Abstract
Stem cells represent a unique population of cells with self-renewal capacity. Although they are important therapeutic targets, the genetic manipulation of tissue-specific stem cells has been limited, which complicates the study and practical application of these cells. Here, we demonstrate successful gene trapping and homologous recombination in spermatogonial stem cells. Cultured spermatogonial stem cells were transfected with gene trap or gene targeting vectors. Mutagenized stem cells were expanded clonally by drug selection. These cells underwent spermatogenesis and produced heterozygous offspring after transplantation into the seminiferous tubules of infertile mouse testes. Heterozygous mutant mice were intercrossed to produce homozygous gene knockouts. Using this strategy, the efficiency of homologous recombination for the occludin gene locus was 1.7% using a nonisogenic DNA construct. These results demonstrate the feasibility of altering genes in tissue-specific stem cells in a manner similar to embryonic stem cells and have important implications for gene therapy and animal transgenesis.
Keywords: spermatogenesis, germ cell, testis, transplantation
Stem cells represent a unique cell population with self-renewal potential (1). Although stem cells are low in number, these cells proliferate extensively to sustain the various self-renewing tissues, such as bone marrow and intestine. Although these tissue-specific stem cells normally divide very slowly, stem cells are the last cell type to be destroyed after cytotoxic damage, and they regenerate the entire tissue in a relatively short time. In addition, stem cells often have migratory activities, and they can be transplanted between animals; transplanted stem cells migrate to a specific niche and regenerate the self-renewing tissue. Because of their unique properties, stem cells have become the attractive target of cell and gene therapies.
Among the many types of tissue-specific stem cells, spermatogonial stem cells are unique in that they have germ-line potential (2, 3). Genetic modification of spermatogonial stem cells creates permanent changes in the germ line, which are transmitted to the offspring by means of fertilization. In contrast to female germ-line cells, which cease to divide after birth, male germ-line cells proliferate continuously and produce sperm throughout the life of the animal. If these stem cells could be cultured and manipulated in a manner similar to embryonic stem (ES) cells (4, 5), they could be used to create knockout animals. As a first step toward this goal, a germ cell transplantation technique was developed in 1994 (6, 7), in which dissociated donor testis cells colonized the seminiferous tubules of infertile recipient testis and produced donor-derived spermatogenesis and offspring. Although this technique was an opportunity to produce offspring from manipulated spermatogonial stem cells, it has been difficult to produce transgenic animals using spermatogonial stem cells, because their number is very low in the testis, and the lack of methods to expand spermatogonial stem cells has restricted genetic manipulation (8, 9).
Recently, we described a culture system for expanding spermatogonial stem cells (10). Germ cells formed colonies of a unique shape in the presence of glial cell line-derived neurotrophic factor (GDNF) (11, 12), a critical factor for the self-renewing division of spermatogonial stem cells (13). The addition of GDNF maintains logarithmic proliferation of spermatogonial stem cells and achieved >1085-fold expansion of the initial cell population (14). The cells retained germ-line potential after 2 years of culture and underwent spermatogenesis and produced offspring after germ cell transplantation. These cells can now be cultured under serum-free or feeder-free conditions (11, 15) and can be transfected to produce transgenic offspring (16). Based on these properties, we designated the cultured spermatogonia as germ-line stem (GS) cells (10). In this work, we describe the derivation of knockout mice using GS cells. GS cells were mutated by gene trap or targeting vectors, both of which are commonly used to create mutant animals using ES cells. Our results have important implications for future gene therapies and for animal mutagenesis.
Results
Mutagenesis of GS Cells Using the Gene Trap Vector.
To induce mutations in the GS cells (Fig. 1A), the cells were infected with ROSAβgeo virus, which has been used to mutagenize ES cells (17). For selection using G418, the transfected GS cells were transferred onto neomycin (neo)-resistant mouse embryonic fibroblasts. Single clones were allowed to proliferate in 96-well plates, and the individual colonies were expanded. From the total of 4.5 × 106 GS cells that were infected, we selected 99 clones for in vitro expansion. Approximately 80% of the resulting neo-resistant colonies stained positively for LacZ expression.
