Abstract
This paper shows that proteins display an unexpectedly wide range of behaviors in buffers containing moderate (0.1–10 mM) concentrations of SDS (complete unfolding, formation of stable intermediate states, specific association with SDS, and various kinetic phenomena); capillary electrophoresis provides a convenient method of examining these behaviors. Examination of the dynamics of the response of proteins to SDS offers a way to differentiate and characterize proteins. Based on a survey of 18 different proteins, we demonstrate that proteins differ in the concentrations of SDS at which they denature, in the rates of unfolding in SDS, and in the profiles of the denaturation pathways. We also demonstrate that these differences can be exploited in the analysis of mixtures.
Keywords: capillary electrophoresis, surfactant, intermediates, kinetics
This manuscript surveys the range of electrophoretic behaviors observed for proteins in solutions containing SDS at concentrations below those used in SDS/PAGE. The aggressive conditions used to prepare proteins for characterization by SDS/PAGE (1) are designed to produce completely denatured, unfolded aggregates of protein and SDS; less forcing conditions have generally been ignored. Because SDS/PAGE uses forcing conditions, it has failed to reveal the wealth of information available from systems of protein and SDS: information about the kinetics of denaturation; about previously undetected, stable aggregates of protein and SDS with reasonably well defined stoichiometry; and about intermediates along the pathway to the fully denatured aggregates of protein and SDS.
Intermediates in the denaturation of some proteins with SDS have been identified: RNase A (2), cytochrome c that had been denatured in acid (3), BSA (4), and mushroom tyrosinase (5). Many proteins have a “low” state, in which the protein binds a few molecules of SDS, and “high” state, in which the protein binds one molecule of SDS per two amino acids (6, 7). These studies have concentrated mostly on single proteins or on the similarities between proteins and have not demonstrated or exploited the wide variability in behavior of proteins as they are denatured in SDS.
We find that proteins show large differences in the concentrations of SDS and in the rates at which they change conformation and unfold in SDS, in the concentrations of SDS at which intermediates form along the unfolding pathway, and in the number of these intermediates. [We use the term “rate” to refer to the kinetics of unfolding of the native protein. With this technique, we can only estimate the time scale for unfolding of a protein at a particular concentration of SDS qualitatively, i.e., estimate whether it is shorter, similar, or longer than the time of the capillary electrophoresis (CE) experiment.] These differences among proteins can be exploited to provide the basis for a method of differentiating (and in some instances separating) proteins and information about the relations between their structure and stabilities. We believe that this procedure will complement universally used techniques (e.g., SDS/PAGE and 2D gel electrophoresis) for analyzing complex mixtures of proteins (8, 9). This method also provides a way of identifying intermediates in the unfolding of proteins in solutions of SDS and in the refolding of proteins when removed from SDS. It does not require knowledge of the molecular details of the intermediates to identify proteins. The strength of the method presented here is that it requires less technical expertise and less expensive tools than many other proteomic tools (e.g., 2D gel electrophoresis or mass spectrometry) and, in some cases, can provide useful information about proteins (singly or in mixtures) in small quantities in ≈10 min.
The relationship between the structure and properties of proteins is complex. The development of new tools to explore this relationship is an important part of the rapidly expanding fields of proteomics and protein chemistry (10–12). We have begun to develop a method that is based on a detailed examination of the interactions (which are themselves incompletely understood) between proteins and surfactants, especially those that underlie SDS/PAGE. This method uses CE to observe the formation of aggregates of proteins with SDS, a process that frequently involves, we presume, the unfolding of the proteins.
SDS/PAGE is widely used to determine the molecular weights of proteins. In SDS/PAGE, the sample is usually boiled in 2% (wt/vol; ≈3.5 mM) SDS, with 0.2 M DTT as a reducing agent, so that proteins are completely denatured and all disulfide bonds are reduced (1). In SDS/PAGE, each protein binds, on the average, one SDS molecule per two amino acids (1.4 g of SDS per gram of protein) (13); in the resulting aggregates, the net charge on most proteins is proportional to their molecular weight. Analysis is accomplished by separation using polyacrylamide gel electrophoresis.
