It was almost 30 years ago when François Jacob declared that evolutionary innovation (the emergence of novel form and function over time) occurred primarily via a process of “tinkering” (1). By tinkering, Jacob essentially meant the creation of novelty through random combinations of preexisting forms. Two fundamental and countervailing notions are implicit in this view of evolution: optimality versus constraint. Were evolution to perform optimally, a more apt metaphor might be that of an engineer. An engineer works according to a plan, with a precise goal for the desired end, and uses material designed specifically toward that end. Evolution, on the other hand, must work without the benefit of foresight and is subject to very real constraints with respect to the material at its disposal; as such, evolutionary biology is replete with examples of suboptimal solutions to functional challenges (2). Similarly, a tinkerer works without a clear plan by using anything and everything at his disposal to produce an entity that possesses some kind of (unanticipated) functional utility. In this issue of PNAS, Cordaux et al. (3) explore an example of tinkering along the human evolutionary lineage, whereby an existing host gene merged with a transposable element (TE) to create a primate-specific chimeric gene.
In the decades since Jacob's exposition, molecular biology studies have produced a deluge of primary data (tens of thousands of three-dimensional protein structures and literally billions of nucleotides of gene sequences, including hundreds of complete genomes in the past few years alone). Comparative studies of the resulting data have underscored the extent to which genome evolution is indeed characterized by tinkering. There are a discrete and finite number of structural folds, protein sequence domains, and gene families (4); new genes evolve through slight modifications and/or recombinations of these preexisting forms. The actual de novo evolution of protein coding sequences is exceedingly rare. For instance, despite the ≈80–100 million years that have elapsed since the human and mouse lineages diverged, the genomes of these two species share >99% homologous genes (5). Clearly, however, mammalian evolution has been marked by substantial functional innovation, and so it must be that the genome-level dynamics underlying this innovation are dominated by creation through rearrangement.
One of the largely unanticipated results of mammalian genome sequencing efforts was the revelation of the extent to which these genomes are made up of sequences derived from TE insertions. The human genome sequence was found to consist of ≈45% TE-derived sequences (6), and this figure is certainly a vast underestimate because many TE-derived human sequences have diverged beyond recognition. In addition to being ubiquitous genomic elements, TEs are also autonomous in the sense that they carry the regulatory and protein coding sequences necessary to catalyze their transposition. The ubiquity of TEs, along with the functional machinery that they encode, makes them ideal genetic building blocks that evolution can tinker with to create novel forms. Indeed, despite the early notion of TEs as being strictly selfish (parasitic) elements that serve no function for their hosts (7), there now exist numerous examples of formerly mobile TE sequences that have been “domesticated” (8) to serve some functional role for the host genomes in which they reside (9, 10). However, there is still a relative paucity of detailed studies that address both the evolutionary dynamics of TE-derived host genes as well as the functional roles of the proteins that they encode. The work of Cordaux et al. (3) on the SETMAR gene represents an important step toward alleviating this knowledge gap.