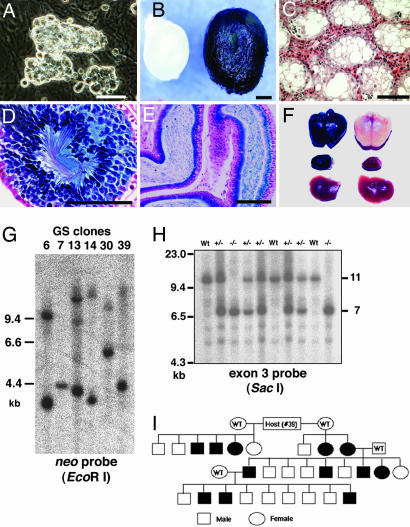
Fig. 1.
Gene trapping in GS cells using the ROSAβgeo vector. (A) Appearance of the GS cells. (B) Appearance of recipient testes after transplantation with GS cells (clone 30) that express the lacZ reporter gene (Right). The testis of a control W mouse (Left) is smaller and does not stain positively for LacZ. (C) The nontransplanted recipient testis shows no spermatogenesis. (D) The recipient testis after transplantation of GS cells (clone 30) shows normal spermatogenesis. (E) LacZ-expressing spermatozoa in the epididymis (clone 30). (F) Macroscopic appearance of X-Gal-stained brain (Top), kidney (Middle), and liver (Bottom) of transgenic (Left) and WT (Right) F1 offspring from clone 30 GS cells. LacZ is expressed in the brain and kidney but not in the liver. (G) Southern blot analysis of F1 animals derived from different clones, showing variable integration patterns. (H) Southern blot analysis of offspring identified as WT animals and heterozygous or homozygous clone 39 (occludin) mutants. (I) Descendents of a recipient W male that was transplanted with clone 39 GS cells. The filled symbols indicate the presence of the transgene. (Scale bars: A, 25 μm; B, 1 mm; C–E, 100 μm.)
The cDNA fragments that contained the junction between the endogenous exons and the site of retroviral insertion were amplified from the RNA using the 5′-rapid amplification of cDNA ends (RACE) method (Table 1). The RACE products were recovered from 89 of 99 (89%) clones. Multiple insertions of the retroviral tag sequence were found in 8% of the samples. Sequence analysis of the RACE cDNAs showed that the splice acceptor sequence had functioned in ≈71% of the clones, resulting in the generation of a single ORF that was in-frame with the ROSAβgeo sequence. The retrovirus insertions were randomly distributed on the chromosomes. The mutations occurred predominantly in the upstream regions of the mouse genes (95%), particularly in the first intron (63%). Sequence analysis identified several classes of mutated genes, including those that encode membrane proteins (ATP synthetase and occludin), metabolic enzymes (pyruvate kinase), DNA repair enzymes (methylguanine DNA methyltransferase and RAD18 homologue), transcription factors (erg and zinc finger protein 111), and proteins associated with the cytoskeleton (Rho GTPase activating protein gene 8 and kinesin-related protein KIFC1). Eleven of the cDNAs obtained with 5′-RACE could not be assigned functions based on database searches (see Table 3, which is published as supporting information on the PNAS web site).
Table 1.