In capillary zone electrophoresis (CZE) (electrophoresis in open capillaries), separation of analytes is based on the ratio of charge to hydrodynamic drag (14). Because charge per unit length and, therefore, charge per drag are nearly constant for all saturated aggregates of proteins with SDS, there is little difference in the mobilities of SDS-denatured proteins in open capillaries. The similarity in mobility of the saturated protein–SDS aggregates makes it very difficult to separate them using CZE. The nature of the structures of these protein·SDS aggregates (aggregates that have approximately the same ratio of numbers of molecules SDS to numbers of amino acids, almost independent of the protein composition and sequence) remains an open question (13), with a general consensus that the protein is associated with micelles of SDS molecules (3, 15).
We hypothesized that the kinetics of denaturation of proteins with SDS and the intermediates formed during denaturation would carry information that was obliterated by the forcing conditions used in SDS/PAGE. We examined the interactions of proteins with SDS under conditions in which differences in sensitivity to formation of aggregates (of whatever structure) could be examined. We used CZE because it is sensitive to both charge and drag; we expect, upon association of proteins with SDS, that the former will dominate. We injected protein (1 mg/ml) as a solution in buffer not containing SDS into running buffer (Tris-Gly: 25 mM Tris/192 mM glycine, pH 8.4) for CE containing concentrations of SDS from 0 to 10 mM (Fig. 1). The critical micelle concentration of SDS in this buffer is 4.3 mM (16).
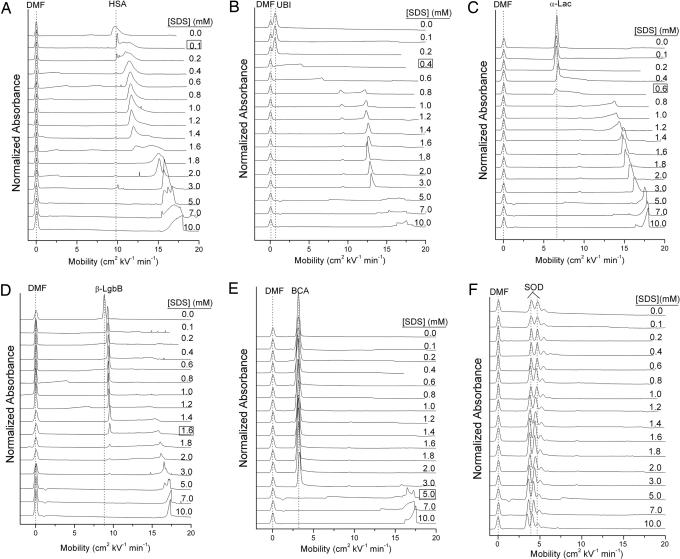
Fig. 1.
Electropherograms showing the denaturation of six representative proteins. (A) Albumin from human serum. (B) Ubiquitin. (C) α-Lactalbumin. (D) β-LgbB. (E) BCA. (F) SOD. The concentration of SDS in the buffer is labeled on each electropherogram, and the concentration of SDS at which the protein denatures on the capillary is boxed and listed in Table 1. These electropherograms were measured in Tris-Gly buffer (25 mM Tris/192 mM glycine, pH 8.4) at 15°C.
A set of electropherograms of the type summarized in Fig. 1 can be acquired efficiently: one electropherogram requires ≈15 min for setup and 10 min for collection of data at each concentration using a capillary 60 cm long. An entire set of 16 electropherograms for one protein requires ≈3 h. This process can be completely automated and, with the Beckman PACE MDQ system we used, the setup for running the entire set of electropherograms for six proteins requires ≈30 min of hands-on time and ≈16 h for data acquisition.
If the protein, either in native conformation or unfolded, forms aggregates with SDS molecules during its passage through the capillary, its mobility shifts relative to that in SDS-free buffer. If the rate of formation of aggregates and of unfolding of the protein is comparable to the time required to migrate from injector to detector (a few minutes), the peak also broadens. The presence of a mixture of aggregates with different mobilities also can result in broad peaks. The rapid (compared with the time scale of the experiment) formation of a new structure having defined composition yields a new, sharp peak in the electropherogram. If the kinetics of formation of protein–SDS aggregate are significantly slower than the time scale of the CE experiment, we observe only the native species.
We used absorption at 214 nm to detect the proteins on the capillary. Because SDS does not adsorb in the UV range, the only peaks we detect are due to the amide bonds and aromatic side chains of protein. We therefore see protein in every form: native and denatured and as aggregates with SDS (structured or unstructured, stable, metastable, or unstable, under the conditions in the CE).
