Abstract
Copy number variation is surprisingly common among humans and can be involved in phenotypic diversity and variable susceptibility to complex diseases, but little is known of the extent of copy number variation in nonhuman primates. We have used two array-based comparative genomic hybridization platforms to identify a total of 355 copy number variants (CNVs) in the genomes of 20 wild-born chimpanzees (Pan troglodytes) and have compared the identified chimpanzee CNVs to known human CNVs from previous studies. Many CNVs were observed in the corresponding regions in both chimpanzees and humans; especially those CNVs of higher frequency. Strikingly, these loci are enriched 20-fold for ancestral segmental duplications, which may facilitate CNV formation through nonallelic homologous recombination mechanisms. Therefore, some of these regions may be unstable “hotspots” for the genesis of copy number variation, with recurrent duplications and deletions occurring across and within species.
Keywords: chimpanzee genome, human evolution, structural genomic variation
Recent studies have unexpectedly shown that structural genomic variation (including copy number variation) is common among normal, healthy human individuals (1–10). Copy number variants (CNVs) are duplications or deletions of several kb or more of segments of nuclear DNA (11). For several gene-containing human CNVs, genomic copy number variability has been shown to correlate with corresponding changes in gene expression levels (7, 12–14). Hence, it is thought that CNVs may be involved in phenotypic variation, including inherent differences in disease susceptibility. For example, Gonzalez and colleagues (15) found that a lower-than-population-average genomic copy number of the CCL3L1 gene correlated with lower protein levels of this ligand for the HIV-1 coreceptor CCR5 and conferred increased susceptibility to HIV-1 infection. More recently, Aitman et al. (16) demonstrated that a reduced genomic copy number of the FCGR3B gene (an activatory Fc receptor for IgG) serves as an increased risk factor in humans for immunologically related glomerulonephritis.
Although there is growing interest in comprehensively identifying human CNVs and investigating their phenotypic and evolutionary significance, very little is known of the location and frequencies of CNVs in other primate species. Comprehensive discovery and characterization of CNVs in the chimpanzee (Pan troglodytes) genome may provide comparative information that would help us to understand the mechanisms leading to the genesis and evolution of these intriguing genomic regions. In an initial analysis of the chimpanzee genome, Newman and colleagues (17) reported the presence of up to 266 deletions, each ≥12 kb in size, within the diploid genome of a single chimpanzee when compared to the human genome. This chimpanzee individual was used for the chimpanzee genome project. Other studies have investigated copy number variation at a limited number of individual loci among the genomes of several chimpanzees to gain insight into the dynamics of corresponding human CNVs (15, 18). However, a genome-wide analysis of CNVs in a chimpanzee sample population is still lacking. Here, we have used genome-wide, array-based comparative genomic hybridization (aCGH) to identify and catalogue CNVs in 20 wild-born chimpanzees and compare these data to currently known human CNVs.
Results
Extensive Copy Number Variation in the Chimpanzee Genome.
In this study, we compared the genomic DNAs of 20 wild-born male western chimpanzees (P. troglodytes verus) with the genomic DNA of the captive-born male donor for the chimpanzee genome sequence (19). We reasoned that this population is ideal for the comparison of CNVs to humans, because levels of nuclear DNA diversity (based on single-nucleotide differences) are thought to be similar among the western chimpanzee subspecies, as within the human species as a whole (19–23). The slides used for aCGH experiments comprised 2,632 large fragments of human DNA (bacterial artificial chromosome or BAC clones), spaced approximately every 1 Mb across the human genome (Spectral Genomics, Houston). This was the same array platform that we used for an earlier study identifying the widespread presence of CNVs in the human genome (1), facilitating a direct comparison of the results between the two studies. Because human–chimpanzee nucleotide sequence divergence for unique regions of the genome is estimated to be <2% (19, 24), the hybridization efficiency of chimpanzee DNA to human BAC clones is sufficient for aCGH experiments (25, 26).
Using this 1-Mb resolution aCGH platform, we initially identified 331 BACs that exhibited gains or losses in the chimpanzee individuals studied, relative to the reference sample (Fig. 1). One hundred fourteen CNVs (34.4%) were observed in two or more individual chimpanzees. It is unclear whether the remaining 217 single-incidence CNVs are low-frequency variants or false positives, although the false-positive error rate is expected to be ≈1 CNV per aCGH experiment (i.e., 2,632 clones) based on self-self-hybridization experiments (ref. 1 and Fig. 4, which is published as supporting information on the PNAS web site). Most CNVs detected in this manner are likely to be <2 Mb in size because they involved isolated individual BAC clones, and, on this array platform, there is ≈1 Mb of DNA to each flanking clone. However, six pairs of CNVs were identified that did not fit such a pattern, with these CNV pairs comprising two consecutive clones on the array (e.g., AC104041.5 and RP11-152F13; human chromosome 15q25.2). These pairs of CNVs may represent large contiguous CNVs that are ≈1–3 Mb in size or separate and smaller CNVs with intervening spacing of up to ≈1 Mb.
