequently leaving the distributions and natural habitats of the species unknown. On the basis of 16S rRNA gene sequences and protein electrophoregrams, the predominant group of N2-fixing Burkholderia isolates associated with maize and sugarcane plants in Brazil could be defined as a novel species. However, we considered that more of these diazotrophic isolates from other varieties of maize and sugarcane plants or from other plants, habitats, or geographical regions would have to be recovered and analyzed with polyphasic taxonomy criteria (47) for validation of the Burkholderia NAR group as a new species, as well as for knowledge about their environmental distribution and natural habitats. Regardless of the taxonomic status of these N2-fixing Burkholderia isolates, they are apparently common plant colonizers in Brazil, since strains SRMrh-20, SRCL-318, and AB48 were found associated with maize, sugarcane, and pineapple, respectively.
Although the cblA and esmR genes, which are related to transmissibility, have been found mainly in clinical isolates of B. cenocepacia as well as in other species of the B. cepacia complex, both genes have also been identified from among environmental isolates of B. cenocepacia and B. cepacia (5, 11, 25, 34). In the present study, cblA and esmR were not detected in any of the plant-associated diazotrophic Burkholderia isolates analyzed, which confirms their restricted presence in clinical and environmental isolates of opportunistic pathogens of the B. cepacia complex. Moreover, the lack of detection of BCESM in these N2-fixing Burkholderia populations also suggests the absence of the genomic island detected in B. cenocepacia (3), which fulfills all the characteristic features of a bacterial pathogenicity island (22). On this basis, the results add new evidence to support the potential for using B. unamae and B. tropica as plant growth-promoting bacteria, since they have the ability to improve maize plant growth (unpublished results). Nevertheless, experiments to directly measure the probability of lateral gene transfer between opportunistic pathogen strains of the B. cepacia complex and other environmental Burkholderia taxa, such as B. unamae and B. tropica, would be necessary to have higher confidence about their safety in agricultural applications. These experiments are in progress.
In conclusion, while B. unamae is predominantly associated with sugarcane, B. tropica more often associates with maize. The isolation of B. unamae from field-grown sugarcane in Brazil and Mexico, as well as the isolation of B. tropica from maize cultivated in Mexico, and the finding of a probably-novel diazotrophic species (the Burkholderia NAR group) in rhizospheric and endophytic association with both maize and sugarcane in Brazil confirm the riches of the genus Burkholderia in nitrogen-fixing plant-associated species as well as its broad environmental and geographic distribution.
Acknowledgments
We acknowledge Mexican-Brazilian scientific and technological cooperation through CONACyT-Mexico and CNPq-Brazil (grant 490172/02-04). This research was partially funded by CONACyT grant 33576.
We thank G. Paredes-Valdez for technical assistance. We are grateful to Michael Dunn (CCG-UNAM) for reading the manuscript. We gratefully acknowledge Peter Vandamme (Universiteit Gent) for supplying type strains of B. hospita, B. terricola, B. fungorum, and B. caledonica. We are also grateful to I. López-Lara (CCG-UNAM) and John LiPuma (University of Michigan Medical School), who kindly provided B. cenocepacia strains J2315T and AU0007, respectively, and to Angela Sessitsch (ARC Seibersdorf Research GmbH) for supplying strain PsJNT of B. phytofirmans.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular microbiology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1999. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J. Clin. Microbiol. 37:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bevivino, A., C. Dalmastri, S. Tabacchioni, L. Chiarini, M. L. Belli, S. Piana, A. Materazzo, P. Vandamme, and G. Manno. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bevivino, A., S. Sarrocco, C. Dalmastri, S. Tabacchioni, C. Cantale, and L. Chiarini. 1998. Characterization of a free-living maize-rhizosphere population of Burkholderia cepacia: effect of seed treatment on disease suppression and growth promotion of maize. FEMS Microbiol. Ecol. 21:225-237. [Google Scholar]
- 7.Brämer, C. O., P. Vandamme, L. F. da Silva, J. G. C. Gomez, and A. Steinbüchel. 2001. Burkholderia sacchari sp. nov., a polyhydroxyalkanoate-accumulating bacterium isolated from soil of a sugar-cane plantation in Brazil. Int. J. Syst. Evol. Microbiol. 51:1709-1713. [DOI] [PubMed] [Google Scholar]
- 8.Burris, R. H. 1972. Nitrogen fixation assay—methods and techniques. Methods Enzymol. 24B:415-431. [DOI] [PubMed] [Google Scholar]
- 9.Caballero-Mellado, J., L. Martínez-Aguilar, G. Paredes-Valdez, and P. Estrada-de los Santos. 2004. Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 54:1165-1172. [DOI] [PubMed] [Google Scholar]
- 10.Chiarini, L., V. Giovannelli, A. Bevivino, C. Dalmastri, and S. Tabacchioni. 2000. Different portions of the maize root system host Burkholderia cepacia populations with different degrees of genetic polymorphism. Environ. Microbiol. 2:111-118. [DOI] [PubMed] [Google Scholar]
- 11.Clode, F. E., M. E. Kaufmann, H. Malnick, and T. L. Pitt. 2000. Distribution of genes encoding putative transmissibility factors among epidemic and nonepidemic strains of Burkholderia cepacia from cystic fibrosis patients in the United Kingdom. J. Clin. Microbiol. 38:1763-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenye, T., D. Henry, D. P. Speert, and P. Vandamme. 2004. Burkholderia phenoliruptrix sp. nov., to accommodate the 2,4,5-trichlorophenoxyacetic acid and halophenol-degrading strain AC1100. Syst. Appl. Microbiol. 27:623-627. [DOI] [PubMed] [Google Scholar]
- 13.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 14.Cruz, L. M., E. M. de Souza, O. B. Weber, J. I. Baldani, J. Döbereiner, and F. O. Pedrosa. 2001. 16S ribosomal DNA characterization of nitrogen-fixing bacteria isolated from banana (Musa spp.) and pineapple (Ananas comosus (L.) Merril). Appl. Environ. Microbiol. 67:2375-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curl, E. A., and B. Truelove. 1986. The rhizosphere. Springer-Verlag, Berlin, Germany.