Examination of the positions and shapes of peaks allows us to estimate the relative stabilities of the proteins to aggregation with SDS (i.e., unfolding), to detect intermediates along the unfolding pathway, and to make qualitative estimates of the rates of interconversion of the various protein-derived species on the time scale of the experiment.
Results and Discussion
We observe an unexpectedly wide range of behaviors for different proteins (Table 1 and Fig. 1; see also Fig. 3, which is published as supporting information on the PNAS web site). In almost all electropherograms, aggregation with SDS increases mobility and shifts peaks to the right; the denatured forms of most proteins in 10 mM SDS have a mobility (μ) of 15–18 cm2 kV−1·min−1. The very slight shift to lower mobilities of superoxide dismutase (SOD) at high concentrations of SDS is probably due to an increase in viscosity of the running buffer (17) and not interaction of the protein with SDS. The fact that all of the proteins [except SOD and streptavidin, which do not denature under our conditions (18)] have approximately the same mobility at high concentrations of SDS is consistent with the claim that all proteins bind SDS in approximately the same ratio of SDS molecules to the number of amino acids. Upon closer inspection of the electropherograms, however, it is apparent that the peaks corresponding to the saturated aggregates of protein and SDS have structures that, we presume, are due to multiple species. These species may have different numbers of molecules of SDS associated with them, may vary in conformation, and may undergo interconversion on a finite time scale; we are presently unable to pinpoint the specific reason for the structure in the peak. This structure is reproducible over multiple injections; it does not arise from random noise associated with the instrument. The finer structure on the peaks in the electropherograms is not observed in SDS/PAGE, either because the resolution in SDS/PAGE is lower or because of the conditions and long times required to prepare proteins for SDS/PAGE and to separate them by using SDS/PAGE.
Table 1.
Proteins surveyed in this work and concentrations of SDS at which they denature
| Protein | Enzyme accession no. | Molecular weight | No. of subunits | pl | [SDS]Δμ |
|---|---|---|---|---|---|
| Albumin, human | P02768 | 66.5 | 1 | 5.7 | 0.1 |
| Hemoglobin A | P69905 and P68871 | 64.5 | 4 (two α- and two β-chains) | 7.2 | 0.2 |
| Alcohol dehydrogenase | P00330 | 146.8 | 4 | 5.6 | 0.2 |
| Pyruvate kinase | P11974 | 231.6 | 4 | 7.6 | 0.2 |
| Ubiquitin | P62990 | 8.6 | 1 | 6.6 | 0.4 |
| Myoglobin | P68082 | 17.0 | 1 | 7.4 | 0.4 |
| α-Lactalbumin | P00711 | 14.2 | 1 | 4.8 | 0.6 |
| β-LgbA* | P02754 | 18.4 | 1 | 4.8 | 1.0 |
| β-LgbB* | P02754 | 18.3 | 1 | 4.8 | 1.6 |
| Ovalbumin | P01012 | 42.8 | 1 | 5.2 | 2.0 |
| Creatine phosphokinase | P00563 | 86.2 | 2 | 6.7 | 3.0 |
| BCA | P00921 | 30.0 | 1 | 5.9 | 5.0 |
| Carboxypeptidase B | P09955 | 34.7 | 1 | 5.7 | 5.0 |
| 4 (two heavy and two light chains, all S-S-linked) | |||||
| IgG | Mixture of all | ≈150.0 | 5.0 | ||
| Bovine antibodies | |||||
| Cytochrome c | 12.4 | 1 | 9.6 | ? | |
| Lysozyme | P00698 | 14.0 | 1 | 10.7 | ? |
| SOD | P00442 | 31.1 | 2 | 5.9 | >10 |
| Streptavidin | P22629 | 65.6 | 4 | 6.9 | >10 |
The molecular weights listed are for the entire protein (not per subunit). All of the data for the molecular weight, number of subunits, and pl come from the Swiss-Prot database. The number of subunits is the number of polypeptide chains comprising the protein, or, equivalently, the number of polypeptide chains that appear in a denaturing and reducing SDS/PAGE experiment. [SDS]Δμ refers to the concentration of SDS at which the peak in the electropherogram corresponding to the native protein disappears in our experiments (60-cm capillary, 30 kV). This value depends on the time the protein spends in the capillary and may change with a capillary of a different length or applied voltage. The critical micelle concentration of SDS in the Tris-Gly buffer used in these experiments (pH 8.3) is 4.3 mM (17). The question marks refer to proteins that, because of their positive charge, cannot be observed until they bind enough molecules of SDS to be negatively charged. We cannot, therefore, determine a concentration of SDS at which the mobility of these proteins begins to change.