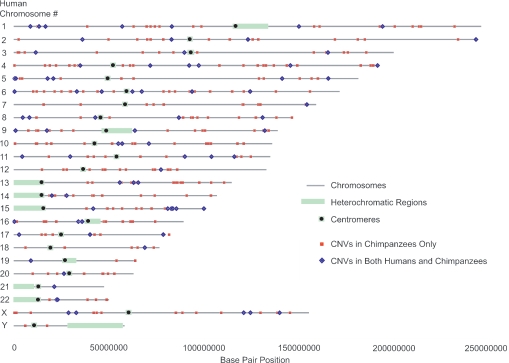
Fig. 1.
Map of CNVs shared between humans and chimpanzees and found in chimpanzees only. The genomic positions of chimpanzee CNVs that do not overlap with any currently known human CNVs are depicted as red squares, and CNVs observed at the same loci in both chimpanzees and humans are depicted as blue diamonds.
Humans and Chimpanzees Share CNV Regions.
The aCGH platform used in this study is the same as that used in a previous study for the discovery of human CNVs (1), allowing us to compare directly levels and patterns of copy number variation between the two species by using a given platform. In the human study, a total of 255 CNVs were identified among 55 individuals (102 CNVs in ≥2 individuals), 76 fewer than in our sample of 20 chimpanzees (1). We found 58 BAC clones that exhibited copy number variation in both chimpanzees and humans, significantly more than expected by chance alone (considering the total number of clones on the array; P < 0.01). The most commonly identified CNVs in one species were also often commonly observed in the other species. For example, there were 11 CNVs observed in >25% of the human individuals. Ten of these CNVs were also observed in chimpanzees, and, for eight of these regions, genomic imbalances were observed in three or more chimpanzees (≥15%). Table 1 lists common CNVs that have been observed in multiple humans and chimpanzees. Comparisons of the chimpanzee CNV loci to human CNVs discovered by using other platforms or methodologies (2–4, 7, 8) resulted in the identification of an additional 16 CNV loci that are shared between the two species (Table 2, which is published as supporting information on the PNAS web site). Altogether, 74 of the 331 identified chimpanzee CNV regions were shared between humans and chimpanzees.
Table 1.
Potential hotspots of copy number variation in humans and chimpanzees
| BAC Name | Location* |
Chimpanzee CNVs incidence† | Human CNVs incidence‡ | RefSeq genes (function)§ | |
|---|---|---|---|---|---|
| Chr. location in humans | Position (Mb) | ||||
| RP1-163M9¶ | 1p36.13 | 16.4 | 2 | 23 | |
| RP6-65F20 | 1p32.2 | 56.8 | 10 | 16 | DAB1 (cell differentiation; nervous system development) |
| RP11-91G12 | 1q31.3 | 193.9 | 2 | 3 | CFH (immune response), CFHL3 (unknown) |
| RP5-963K6¶ | 4q35.2 | 191.3 | 2 | 7 | |
| RP11-88L18¶ | 5p15.1 | 17.5 | 17 | 10 | |
| AL035696.14¶ | 6p25.3 | 0.1 | 3 | 13 | |
| RP11-96G1¶ | 8q21.2 | 86.6 | 3 | 18 | REXO1L1 (exonuclease and hydrolase activity) |
| RP11-130C19 | 9p24.3 | 0.6 | 2 | 3 | ANKRD15 (cell cycle and growth) |
| RP11-100C24 | 13q21.1 | 55.5 | 5 | 29 | FLJ40296 (unknown) |
| RP11-499D5¶ | 16p11.2 | 33.7 | 4 | 11 | TP53TG3 (unknown) |
| C197.4 | 17p13.3 | 2.5 | 2 | 3 | RUTBC1 (unknown) |
| MNT (transcription factor; development) | |||||
| RP11-79F15¶ | 19p13.2 | 8.7 | 3 | 34 | ZNF558 (transcription factor), MBD3L1 (transcription factor) |
| RP11-49J9 | 21q21.1 | 21.0 | 2 | 2 | |
| RP6-27C10 | Xp21.3 | 28.5 | 12 | 16 | IL1RAPL1 (learning and/or memory; signal transduction) |
| AL031643.1 | Xp21.1 | 32.6 | 10 | 21 | DMD (cytoskeleton; muscle activity) |
| RP6-64P14 | Xq25 | 120.7 | 9 | 16 | |
| RP6-232G24 | Xq27.2 | 139.7 | 13 | 18 | MAGEC3 (unknown), MAGEC1 (unknown) |
*Cytogenetic location and physical position (in Mb) of BAC clones, based on the human reference genome sequence (Build 34).