Examples of fusion transcripts identified by 5′ RACE from the gene trap GS cell lines
| No. | Sequences of the trapped gene | Homology (%) | GenBank | Symbol | Chromosome | Insertion | Definition |
|---|---|---|---|---|---|---|---|
| 39 | ggtgagcacc ttgggattcc ggccgccaag ctcgcgg | 100/100 (100) | NM_008756 | Ocln | 13 | First intron | Occludin |
| 67 | gctgctcagt gtggtggatc atttcaaccg ctcgcgg | 100/100 (100) | NM_010817 | Psmd7 | 8 | First intron | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| 78 | nngaaagact tggtaatggc gacgggtttg ctcgcgg | 28/28 (100) | NM_177993 | Hbp1 | 12 | First intron | High-mobility group box transcription factor 1 |
| 97 | attcccacct gctgacttaa gtgcgcccag ctcgcgg | 100/100 (100) | NM_009686 | Apbb2 | 5 | Second intron | Amyloid β (A4) precursor protein-binding, family B, member 2 |
| 101 | ccgccgcgcc cggccccgcg cgcgacccgg ctcgcgg | 100/100 (100) | NM_010757 | Mafk | 5 | First intron | V-maf musculoaponeurotic fibrosarcoma oncogene family, protein K |
| 104 | ccggtgcggc ccggatccgt ggcacgggag ctcgcgg | 98/100 (98) | NM_030730 | Srisnf21 | 9 | First intron | Steroid receptor-interacting SNF2 domain protein |
| 112 | tctgtacctg acccagcagn tnatattaan ctcgcgg | 88/100 (88) | NM_133659 | Erg | 16 | First intron | Avian erythroblastosis virus E-26 (v-ets) oncogene related |
| 114 | tcttcaagta catcatcatc ggcgacacag ctcgcgg | 99/100 (99) | NM_021518 | Rab2 | 4 | First intron | RAB2, member RAS oncogene family |
Gene Targeting in GS and mGS Cells.
To generate targeted mutations (18, 19), we chose the occludin gene (20), which is one of the genes that was successfully trapped in the previous section (Fig. 2A). Occludin is one of the tight junction proteins and is expressed in various organs (20). This gene was chosen for two reasons. First, we assumed that gene targeting in GS cells would occur at a higher rate in a transcriptionally active locus (Fig. 2B). Second, occludin knockout mice have been produced by ES cell technology (20), which allows comparisons of the efficiency of homologous recombination. Homologous recombination between the vector and the occludin gene would result in the deletion of exon 3. We electroporated the knockout vector into enhanced green fluorescent protein (EGFP)-expressing GS cells. The transfected cells were plated on neo-resistant mouse embryonic fibroblasts and exposed to G418 to select for neo-resistant clones. Of the total of 2.4 × 108 GS cells, selection with G418 yielded 120 neo-resistant colonies. Approximately 12–14 weeks of culture elapsed between the transfection and collection of ≈2 × 106 cells for cryopreservation and PCR analysis. In this gene-targeting experiment, we also used ES-like cells (mGS cells) (Fig. 3A), which could be generated from the neonatal testis cell culture (21). The same occludin gene targeting vector that was used for the GS cells was transfected into 1.9 × 108 cells, and neo selection was conducted on neo-resistant mouse embryonic fibroblasts. The frequency of neo-resistant colonies was ≈10−5 to 10−6. Of the neo-resistant colonies, a total of 139 clones were chosen, and 1–2 × 106 cells were recovered within 1 month of transfection.
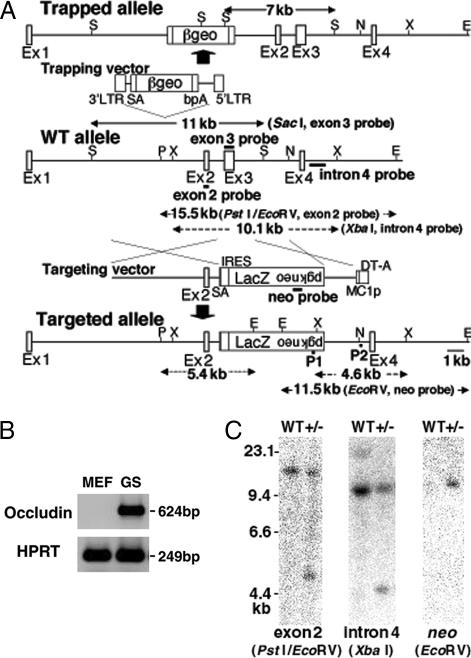
Fig. 2.