The concentrations of SDS at which the proteins began to change mobility varied by more than two orders of magnitude (0.1 mM SDS for human serum albumin to >10 mM SDS for SOD and streptavidin). Some proteins seem to denature on the capillary well below the critical micelle concentration (e.g., albumin and ubiquitin), some above the critical micelle concentration [e.g., carbonic anhydrase (BCA) and IgGs], and some do not denature in any concentration of SDS we examined (e.g., streptavidin and SOD). We have not established the nature of the changes that result in the shifts of the mobilities of the protein peaks. We hypothesize, for the discussion here, that small changes in the mobility that leave the peak shapes similar to those of the native protein [e.g., β-lactoglobulin B (β-LgbB)] probably reflect specific, nondenaturing complex formation (and, presumably, a small but defined number of specifically bound SDS molecules), and that large changes in mobility (e.g., BCA) reflect unfolding (and, presumably, a large number – hundreds – of associated SDS molecules). We cannot characterize species with shifts of intermediate magnitude (e.g., human serum albumin at [SDS] = 0.4–1.2 mM) without further information, although we speculate that these are partially unfolded aggregates of proteins with a subsaturating number of associated SDS molecules. These species (for example, the two aggregates formed by association of ubiquitin and SDS at concentrations between 0.8 and 3.0 mM SDS) seem to be previously undescribed.
The electropherograms collected with this procedure do not, at the level of analysis we describe here, provide quantitative information about the kinetics and thermodynamics of unfolding, the lifetime of the intermediates, or the number of SDS molecules bound at any state. These electropherograms may provide an opportunity to begin to put bounds on the rates of denaturation and to constrain models of interaction of proteins with SDS.
Specific proteins in Figs 1 and 3 illustrate the range of behaviors we observe; the variety of these behaviors and the speed at which the data are gathered using CE are reasons for believing that these information-rich experiments will provide a useful method for differentiating proteins.
SOD (Fig. 1F).
This protein does not denature at any concentration of SDS surveyed in this work. Manning and Colón (18) hypothesized that the kinetic stability of this proteins was due to extensive β-sheet structure.
BCA (Fig. 1E).
The large change in μ that we associate with complete unfolding occurs at [SDS] = 5 mM at 15°C. At this concentration of SDS, the rate of unfolding of the protein becomes comparable with the time required for the protein to move through the capillary at this concentration of SDS.
β-LgbB (Fig. 1D).
This protein appears to associate with one equivalent of SDS at 0.1 mM (as judged by the shift in mobility). Because β-LgbA (Fig. 3D) has one more negatively charged residue than does β-LgbB (Asp-64 in β-LgbA is Gly-64 in β-LgbB) (19, 20), we can compare the shift in mobility upon adding SDS to β-LgbB (ΔμSDS = 0.52 cm2·kV−1·min−1) to the shift in mobility upon removal of one negatively charged amino acid (μβ-LgbA − μβ-LgbB = 0.61 cm2·kV−1·s−1) (Figs. 1D and 3D). The agreement between their values, ΔμSDS ≈ 0.9·(μβ-LgbA − μβ-LgbB), is sufficient to associate the intermediate observed in Fig. 1D with approximately one unit of charge and, hence, one molecule of SDS. At 1.6 mM SDS, β-LgbB begins to denature.
Lac (Fig. 1C).
The substantial broadening of the peak due to α-lactalbumin at [SDS] = 0.6 mM corresponds to the conversion of native protein to an aggregate involving a substantial number of SDS molecules. From 0.8 to 5 mM SDS, there is a gradual increase in mobility that corresponds, we believe, to a further increase in the number of associated molecules of SDS.
Ubiquitin (Fig. 1B).
The electropherograms of ubiquitin show broadening at [SDS] = 0.4 mM, with formation of two stable intermediates (we judge their stability from the sharpness of peaks), whose relative concentrations varies with SDS in the range 0.8–3.0. At [SDS] > 3.0, these intermediates convert into a mixture of aggregates whose mobilities suggest they are completely denatured.