†Number of chimpanzees (of 20) for whom gains/losses (relative to the reference chimpanzee individual, Clint) were detected in this study using the 1-Mb aCGH platform.
‡Total number of human individuals for whom gains/losses were detected for the regions overlapping/encompassing a given BAC clone. Human CNV data were collected from different studies (1–4, 7, 8).
§RefSeq genes partially or completely contained within the BAC sequence and gene function based on GO categories.
¶These seven clones overlap with ancestral segmental duplications (as classified by ref. 33).
Previous studies have shown that human CNV loci are enriched for genes involved in immunity and environmental responses (11, 27). We performed an analysis based on Gene Ontology (GO) categories (28) for genes that mapped to chimpanzee CNV loci and found a similar pattern for immunity and environmental response-related genes (P < 0.001) (Table 3, which is published as supporting information on the PNAS web site). Several other GO categories were also overrepresented. We then performed a similar analysis for the CNVs that are shared between humans and chimpanzees and found that these loci are enriched for genes in the GO categories of organismal physiological processes (P < 0.01), defense response (P < 0.01), receptor activity (P < 0.001), non-membrane-bound organelles (e.g., cytoskeletal proteins with integral roles in cell structure and stability; P < 0.0001), structural molecule activity (P < 0.0001), and unknown biological processes (P < 0.01). It is possible that CNVs containing genes in these functional categories are favored and maintained by natural selection in both species. Alternatively, the observed enrichment may reflect a relative relaxation of selective pressure on copy number for these genes (i.e., stronger purifying selection against copy number variation involving genes in nonenriched categories).
The first level of validation involved the identification of 114 of these CNV loci in more than one chimpanzee individual. Moreover, we used real-time quantitative PCR (qPCR) of genomic DNA to confirm the genomic imbalances at four different genomic regions in both humans and chimpanzees and in a further six regions in chimpanzees only. In each case, the qPCR results were consistent with results from the aCGH experiments (Fig. 2; and see Fig. 5, which is published as supporting information on the PNAS web site). Finally, we performed aCGH experiments on the platform used previously by Sharp and colleagues (4) to identify 160 CNVs among 47 humans. This particular array contains human BAC clones selected to specifically cover known low-copy repeat sequences (segmental duplications) in the human genome (29). By using genomic DNAs from 5 of the same 20 chimpanzees studied, 28 CNV regions were identified (see Table 4, which is published as supporting information on the PNAS web site). Four of these CNV regions overlapped with CNV regions identified by using the 1-Mb resolution arrays (Fig. 6, which is published as supporting information on the PNAS web site), and 11 of the 28 CNV regions corresponded to human CNVs previously identified by using this same segmental duplication-enriched array platform (4), significantly more than expected by chance alone (P < 0.05).
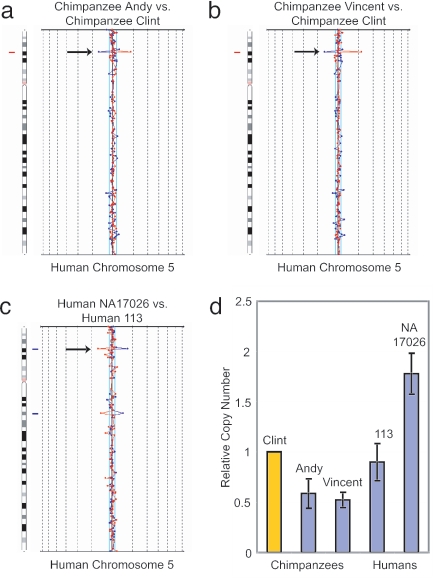
Fig. 2.
Copy number variation at the RP11-88L18 locus. The RP11-88L18 BAC clone (human chromosome 5p15.1) is an example of a CNV that is shared by humans and chimpanzees. Arrows on the aCGH chromosome-specific plots depict (a) a genomic loss for chimpanzee Andy, relative to the reference chimpanzee Clint; (b) a genomic loss for chimpanzee Vincent, relative to Clint; and (c) a genomic gain for human NA17026, relative to human individual 113. (d) These observations were confirmed by qPCR. Genomic DNA from the reference chimpanzee Clint (relative quantity = 1) was used for the standard curve. Error bars depict ±2 SD (95% confidence interval).
Shared CNV Regions Are Enriched for Ancestral Segmental Duplications.
Segmental duplications may facilitate the formation of some CNVs through the occurrence of nonallelic homologous recombination mechanisms (30–32). Studies in humans have shown that copy-number-variable loci are enriched for segmental duplications (1–4), and we found that chimpanzee CNVs are similarly enriched for segmental duplications: 2.8% of all clones on the 1-Mb aCGH platform contain a segmental duplication in the chimpanzee genome, compared with 7.5% of all chimpanzee CNV loci (P < 0.0001) and 17.6% of all multiple-incidence chimpanzee CNV loci (P < 0.000001). Therefore, the presence of ancestral segmental duplications that arose before the divergence of the human and chimpanzee lineages, which are now shared by both species, could partially explain the finding that more CNVs are found in these same regions in both humans and chimpanzees than expected by chance alone.