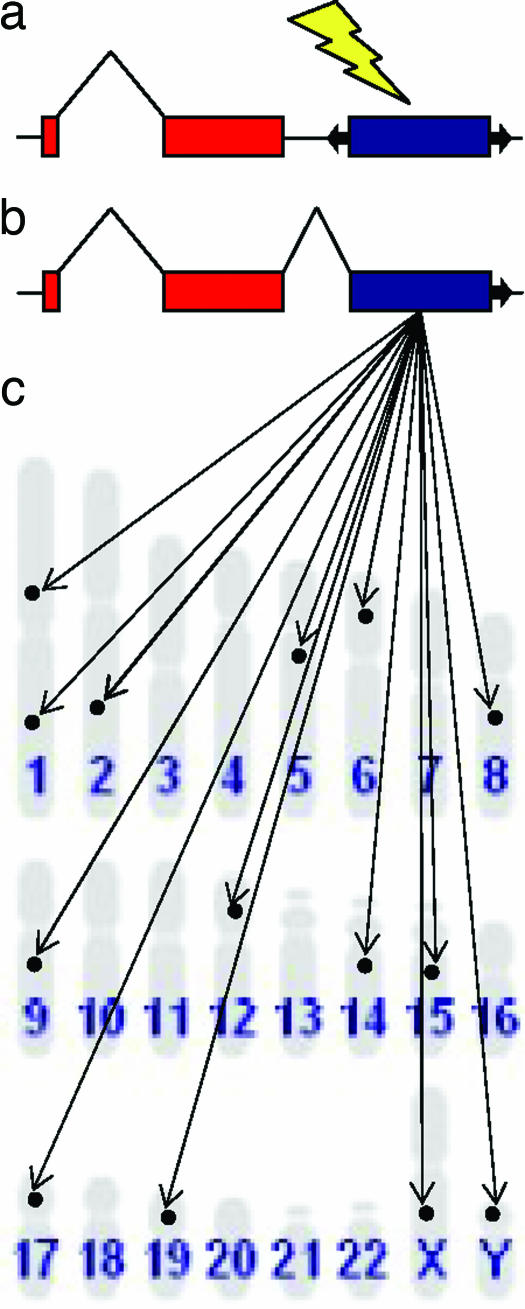
SETMAR, originally discovered by Robertson and Zumpano (11), is a chimeric gene made up of a SET histone methyltransferase transcript fused to the transposase domain of a formerly mobile TE sequence. The transposase domain in question comes from a member of the Hsmar1 mariner-like family of elements. Mariner-like elements are more commonly found in insects, and Hsmar1 was the first TE of this type found in the human genome. Hsmar1 elements are class II, or DNA elements, that have terminal inverted repeats (TIRs) flanking an ORF that encodes a transposase. Class II elements transpose via a cut-and-paste mechanism catalyzed by the transposase, which binds to the TIRs, excises the element, and then inserts it in a new location. Class I elements, or retrotransposons, which transpose via the reverse transcription of an RNA intermediate, are actually far more common than DNA elements in the human genome. The so-called long- and short-interspersed nuclear elements, LINEs and SINEs, respectively, make up ≈25% of the human genome. However, for as yet unknown reasons, DNA elements like Hsmar1 are overrepresented among host genes with TE-derived coding sequences. This overrepresentation may be because of the broad utility of the DNA-binding properties encoded by the transposase ORF. In fact, there is a distinct possibility that, as these kinds of chimeric genes are born, they are able to bind to multiple dispersed sites around the genome (those occupied by their cognate TIRs), resulting in the emergence of complex regulatory networks (Fig. 1). Britten and Davidson (12) articulated a very similar model for the evolution of cis-regulatory networks based on repetitive DNA. Recruitment of DNA-type element sequences into host genes may also represent a distinctly mutualistic evolutionary strategy that these relatively low-frequency elements employ on occasion to help ensure their long-term survival in the genome.
Fig. 1.
Establishment of a TE-derived genetic regulatory network. (a) DNA type, class II, TE (blue) inserts downstream of host gene exons (red). (b) TE binding domain fuses with host gene transcript. (c) The chimeric gene can now regulate multiple cognate binding site-containing locations around the genome.
Cordaux et al. (3) began their study by performing a series of sequence analyses aimed at elucidating the evolutionary dynamics and potential function of the SETMAR gene. First of all, they were able to identify SETMAR orthologs computationally among a fairly diverse set of vertebrate genomes ranging from mouse to zebrafish. All of these orthologs were shown to possess only the two SET exons, and none of them is flanked by an Hsmar1 element insertion. A more detailed analysis of orthologous regions cloned and sequenced from eight primate genomes was then used to precisely determine when SETMAR emerged along the evolutionary lineage leading to humans. Based on presence/absence patterns, they were able to determine that an Hsmar1 element inserted in the SET locus 40–58 million years ago. Interestingly, this time span is around the same time that an Alu (SINE) element inserted in the Hsmar1 5′ TIR, rendering the element immobile. After the Hsmar1 insertion, an exon capture event resulting in the fusion of the transposase encoding domain to the preexisting SET transcript was facilitated by a 27-bp deletion that removed the original SET stop codon and also activated a downstream cryptic 5′ donor splice site. This 5′ splice site presumably became activated together with a cryptic 3′ splice acceptor site in the Hsmar1 sequence, resulting in the formation of a novel intron/exon structure.