The residence time of the protein in the capillary can be varied either by increasing the length of the capillary or by decreasing the applied voltage. We tested two cases, ubiquitin at 0.6 mM SDS and BCA at 3 mM SDS, at 25°C, to see whether the observed electropherograms are sensitive to the applied voltage and, thus, residence time on the capillary (Fig. 4, which is published as supporting information on the PNAS web site). The shape of the peak of ubiquitin remained invariant with decreasing voltage; this observation suggests that the formation of ubiquitin–SDS aggregate at 0.6 mM SDS is rapid relative to the time scale of the CE separation. In the case of BCA at 3 mM SDS, the area of native BCA peak decreased as the residence time of the protein in the capillary increased; we infer that denaturation of BCA at 3 mM SDS occurs on a time scale similar to that of the CE separation (21).
We also analyzed the intermediates in the renaturation of the proteins upon removal of SDS from the solution (Fig. 5, which is published as supporting information on the PNAS web site) (22). Most of the proteins showed a gradual shift toward the native mobility with decreasing concentrations of SDS in the running buffer. In the time scale of these experiments, none of the proteins return to the mobility of the folded protein when SDS was removed from the buffer. (SOD and streptavidin are exceptions; they simply do not denature in our experiments.) Refolding is clearly slower than unfolding under these conditions. We have studied BCA independently (21, 23) and know that it does refold completely with a time constant of ≈10 min when the SDS is removed; we cannot predict the long-term behavior of the other proteins.
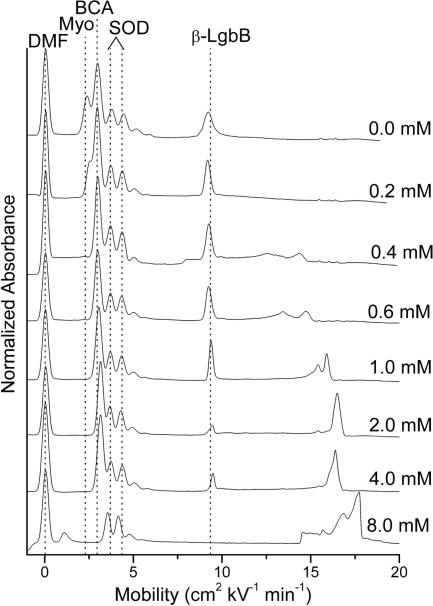
The wide range of behaviors we have observed suggests strategies for differentiating proteins in mixtures based on uses of SDS. For example, one plausible immediate application of this technique would be to the simplification of the electropherograms of relatively simple mixtures (perhaps obtained as intermediate mixtures in purifications); Fig. 2 shows an example. In the electrophoresis buffer with no SDS, the peaks corresponding to native myoglobin, BCA, and SOD overlap. Because the proteins here have very different stabilities to SDS, running the sample with enough SDS to denature one or more type of protein allows better quantitation of all of the proteins in the sample.
Fig. 2.
A mixture of myoglobin, BCA, SOD, and α-LgbB in increasing concentrations of SDS in the separation buffer in the CE. The order of denaturation in increasing concentrations of SDS is myoglobin, β-LgbB, BCA, and SOD, which does not denature at the concentrations of SDS run in this experiment.
We also examined the application of this technique to unfractionated serum (Fig. 6, which is published as supporting information on the PNAS web site). Serum is a complicated mixture, and the results are complicated. There are observable changes to the electropherograms as the concentration of SDS changes, and the peaks due to the most abundant proteins (including albumin and γ-globulins) shift and reveal many new, previously obscured peaks. The changes, however, are too complex to interpret without more knowledge of the contributing proteins.
Proteins in solutions of SDS therefore show a rich variety of characteristic behaviors unanticipated from experience with SDS/PAGE. CZE is a convenient, high-resolution analytical tool that can provide the number of intermediates in interaction of SDS with proteins, some information about the composition of these intermediates, and semiquantitative information about the kinetics of their interconversion, all in a single class of easily performed experiments. Although we have focused on SDS, we believe that other surfactants, especially other charged surfactants, will provide additional and complementary information.
To what use in proteomics and protein chemistry can this wealth of information be put? We can speculate. It may be possible to (i) separate one protein from another based on differential formation of aggregates or unfolding/denaturation; (ii) shift overlapping peaks in CZE to improve analytical resolution; (iii) use information derived from shifts in mobility as a function of SDS (after calibration) to infer information about the stability, and perhaps structure, of proteins and protein·SDS aggregates. Although most of the details of this technique remain to be developed, the combination of CE and charged surfactants (perhaps combined with uncharged ones) has the potential to provide a variety of information about proteins and to do so using procedures that are widely accessible to protein biochemists. In protein chemistry, the complexity of the problems is such that new tools for analysis and separation are always useful.