To evaluate this hypothesis, we compared the locations of the 74 chimpanzee/human shared CNVs with a database of human-specific, chimpanzee-specific, and shared human/chimpanzee segmental duplications (33). We found that 11 of these 74 (14.9%) chimpanzee/human-shared CNV regions contained segmental duplications that existed in both species (ancestral segmental duplications) versus only 2.0% for all clones on the array (P < 0.00001). Among the 17 regions that were found to contain CNVs in multiple individuals in chimpanzees and humans (Table 1), seven (41.2%) overlapped with ancestral segmental duplications (P < 0.000001), representing a >20-fold enrichment relative to all clones on the 1-Mb array. Because the comprehensive identification of segmental duplications in the chimpanzee genome is limited by the current draft of the chimpanzee genome sequence (33), it is likely that there is an even greater percentage of shared CNVs that occur in close proximity to ancestral segmental duplications than is currently estimated.
Discussion
We have identified a total of 355 CNVs (331 on the 1-Mb-resolution aCGH platform and an additional 24 on the segmental duplication-enriched aCGH platform) among the genomes of 20 unrelated chimpanzees from the western subspecies. Because both aCGH platforms actually sample ≤12% of the current reference human genome, the number of CNVs identified in this study is likely to be a gross underestimation of the true number of CNVs in the chimpanzee genome. By using the same aCGH platform, more CNVs were identified among the 20 western chimpanzees tested than among 55 ethnically diverse humans. We have identified an average of 31 CNVs in each of the 20 chimpanzees studied, compared to an average of 12.4 CNVs per person among the 55 human genomes interrogated in the Iafrate et al. (1) study. This finding implies that overall chimpanzee genetic diversity may be more extensive than was previously thought; a notion that is based on estimates of general nuclear DNA sequence diversity among the western chimpanzee subspecies being similar to that of the human species as a whole (19–23). It is unclear whether this unexpected difference between chimpanzees and humans reflects species-level differences in selective pressures on copy number variation or higher duplication/deletion mutation rates in chimpanzees or both. Detailed studies on the evolutionary histories of specific CNV loci in both humans and chimpanzees may help resolve this issue.
A subset of CNVs was found to be shared in both chimpanzees and humans. Interestingly, these CNVs appear to be relatively common in both species. If one considers the theoretical and empirical estimates of coalescence times (i.e., the most recent common ancestor of all currently known alleles at a given locus based on effective population sizes, with coalescence times for nearly all human loci being <2 million years) along with a ≈6-million-year divergence of the human and chimpanzee lineages, it seems unlikely that genetic polymorphisms present in our common ancestor would have been maintained in extant humans and chimpanzees (22, 23, 34–39). This expectation is substantiated by several empirical studies (40–44), with exceptions only in rare cases of strong and long-term balancing selection, under which a fitness advantage is conferred to heterozygous individuals (e.g., the MHC locus, ref. 45). Therefore, CNVs found in the same regions in both species likely represent recurrent gains and losses at the same loci. Because the underlying fine structure of the duplication/deletion regions are not yet known, we cannot determine whether the CNVs are structurally similar (i.e., similar breakpoints) in both humans and chimpanzees. Regardless, this finding suggests that these regions may contain features shared between the human and chimpanzee genomes that facilitate frequent CNV genesis.
Many common CNVs may be located in duplication and deletion “hotspots” that are inherently and, perhaps, continually unstable (e.g., ref. 10), which may explain why we found the most common CNVs for one species to be nearly always commonly observed in the other species. In support of this theory, Conrad and colleagues (8) recently observed several deletion variants in humans for which there were dissimilar breakpoints and/or SNP haplotype backgrounds among different individuals. One of these deletion variants is mapped to the same region as BAC clone RP11-130C19 (human chromosome 9p24.3), which was identified as a CNV in multiple chimpanzees in this study (Table 1). Different mechanisms are likely to act on different types of common CNVs. For example, unlike the deletion variants with dissimilar breakpoints (8), other common human CNVs are reportedly in complete linkage disequilibrium with flanking SNPs (7, 9), implying that these particular CNVs have a single origin. Attaining breakpoint and SNP haplotype data on other common CNVs, including those that are observed in both humans and chimpanzees, will help us to stratify classes of CNVs and more comprehensively delineate the molecular mechanisms involved in their generation and maintenance (including potential within-species recurrence).