Disruption of the occludin gene. (A) Insertion of the gene trap and gene-targeting vectors in the mouse occludin locus. SA, splice acceptor; bpA, bovine growth hormone gene polyadenylation signal; E, EcoRV; N, NheI; P, PstI; S, SacI; X, XbaI. P1 and P2 represent the primers used for PCR. (B) Expression of occludin in GS cells by RT-PCR. (C) Southern blot analysis of a GS cell clone (clone 101). Genomic DNA was digested with the indicated restriction enzymes and hybridized with three different probes.
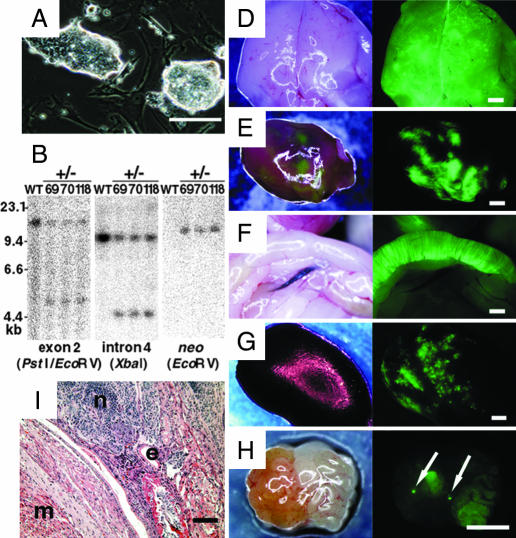
Fig. 3.
Production of chimeric animals by heterozygous occludin knockout mGS cells. (A) Appearance of the mGS cells. (B) Southern blot analysis of the mGS cell lines. (C) Section of a teratoma from the mGS cells after gene targeting. Note the presence of tissues from three germ layers, which include muscle (m), neural (n), and epithelial (e) tissues. (D–H) Macroscopic appearance of the brain (D), heart (E), gut (F), kidney (G), and ovary (H), showing fluorescence under UV light. Arrows indicate oocyte-like cells in the ovary. (Scale bars: A and C, 100 μm; D–H, 1 mm.)
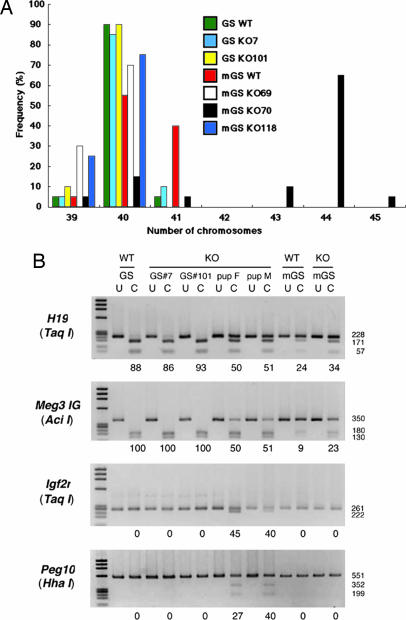
To check for homologous recombination, DNA samples were screened by PCR, which was designed to amplify the 2.4-kb fragment spanning the 3′ junction of the targeted locus. By using this strategy, we demonstrated that 1.7% (2/120) or 2.2% (3/139) of the neo-resistant GS or mGS cell colonies contained correctly targeted cells (Figs. 2C and 3B). These rates were somewhat lower than that (4.5%) reported for experiments with ES cells (20). The probe for the neo-resistance gene as well as those for exon 2 (internal) and intron 4 (external) hybridized to fragments of the correct sizes. Further confirmation of successful gene targeting was obtained by sequencing the PCR products (data not shown). Cytogenetic analysis using quinacrine plus Hoechst 33258 staining showed that 85–90% of the GS cells retained the normal karyotype, whereas the frequency of euploidity in the mGS cells ranged from 15–75% after genetic selection (Fig. 4A). Furthermore, combined bisulfite restriction analysis (COBRA) revealed that the androgenetic imprint patterns were stable in the GS cells, whereas those in the mGS cells changed dramatically (Fig. 4B).