- 16.Dalton, D. A., S. Kramer, N. Azios, S. Fusaro, E. Cahill, and C. Kennedy. 2004. Endophytic nitrogen fixation in dune grasses (Ammophilia arenaria and Elymus mollis) from Oregon. FEMS Microbiol. Ecol. 49:469-479. [DOI] [PubMed] [Google Scholar]
- 17.Di Cello, F., A. Bevivino, L. Chiarini, R. Fani, D. Paffetti, S. Tabacchioni, and C. Dalmastri. 1997. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63:4485-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estrada-de los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiore, A., S. Laevens, A. Bevivino, C. Dalmastri, S. Tabacchioni, P. Vandamme, and L. Chiarini. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 20.Gillis, M., V. Tran Van, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kersters, T. Heulin, and M. P. Fernandez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 21.Goris, J., P. De Vos, J. Caballero-Mellado, J.-H. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the PCB- and biphenyl-degrading strain LB400 and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 22.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 23.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Arizona State University, Tempe. [DOI] [PubMed]
- 25.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious multireplicon Burkholderia cepacia complex. Nat. Rev. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 27.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 28.Muñoz-Rojas, J., L. E. Fuentes-Ramírez, and J. Caballero-Mellado. 2005. Antagonism among Gluconacetobacter diazotrophicus strains in culture media and in endophytic association. FEMS Microb. Ecol. 54:57-66. [DOI] [PubMed] [Google Scholar]
- 29.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variations in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poly, F., L. Jocteur-Monrozier, and R. Bally. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152:95-103. [DOI] [PubMed] [Google Scholar]
- 31.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reik, R., T. Spilker, and J. J. LiPuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reis, V. M., P. Estrada-de los Santos, S. Tenorio-Salgado, J. Vogel, M. Stoffels, S. Guyon, P. Mavingui, V. L. D. Baldani, M. Schmid, J. I. Baldani, J. Balandreau, A. Hartmann, and J. Caballero-Mellado. 2004. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 54:2155-2162. [DOI] [PubMed] [Google Scholar]
- 34.Richardson, J., D. E. Stead, and R. H. A. Coutts. 2001. Incidence of the cblA major subunit pilin gene amongst Burkholderia species. FEMS Microbiol. Lett. 196:61-66. [DOI] [PubMed] [Google Scholar]
- 35.Roselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salles, J. F., F. A. De Souza, and J. D. van Elsas. 2002. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl. Environ. Microbiol. 68:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segonds, C., T. Heulin, N. Marty, and G. Chabanon. 1999. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 37:2201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 41.Tabacchioni, S., L. Chiarini, A. Bevivino, C. Cantale, and C. Dalmastri. 2000. Bias caused by using different isolation media for assessing the genetic diversity of a natural microbial population. Microb. Ecol. 40:169-176. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trân Van, V., O. Berge, S. Ngo Ke, J. Balandreau, and T. Heulin. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield component in low fertility sulphate acid soils of Vietnam. Plant Soil 218:273-284. [Google Scholar]
- 44.Valdés, M., N. O. Pérez, P. Estrada-de los Santos, J. Caballero-Mellado, J. J. Peña-Cabriales, P. Normand, and A. M. Hirsch. 2005. Non-Frankia actinomycetes isolated from surface-sterilized roots of Casuarina equisetifolia fix nitrogen. Appl. Environ. Microbiol. 71:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vandamme, P., J. Goris, W.-M. Chen, P. de Vos, and A. Willems. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507-512. [DOI] [PubMed] [Google Scholar]
- 46.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. W. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 47.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viallard, V., I. Poirier, B. Cournoyer, J. Haurat, S. Wiebkin, K. Ophel-Keller, and J. Balandreau. 1998. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of (Pseudomonas) phenazinium, (Pseudomonas) pyrrocinia and (Pseudomonas) glathei as Burkholderia. Int. J. Syst. Bacteriol. 48:549-563. [DOI] [PubMed] [Google Scholar]
- 49.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, H., S. Hanada, T. Shigematsu, K. Shibuya, Y. Kamagata, T. Kanagawa, and R. Kurane. 2000. Burkholderia kururiensis sp. nov. a trichloroethylene (TCE)-degrading bacterium isolated from aquifer polluted with TCE. Int. J. Syst. Evol. Microbiol. 50:743-749. [DOI] [PubMed] [Google Scholar]