We propose that inherently unstable CNV hotspots are shared across the human and chimpanzee genomes and that this phenomenon is driven in part by frequent nonallelic homologous recombination of ancestral segmental duplications (Fig. 3), a hypothesis supported by our finding that CNVs observed at the same loci in multiple humans and chimpanzees are enriched 20-fold for ancestral segmental duplications. However, it is also of interest that not all regions with ancestral segmental duplications are currently known to harbor CNVs. Future studies can evaluate copy number variation frequencies and patterns across multiple species to evaluate what genomic features besides segmental duplications (e.g., recombination rate, G+C content) influence localized rates of CNV mutation and evolution (46). When CNVs are common in one species but not observed in the other, species-specific segmental duplications (i.e., the segmental duplications that arose after divergence of the human and chimpanzee lineages) may be involved. For example, locus RP11-315O22 on human chromosome 2q11.2 contains chimpanzee-only segmental duplications (33) and was found to be copy-number variable in >25% of chimpanzees but has not yet been identified as a CNV in any human study. Also of evolutionary interest are regions for which CNVs are common in one species but not observed in the other, despite the presence of ancestral segmental duplications. Such patterns may reflect lineage-specific changes in other genomic features (e.g., recent studies have shown that recombination hotspots are not always shared between humans and chimpanzees) (47–49) or recent selective pressure in one lineage.
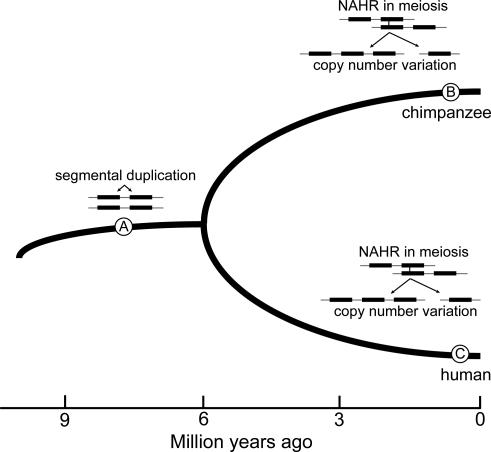
Fig. 3.
Model for evolution of CNV hotspots. Certain segmental duplications that arose in a human–chimpanzee common ancestor (depicted at point A) may facilitate separate nonallelic homologous recombination (NAHR) in both chimpanzees (B) and humans (C), leading to the genesis of CNVs in both species. If NAHR in these regions occurs frequently, it may be expected to lead to the maintenance of common CNVs by way of recurrent duplications and deletions.
In summary, we have shown that intraspecific copy number variation is common in the chimpanzee genome, at a level perhaps exceeding that of humans. The evolution of copy number variation appears to be a dynamic process, with a subset of duplication and deletion events possibly reoccurring across, and within, species. Such regions may be inherently unstable and serve as hotspots of structural genomic rearrangements. CNVs observed at the same loci in both humans and chimpanzees will be an important focus of future investigation and may provide interesting opportunities for testing hypotheses concerning the involvement of copy number variation in phenotypic diversity.
Materials and Methods
The wild-born chimpanzee males included in this study were housed at various research facilities and zoological institutions. Whole blood was collected during routine veterinary examinations, and genomic DNA was isolated with a standard phenol-chloroform extraction method (50). Subspecies designations were determined/confirmed by mitochondrial DNA and Y chromosome nucleotide sequencing and comparisons with the sequences of wild-born individuals with known capture or sample-collection location (20). For the reference sample, we obtained a B lymphoblast cell line (S006006) of the captive-born donor for the chimpanzee genome sequence, Clint, from the Coriell Cell Repositories (Camden, NJ). Based on sequence and pedigree data, Clint is likely a western chimpanzee, although the possibility that his genome reflects limited captive admixture with other subspecies cannot be excluded (19). DNA was isolated from the cell line with the Puregene DNA Purification kit (Gentra Systems, Minneapolis).
Chimpanzee CNVs were first detected by aCGH (51) by using SpectralChip 2600 microarray slides (Spectral Genomics, Houston). Each slide is spotted with 2,632 human BAC clones, spaced at ≈1-Mb intervals throughout the genome, for ≈12% coverage of the genome. Two slides were used per experiment in a dye-swap design, to reduce the false-positive error rate. DNA sample labeling, hybridization, washes, and normalization were performed as described in ref. 1, and resulting threshold values of two standard deviations from the population mean ratios of the relative intensity differences were established. aCGH experiments were also performed by using arrays spotted with 2,194 human BAC clones, selected to specifically target known segmental duplication regions in the human genome (4). DNAs from five of the same chimpanzees used in the 1-Mb aCGH experiments were compared with genomic DNA from the same reference chimpanzee. A dye-swap design was also used for the latter aCGH experiments, and the threshold value was again set to two standard deviations from the population mean ratios. For analyses, clones overlapping the Ig heavy-chain locus on human chromosome 14q32.33 were excluded (i.e., one clone from the 1-Mb array and four clones from the segmental duplication array), because a somatic deletion removes portions of this locus in B lymphoblast cells (52, 53).