In addition to detailing how the SETMAR gene fusion occurred, Cordaux et al. (3) took the critical step of demonstrating that this chimeric gene is actually functional. SETMAR function was demonstrated by (i) showing that the gene is widely expressed and (ii) demonstrating that the SETMAR coding sequences are evolving under selective constraint. The latter conclusion is based on a pattern of elevated synonymous (KS) versus nonsynonymous (KA) substitution rates. KS ≫ KA is consistent with purifying (negative) selection because of functional constraint (13). The relative differences of KS vs. KA along the coding sequence were also taken to suggest that the DNA-binding capability of the N-terminal MAR domain is being conserved, whereas the catalytic activity located in the C-terminal domain has been lost. The substitution of the characteristic transposase catalytic sequence motif in the C-terminal MAR domain is also consistent with the absence of catalytic activity.
SETMAR coding sequences are evolving under selective constraint.
The sequence-based evidence described earlier, together with previously conducted experimental work demonstrating SETMAR methyltransferase activity (14), make up a compelling and fairly detailed story of the birth of the TE-derived chimeric gene. However, Cordaux et al. (3) did not stop there; they went on to biochemically characterize the MAR domain's ability to bind TIR-like DNA sequences, as well as its potential to encode an active transposase. The experimental assays conducted followed directly from the sequence analyses that suggested conservation of the binding domain and loss of the catalytic domain. Indeed, the experiments bear these predictions out because the MAR peptide was shown to be able to bind TIR sequences but could not catalyze transposition by using a standard in vivo assay. The tight integration of sequence analysis and experimental work is one of the distinguishing features of the article by Cordaux et al.; the sequence analyses yielded specific predictions that were then experimentally confirmed. Moreover, the binding experiments can be taken to suggest specific sequence analyses that could be used to characterize the distribution and evolutionary conservation of SETMAR binding sites in the human genome. One can easily imagine further experiments that could uncover the regulatory properties of the SETMAR gene. Such an approach could help to illuminate the most provocative aspect of this study: the suggestion of a specific mechanism for the rapid evolution of a genetic regulatory network composed of a domesticated transposase domain and its cognate binding sites dispersed throughout the genome (Fig. 1).
Conflict of interest statement: No conflicts declared.
See companion article on page 8101.
References
- 1.Jacob F. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 2.Darwin C. On the Origin of Species. London: John Murray; 1859. [Google Scholar]
- 3.Cordaux R., Udit S., Batzer M. A., Feschotte C. Proc. Natl. Acad. Sci. USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chothia C. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 5.Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 6.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle W. F., Sapienza C. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 8.Miller W. J., Hagemann S., Reiter E., Pinsker W. Proc. Natl. Acad. Sci. USA. 1992;89:4018–4022. doi: 10.1073/pnas.89.9.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kidwell M. G., Lisch D. R. Evolution Int. J. Org. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 10.Smit A. F. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 11.Robertson H. M., Zumpano K. L. Gene. 1997;205:203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- 12.Britten R. J., Davidson E. H. Q. Rev. Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 13.Sharp P. M. Nature. 1997;385:111–112. doi: 10.1038/385111a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee S. H., Oshige M., Durant S. T., Rasila K. K., Williamson E. A., Ramsey H., Kwan L., Nickoloff J. A., Hromas R. Proc. Natl. Acad. Sci. USA. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]