Materials and Methods
Sources of Chemicals and Reagents.
All chemicals were reagent-grade unless stated otherwise. All proteins, human serum (from human male AB plasma, sterile-filtered) and 10× Tris-Gly concentrate were all purchased from Sigma–Aldrich and used without further purification. SDS was purchased from J. T. Baker. SDS was recrystallized in hot ethanol three times (24), then dried and stored at −20°C until use. No impurities (in particular, no signal from dodecanol) were observed by NMR. SDS was discarded or repurified after 2 months. Tris-Gly buffer was made by diluting 100 ml of the 10× concentrate with 900 ml of freshly distilled, deionized water. It was filtered with a 0.22-μm filter (Pall) before use.
CE.
CE experiments were carried out in a Beckman PACE-MDQ system, with a capillary having an inner diameter of 50 μm and a total length of 60.2 cm (50 cm to the detector) and with Tris-Gly as the running buffer, using an applied voltage of 30 kV, unless stated otherwise. Each sample contained 0.65 mM dimethylformamide as a neutral marker to monitor the electroosmotic flow (25, 26). The solutions of proteins (≈1 mg/ml) were injected on the capillary at 0.5 psi for 20 s. This procedure generates an injected plug of ≈15 nl. The injection volume and protein concentration were chosen to provide good signal-to-noise ratio for all species.
Analysis of Electropherograms Showing Denaturation.
Fig. 3 shows electropherograms of 12 additional proteins in buffer containing SDS. The experiments with lysozyme shows the behavior of a protein that has a net positive charge in its native form in our buffer system (Tris-Gly) and does not appear when using an uncoated silica capillary. Lysozyme is not observed, therefore, at low concentrations of SDS. Above 5 mM SDS, lysozyme complexes with enough SDS that it becomes negatively charged and can be observed. Similar behavior is observed with cytochrome c.
Analysis of Renaturation Electropherograms.
We measured the intermediates in renaturation of the proteins by incubating them in 10 mM SDS for 24 h at room temperature and then injecting the resulting solutions onto the CE using a separation buffer containing concentrations of SDS from 0 to 10 mM (Fig. 5). SOD did not denature in 10 mM SDS, even after 24 h. The other proteins analyzed here (albumin from human serum, ubiquitin, α-lactalbumin, β-LgbB, and BCA) showed a gradual shift in mobility toward lower values, with decreasing concentrations of SDS in the running buffer. The results with lysozyme are uninterpretable because, at [SDS] < 2 mM in the running buffer, no protein peaks are observed. None of the proteins return to the mobility of the native protein in buffer containing no SDS (Fig. 1), except SOD, which does not denature in our experiments.
Analysis of Human Serum.
We used the technique described in the text to analyze a complex mixture of proteins: human serum. Fig. 6 shows the electropherograms of human serum with increasing concentrations of SDS in the separation buffer.
As the SDS concentration increases, many proteins (e.g., albumin and IgGs) bind SDS and move to higher mobilities; small, structured peaks can be observed at intermediate SDS concentrations. It is likely that these peaks represent peptides or perhaps nonpeptidic metabolites, because they do not interact as strongly with SDS but without further experimental evidence (e.g., CE mass spectrometry or comparison with pure samples of known materials) conclusive identification of these peaks is impossible.
Supplementary Material
Acknowledgments
We thank Andrew Lee, Gregory Schneider, and Demetri Moustakas for helpful discussions. This work was supported by National Institutes of Health Grant GM51559.