Fig. 4.
Effects of genetic manipulation on karyotype and imprint patterns in GS and mGS cells. (A) Karyotype analysis of the heterozygous knockout GS and mGS cells before and after genetic selection. At least 20 cells were counted. (B) Combined bisulfite restriction analysis (COBRA) of imprinted genes. DNA was isolated from heterozygous knockout GS (clones 7 and 101) and mGS (clone 118) cells. Heterozygous F1 offspring were produced from GS clone 101. PCR products from differentially methylated regions were digested with enzymes with a recognition sequence containing CpG in the original unconverted DNA. The percentage methylation is indicated below the gel. Enzymes used to cleave each locus are indicated in parentheses. U, uncleaved; C, cleaved. F, female; M, male.
Germ-Line Potential of Mutated GS Cells.
To confirm the germ-line potential of the mutated GS cells, eight clones, which included seven randomly chosen trapped clones (clone 6, 7, 13, 14, 27, 30, and 39) and one of targeted clones (clone 101), were microinjected into the seminiferous tubules of three to nine infertile WBB6F1-W/Wv (W) mice. Because the germ cells of W mice are unable to differentiate beyond the spermatogonium stage owing to mutations in the c-kit gene, spermatogenesis in the recipient testis must originate from transplanted cells (22). After transplantation, the testes grew significantly (Fig. 1B), and normal spermatogenesis was confirmed (Fig. 1 C–E). The recipients began to produce offspring by natural mating with wild-type (WT) females as early as 77 days after GS cell transplantation; 18 of the 48 (37.5%) recipient mice became fertile within 4 months of transplantation (Table 2). When in vitro microinsemination was used, heterozygous offspring were obtained as early as 69 days after transplantation.
Table 2.
Germ-line transmission of mutagenized GS cell lines by germ cell transplantation
| Type of experiments | Clone | LacZ stain | Days from transfection to transplantation* | Days to first transgenic† | No. (%) of fertile out of total recipients | No. of litters analyzed | No. (%) of transgenic out of total offspring‡ |
|---|---|---|---|---|---|---|---|
| Gene trap | 6 | + | 64 | 109 | 2/7 (28.6) | 2 | 9/12 (75.0) |
| 7 | + | 64 | 82 | 1/7 (14.3) | 5 | 10/21 (47.6) | |
| 13 | + | 52 | 77 | 3/5 (60.0) | 6 | 38/46 (82.6) | |
| 14 | − | 78 | 85 | 3/8 (37.5) | 3 | 6/15 (40.0) | |
| 27 | + | 52 | 80 | 2/3 (66.7) | 5 | 26/35 (74.3) | |
| 30 | − | 57 | 89 | 2/9 (22.2) | 5 | 15/24 (62.5) | |
| 39 | − | 57 | 89 | 2/4 (50.0) | 4 | 12/27 (44.4) | |
| Gene targeting | 101 | NA | 109 | 109 (69)§ | 3/5 (60.0) | 2 | 10/14 (71.4) |
Results at 4 months after transplantation. NA, not applicable.
*Time in days from transfection of GS cells to transplantation into infertile recipients.
†Time in days from transplantation of donor cells to birth of first transgenic progeny.
‡Numerator is the number of transgenic progeny; denominator is the total number of progeny from all recipient mice.
§In parentheses is the number of days for first offspring by in vitro microinsemination.
To date, we have generated F1 offspring from all of the injected clones, and these offspring have produced F2 progeny. The F1 offspring from the fertile males were examined for the presence of transgenes in their genomes by PCR or Southern blot analysis using a neo-specific probe. In total, 65% (115/178) and 71% (10/14) of the F1 mice were heterozygous for the transgenes for trapped and targeted clones, respectively. The patterns of retrovirus integration and LacZ staining in the offspring differed among the clones (Fig. 1 F and G). The transgene was transmitted in a Mendelian fashion (Fig. 1 H and I). The F1 offspring from targeted GS cells showed the normal imprint pattern (Fig. 4B).