We obtained position information from public sources (Build 34 of the human genome sequence, http://genome.ucsc.edu, and www.ensembl.org) for the BAC clones on the 1-Mb array. To assess the association of CNVs with segmental duplications, we accessed a database of chimpanzee-only, human-only, and shared chimpanzee and human segmental duplications provided by Cheng and colleagues (33) and noted when there was overlap between a segmental duplication and a given BAC clone. Their analysis was based on data from the chimpanzee genome sequencing project (19). We also performed a clustering analysis to determine whether any two BAC clones overlapped paralogous segmental duplication DNA segments. If one such BAC contained a CNV, then this could have led to the erroneous identification of a CNV at the second locus (i.e., a segmental duplication “shadowing” effect). However, none of the BAC clones contained known paralogous segmental duplication copies. Fisher’s exact test was used for all statistical comparisons.
GO analyses were performed by using the program gotree machine (54). To determine whether any GO categories are significantly overrepresented among genes mapped to chimpanzee CNV loci, we compared this subset of genes with the total set of genes that overlap one of the 2,632 BAC clones on the 1-Mb array. A similar analysis was performed for shared human and chimpanzee CNV loci. Shared human and chimpanzee CNV loci were determined based on overlap between identified chimpanzee CNV loci in this study and identified human CNVs in the Database of Genomic Variants (http://projects.tcag.ca/variation) as of January 7, 2006. Several CNV-containing BACs encompass multiple genes of similar function (i.e., gene families), introducing a potential bias into our GO analyses. For example, BAC RP11-315O22 (with a genomic loss detected in eight chimpanzees) is mapped to the same region as four genes, all with immune and environmental response functions: IL1RL2, IL1RL1, IL18R1, and IL18RAP. It is unclear whether all genes are copy-number-variable and, if so, whether any phenotypic effects of copy number variation at such loci are gene-specific or more general to the gene family as a whole. To investigate the potential effects of tandemly arranged gene families on our GO results, we eliminated all but one gene from each BAC (for all clones on the 1-Mb array, including the CNV-containing BACs) in the immune response and defense response GO categories and then repeated the chimpanzee CNV GO analysis. Although additional investigation is needed into the phenotypic consequences of copy number variation of gene families, the result of our a posteriori analysis showed that, even under this very conservative approach, chimpanzee CNV loci are significantly enriched for genes with function in immune and defense response (immune response: observed genes, n = 24; expected, n = 14.45; P < 0.01; defense response: observed genes, n = 26; expected, n = 16.23; P < 0.01).
For qPCR experiments, primers were designed by using the program primer3 (55) to amplify 100- to 150-bp fragments. These fragments were positioned within or between segmental duplications. We referred to an alignment of the human (Build 34) and chimpanzee (Build 1.1) genome sequences (http://genome.ucsc.edu) to ensure that primers would amplify the intended fragments in both species. For each locus, the primers (all given 5′–3′) used were RP11-88L18 (human chromosome 5p15.1) forward (F) GGGTCTGTTTGTGCAGGAAT and reverse (R) TTCATCCAGGTAAGGGCAAC, RP11-81P11 (1q21.2) (F) GTTAGGGTCACCATGTCCATTT and (R) TCAGAGGAAGACCAAGAAAAGC, RP11-96G1 (8q21.2) (F) TGGTGTTGTAGTCCACGATCTC and (R) GACGCTTATCCAGGAATCTACG, RP11–100C24 (13q14.3) (F) TGTTCTCCATTCATATCGCATC and (R) CCTGCCTGGACCTTATAGTCAC, RP11-315O22 (2q11.2) (F) TGAACGTGTTCTTTTGTGCTCT and (R) ACTTTACCACCCTCGCTAACAA, RP11-130C19 (9p24.3) (F) TGTTTCCCCTCTTATTTCCAGA and (R) GAGGGCACTGTGATCCTAAAAC, RP11-79F15 (19p13.2) (F) GAAAACTCTCCTTGGGTGTGAG and (R) ATTCGAATCCTAAGACCCCATT, RP11-79I15 (16p13.11) (F) TGTAACCCTTCTCTTGCCAAAT and (R) CTAGCAGCCCTCATCTACCACT, RP11-499D5 (16p11.2) (F) CATGGGTATCAGAGACACTGGA and (R) TCTTTATCCACTCCCTGCAGTT, and RP11-28G16 (14q32.12) (F) TGACCTCTGATTTTTCCCTCAT and (R) AATGAATGTGATTTCCCCAAAC. A fragment from a reference gene, TP53 (chr17p13.1) (F) CCCTTCCCAGAAAACCTACC and (R) CAGGCATTGAAGTCTCATGG, was amplified as a calibrator to adjust for any minor variance among the DNA dilution quantities of different samples. Samples were run in 25-μl reactions using iQ SYBR Green Supermix (Bio-Rad) in a 96-well plate on a Bio-Rad iCycler Thermal Cycler with an initial denaturation of 93°C for 2 min, followed by 40 cycles of 93°C for 15 sec and 60°C for 30 sec. For each test sample replicate, 2 ng of genomic DNA was used. Standard