Abbreviations
- CE
capillary electrophoresis
- CZE
capillary zone electrophoresis
- β-Lgb
β-lactoglobulin
- SOD
superoxide dismutase
- BCA
bovine carbonic anhydrase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Gallagher S. R. In: Current Protocols in Molecular Biology. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K, editors. New York: Wiley; 2003. [Google Scholar]
- 2.Moosavi-Movahedi A. A., Gharanfoli M., Nazari K., Shamsipur M., Chamani J., Hemmateenejad B., Alavi M., Shokrollahi A., Habibi-Rezaei M., Sorenson C., Sheibani N. Colloids Surf. B. 2005;43:150–157. doi: 10.1016/j.colsurfb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Xu Q., Keiderling T. A. Protein Sci. 2004;13:2949–2959. doi: 10.1110/ps.04827604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda K., Moriyama Y. Curr. Top. Colloid Interface Sci. 1997;1:109–135. [Google Scholar]
- 5.Park Y.-D., Jung J.-Y., Kim D.-W., Kim W.-S., Hahn M.-J., Yang J.-M. J. Protein Chem. 2003;22:463–471. doi: 10.1023/b:jopc.0000005462.05642.89. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds J. A., Tanford C. J. Biol. Chem. 1970;245:5161–5165. [PubMed] [Google Scholar]
- 7.Takagi T., Tsujii K., Shirahama K. J. Biochem. (Tokyo) 1975;77:939–947. doi: 10.1093/oxfordjournals.jbchem.a130818. [DOI] [PubMed] [Google Scholar]
- 8.Loo J. A. Adv. Protein Chem. 2003;65:25–56. doi: 10.1016/s0065-3233(03)01015-5. [DOI] [PubMed] [Google Scholar]
- 9.Herbert B. R., Sanchez J.-C., Bini L. In: Proteome Research: New Frontiers in Functional Genomics. Wilkins M. R., Williams K. L., Appel R. D., Hochstrasser D. F., editors. Berlin: Springer; 1997. pp. 13–34. [Google Scholar]
- 10.Xu Q., Lam K. S. J. Biomed. Biotechnol. 2003;5:257–266. doi: 10.1155/S1110724303209220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K. H., Reardon K. F. AIChE J. 2003;49:2682–2686. [Google Scholar]
- 12.Cramer R., Gobom J., Nordhoff E. Exp. Rev. Proteomics. 2005;2:407–420. doi: 10.1586/14789450.2.3.407. [DOI] [PubMed] [Google Scholar]
- 13.Jones M. N. Chem. Soc. Rev. 1992;21:127–136. [Google Scholar]
- 14.Manabe T. Electrophoresis. 1999;20:3116–3121. doi: 10.1002/(SICI)1522-2683(19991001)20:15/16<3116::AID-ELPS3116>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Turro N. J., Lei X.-G., Ananthapadmanabhan K. P., Aronson M. Langmuir. 1995;11:2525–2533. [Google Scholar]
- 16.Gudiksen K. L., Gitlin I., Yang J., Urbach A. R., Moustakas D. T., Whitesides G. M. J. Am. Chem. Soc. 2005;127:4707–4714. doi: 10.1021/ja043804d. [DOI] [PubMed] [Google Scholar]
- 17.Shen A. Q., Gleason B., McKinley G. H., Stone H. A. Phys. Fluids. 2002;14:4055–4068. [Google Scholar]
- 18.Manning M., Colon W. Biochemistry. 2004;43:11248–11254. doi: 10.1021/bi0491898. [DOI] [PubMed] [Google Scholar]
- 19.Braunitzer G., Chen R., Schrank B., Stangl A. Hoppe-Seyler’s Z. Physiol. Chem. 1973;354:867–878. [PubMed] [Google Scholar]
- 20.Wong D. W. S., Camirand W. M., Pavlath A. E. Crit. Rev. Food Sci. Nutr. 1996;36:807–844. doi: 10.1080/10408399609527751. [DOI] [PubMed] [Google Scholar]
- 21.Gitlin I., Gudiksen K. L., Whitesides G. M. J. Phys. Chem. B. 2006;110:2372–2377. doi: 10.1021/jp055699f. [DOI] [PubMed] [Google Scholar]
- 22.Stutz H., Wallner M., Malissa H., Jr., Bordin G., Rodriguez A. R. Electrophoresis. 2005;26:1089–1105. doi: 10.1002/elps.200406195. [DOI] [PubMed] [Google Scholar]
- 23.Gudiksen K. L., Urbach A. R., Gitlin I., Yang J., Vazquez J. A., Costello C. E., Whitesides G. M. Anal. Chem. 2004;76:7151–7161. doi: 10.1021/ac0488560. [DOI] [PubMed] [Google Scholar]
- 24.Merta J., Torkkeli M., Ikonen T., Serimaa R., Stenius P. Macromolecules. 2001;34:2937–2946. [Google Scholar]
- 25.Carbeck J. D., Colton I. J., Gao J., Whitesides G. M. Acc. Chem. Res. 1998;31:343–350. [Google Scholar]
- 26.Mammen M., Colton I. J., Carbeck J., Bradley R., Whitesides G. M. Anal. Chem. 1997;69:2165–2170. doi: 10.1021/ac961123y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.