Failure to Produce Germ-Line Chimeras Using Mutated mGS Cells.
To generate offspring from mGS cells, we microinjected one of the mGS cell clones (clone 118) into blastocysts. This clone was able to produce teratomas with three germ layers when injected s.c. (Fig. 3C). After microinjection into the blastocyst, 192 chimeric embryos were derived. Of these, 58 offspring were born, 43 of which developed into mature normal adults, 14 males and 29 females. To determine the level of chimerism, we killed 7 males and 15 females and examined the contribution of the EGFP-positive mGS cells to the mature adult animals. In all, 18% (4/22) of the animals were chimeric. The EGFP-positive cells were distributed throughout the body, i.e., in the gut, stomach, liver, lung, brain, heart, pancreas, muscle, and kidney (Fig. 3 D–G). Significantly, EGFP-positive cells also were found in the ovaries of two individual females (Fig. 3H), which suggests their contribution to the germ-line. However, no EGFP-positive offspring were produced from the remainder of the chimeras, and fluorescence was not found in the testes.
Phenotype of occludin-Deficient Mice Derived from Mutated GS Cells.
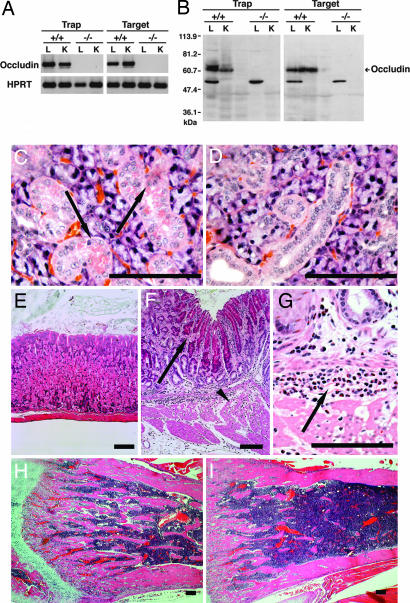
To examine whether the retroviral insertion and gene targeting in GS cells disrupted the occludin gene, tissue samples were prepared from the lungs and kidneys of the WT and homozygous mutant F2 offspring. RT-PCR and Western blot analysis showed that the both types of homozygous mutant mouse lack the occludin mRNA and protein (Fig. 5A and B).
Fig. 5.
Phenotype of the homozygous knockout mice. (A) RT-PCR analysis of RNA samples from the lungs (L) and kidneys (K) of the WT and homozygous knockout mice. Homozygous knockout offspring lack expression of occludin mRNA (624-bp fragment). (B) Western blot analysis with anti-occludin polyclonal antibody of samples from the lungs and kidneys of the WT and homozygous knockout mice. The occludin protein (≈66 kDa) is absent in the homozygous knockout mouse. (C and D) Histological analysis of the salivary glands of 6-week-old WT (C) and homozygous (D) mice produced by gene targeting, showing a loss of cytoplasmic granules in the mutant, which can be found in the WT (C, arrow). (E–G) Histological analysis of the gastric mucosa of 4-week-old WT (E) and homozygous (F and G) mice produced by gene trapping. The predominant hyperplasia of the foveolar cells (F, arrow) is accompanied by a significant loss of chief and parietal cells and the degeneration of the proper muscle of the stomach with chronic inflammation (F, arrowhead). Infiltration of neutrophils and lymphocytes is widely apparent (G, arrow). Similar histology was found in the homozygous mutant made by gene targeting. (H and I) Histological analysis of the femoral bones of 6-week-old WT (H) and homozygous (I) mice produced by gene targeting. Trabecular development is decreased in the mutant. Hematoxylin and eosin staining was used. (Scale bars: C–I, 100 μm.)