curves were created by using dilutions of genomic DNA from the chimpanzee reference individual (S006006; Clint). Test samples were run in triplicate and standards run in duplicate. Results were analyzed, and the final standard deviation was calculated from the standard deviations of the test gene and the reference gene according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank S. Dallaire and Z. Cheng for laboratory and bioinformatics assistance, respectively; L. Feuk, M. E. Hurles, and S. A. McCarroll for critical reading and helpful comments on this manuscript; and the following institutions, research facilities, and zoological parks for the chimpanzee samples used in this study: Coriell Institute for Medical Research, Camden NJ; New Iberia Research Center, New Iberia, LA; Primate Foundation of Arizona, Mesa, AZ; Southwest Foundation for Biomedical Research, San Antonio, TX; Yerkes National Primate Research Center, Atlanta; the Lincoln Park Zoo, Chicago; and the Riverside Zoo, Scottsbluff, NE. This work was supported in part by National Institutes of Health Grant HD004385 (to E.E.E.), a grant from the Leukemia and Lymphoma Society and the Department of Pathology, Brigham and Women’s Hospital (to C.L.), and National Institutes of Health National Center for Research Resources Grant 3 U42 RR015087-05S1 to the University of Louisiana at Lafayette New Iberia Research Center. Chimpanzee sampling and subspecies identification analyses were supported by National Science Foundation Grant BCS-0073871 (to A.C.S.). C.T.S. was supported by the Wellcome Trust, S.W.S is an investigator of the Canadian Institutes of Health Research and International Scholar of the Howard Hughes Medical Institute, and E.E.E. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- aCGH
array-based comparative genomic hybridization
- BAC
bacterial artificial chromosome
- CNV
copy number variant
- GO
Gene Ontology
- qPCR
quantita-tive PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Iafrate A. J., Feuk L., Rivera M. N., Listewnik M. L., Donahoe P. K., Qi Y., Scherer S. W., Lee C. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Tuzun E., Sharp A. J., Bailey J. A., Kaul R., Morrison V. A., Pertz L. M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 4.Sharp A. J., Locke D. P., McGrath S. D., Cheng Z., Bailey J. A., Vallente R. U., Pertz L. M., Clark R. A., Schwartz S., Segraves R., et al. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefansson H., Helgason A., Thorleifsson G., Steinhorsdottir V., Masson G., Barnard J., Baker A., Jonasdottir A., Ingason A., Gudnadottir V. G., et al. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 6.Feuk L., Macdonald J. R., Tang T., Carson A. R., Li M., Rao G., Khaja R., Scherer S. W. PLoS Genet. 2005;1:e56. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarroll S. A., Hadnot T. N., Perry G. H., Sabeti P. C., Zody M. C., Barrett J. C., Dallaire S., Gabriel S. B., Lee C., Daly M. J., et al. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 8.Conrad D. F., Andrews T. D., Carter N. P., Hurles M. E., Pritchard J. K. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 9.Hinds D. A., Kloek A. P., Jen M., Chen X., Frazer K. A. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 10.Repping S., van Daalen S. K., Brown L. G., Korver C. M., Lange J., Marszalek J. D., Pyntikova T., van der Veen F., Skaletsky H., Page D. C., et al. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 11.Feuk L., Carson A. R., Scherer S. W. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 12.Aldred P. M., Hollox E. J., Armour J. A. Hum. Mol. Genet. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- 13.Linzmeier R. M., Ganz T. Genomics. 2006;86:423–430. doi: 10.1016/j.ygeno.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Hollox E. J., Armour J. A., Barber J. C. Am. J. Hum. Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R. J., Freedman B. I., Quinones M. P., Bamshad M. J., et al. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 16.Aitman T. J., Dong R., Vyse T. J., Norsworthy P. J., Johnson M. D., Smith J., Mangion J., Roberton-Lowe C., Marshall A. J., Petretto E., et al. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 17.Newman T. L., Tuzun E., Morrison V. A., Hayden K. E., Ventura M., McGrath S. D., Rocchi M., Eichler E. E. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suto Y., Ishikawa Y., Hyodo H., Ishida T., Kasai F., Tanoue T., Hayasaka I., Uchikawa M., Juji T., Hirai M. Cytogenet. Genome Res. 2003;101:161–165. doi: 10.1159/000074173. [DOI] [PubMed] [Google Scholar]
- 19.Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 20.Stone A. C., Griffiths R. C., Zegura S. L., Hammer M. F. Proc. Natl. Acad. Sci. USA. 2002;99:43–48. doi: 10.1073/pnas.012364999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaessmann H., Wiebe V., Paabo S. Science. 1999;286:1159–1162. doi: 10.1126/science.286.5442.1159. [DOI] [PubMed] [Google Scholar]
- 22.Yu N., Jensen-Seaman M. I., Chemnick L., Kidd J. R., Deinard A. S., Ryder O., Kidd K. K., Li W. H. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer A., Wiebe V., Paabo S., Przeworski M. Mol. Biol. Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe H., Fujiyama A., Hattori M., Taylor T. D., Toyoda A., Kuroki Y., Noguchi H., BenKahla A., Lehrach H., Sudbrak R., et al. Nature. 2004;429:382–388. doi: 10.1038/nature02564. [DOI] [PubMed] [Google Scholar]
- 25.Locke D. P., Segraves R., Carbone L., Archidiacono N., Albertson D. G., Pinkel D., Eichler E. E. Genome Res. 2003;13:347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson G. M., Flibotte S., Missirlis P. I., Marra M. A., Jones S., Thornton K., Clark A. G., Holt R. A. Genome Res. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen D. Q., Webber C., Ponting C. P. PLoS Genet. 2006;2:e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., et al. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey J. A., Gu Z., Clark R. A., Reinert K., Samonte R. V., Schwartz S., Adams M. D., Myers E. W., Li P. W., Eichler E. E. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 30.Smith G. P. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 31.Samonte R. V., Eichler E. E. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- 32.Inoue K., Lupski J. R. Annu. Rev. Genomics Hum. Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R. K., Paabo S., et al. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 34.Clark A. G. Proc. Natl. Acad. Sci. USA. 1997;94:7730–7734. doi: 10.1073/pnas.94.15.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harding R. M., Fullerton S. M., Griffiths R. C., Bond J., Cox M. J., Schneider J. A., Moulin D. S., Clegg J. B. Am. J. Hum. Genet. 1997;60:772–789. [PMC free article] [PubMed] [Google Scholar]
- 36.Wall J. D. Genetics. 2003;163:395–404. doi: 10.1093/genetics/163.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Won Y. J., Hey J. Mol. Biol. Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- 38.Harding R. M., McVean G. Curr. Opin. Genet. Dev. 2004;14:667–674. doi: 10.1016/j.gde.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Excoffier L. Curr. Opin. Genet. Dev. 2002;12:675–682. doi: 10.1016/s0959-437x(02)00350-7. [DOI] [PubMed] [Google Scholar]
- 40.Hacia J. G., Fan J. B., Ryder O., Jin L., Edgemon K., Ghandour G., Mayer R. A., Sun B., Hsie L., Robbins C. M., et al. Nat. Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- 41.Carroll M. L., Roy-Engel A. M., Nguyen S. V., Salem A. H., Vogel E., Vincent B., Myers J., Ahmad Z., Nguyen L., Sammarco M., et al. J. Mol. Biol. 2001;311:17–40. doi: 10.1006/jmbi.2001.4847. [DOI] [PubMed] [Google Scholar]
- 42.Weber J. L., David D., Heil J., Fan Y., Zhao C., Marth G. Am. J. Hum. Genet. 2002;71:854–862. doi: 10.1086/342727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hedges D. J., Callinan P. A., Cordaux R., Xing J., Barnes E., Batzer M. A. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asthana S., Schmidt S., Sunyaev S. Trends Genet. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Lawlor D. A., Ward F. E., Ennis P. D., Jackson A. P., Parham P. Nature. 1988;335:268–271. doi: 10.1038/335268a0. [DOI] [PubMed] [Google Scholar]
- 46.Lupski J. R. Genome Biol. 2004;5:242. doi: 10.1186/gb-2004-5-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ptak S. E., Roeder A. D., Stephens M., Gilad Y., Paabo S., Przeworski M. PLoS Biol. 2004;2:e155. doi: 10.1371/journal.pbio.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ptak S. E., Hinds D. A., Koehler K., Nickel B., Patil N., Ballinger D. G., Przeworski M., Frazer K. A., Paabo S. Nat. Genet. 2005;37:429–434. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- 49.Winckler W., Myers S. R., Richter D. J., Onofrio R. C., McDonald G. J., Bontrop R. E., McVean G. A., Gabriel S. B., Reich D., Donnelly P., et al. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 51.Pinkel D., Albertson D. G. Annu. Rev. Genomics Hum. Genet. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- 52.Honjo T., Kataoka T. Proc. Natl. Acad. Sci. USA. 1978;75:2140–2144. doi: 10.1073/pnas.75.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jack H. M., McDowell M., Steinberg C. M., Wabl M. Proc. Natl. Acad. Sci. USA. 1988;85:1581–1585. doi: 10.1073/pnas.85.5.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B., Schmoyer D., Kirov S., Snoddy J. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rozen S., Skaletsky H. J. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S., Misener S., editors. Totowa, NJ: Humana; 2000. pp. 365–386. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.