We prepared several histological sections from the salivary gland, stomach, and femoral bone of the WT and homozygous mutant mice. In the salivary gland, histological analysis showed that striated ductal cells of the homozygous mutant mice lacked the characteristic cytoplasmic granules (Fig. 5 C and D). Defects in the stomach were more dramatic: most of the gastric mucosa of the homozygous mice consisted of regenerating foveolar cells with infiltrating neutrophils and lymphocytes (Fig. 5 E–G); there was a marked decrease in the number of parietal as well as chief cells. Degeneration of the proper muscle of the stomach also was observed. The femoral bones showed signs of osteoporosis, revealing decreased thickness of bone trabeculae in the mutant animals (Fig. 5 H and I). We could not observe any other abnormalities including brain and testis by morphological analyses during the observation period (≈6 weeks), probably because these abnormalities become apparent at later stages. These findings confirm the previous findings that occludin-deficient mice derived by gene targeting in ES cells are viable and have chronic gastritis (20).
Discussion
Ideally, a pure population of stem cells could be transfected to manipulate specific genes and produce mature differentiated cells. However, although site-specific homologous recombination has been achieved by using hematopoietic progenitor cells in one study (23), it has not been possible to achieve this goal using tissue-specific stem cells. The present study demonstrates that it is possible to apply the concept and techniques of ES cell manipulation to spermatogonial stem cells (24). Currently, knockout animals are generally produced by microinjection of mutated ES cells into blastocysts or by nuclear transfer of mutated somatic cells into oocytes (24, 25). Our results now demonstrate a third method to create mutant animals and suggest that the genomes of other stem cells can be similarly manipulated.
There has been uncertainty that homologous recombination can occur at a usable frequency in tissue-specific stem cells (23). Several studies have indicated a low mutation frequency in spermatogonial stem cells. For example, sister chromatid exchange, a measure of homologous recombination (26), occurs less frequently in spermatogonia than in other somatic cell types (27). In addition, there are indications that the level of mutations induced by ethylnitrosourea in spermatogonia is similar to or lower than that induced in ES cells (28). Nevertheless, the frequency of homologous recombinants was within the range reported for ES and other somatic cells (25). In contrast, the frequency of neo-resistant stable clones was low (1 of ≈2 × 106 transfected cells), and it influenced the efficiency of our experiments. This result was probably caused by the relatively low concentrations of stem cells in GS cell cultures; only up to 1–2% of the cells have repopulation potential (15). Although the current degree of success is modest, our protocol is open to several improvements. For example, the enrichment of stem cells before transfection or changes in the culture condition would probably improve transfection efficiency (29). Although we used a nonisogenic construct in the current study (129 vs. DBA/2), the use of an isogenic construct would further enhance (10- to 50-fold) the gene targeting frequency (30).
Although mutagenesis in GS cells led to the production of knockout animals, we could not generate knockout mice using mGS cells. Although the gene-targeting efficiency in mGS cells was comparable with that in GS cells (2.2% vs. 1.7%), the current study revealed unstable germ-line potential in culture. This situation is somewhat similar to ES cells (31–33), which suggests that the multipotentiality of germ-line cells is maintained at the expense of stability. However, although ES cells acquire aberrant karyotypes during repeated passages (31, 32), they are commonly used for gene targeting, which indicates that they have an acceptable level of instability. In contrast, no targeted mutations have been reported using embryonic germ (EG) cells (34, 35), which suggests that EG cells are less stable than ES cells in germ-line potential. Nevertheless, it will be necessary to test independent mGS cell lines before their utility for germ-line modification can be assessed.
Although the strategies for generating knockout mice from GS and ES cells differ significantly, they require comparable amounts of time. In the present case, transfected cells were expanded for 3–4 months before transplantation. The recipients produced heterozygous progeny 2–3 months after germ cell transplantation, and these animals could be mated to produce homozygous offspring in 3–4 months. Thus, the knockout mice were obtained ≈8–11 months after DNA transfection. In contrast, because ES cells depend on chimera formation with cells of embryos, the donor cells distribute in a mosaic pattern in the F1 offspring. Because it is necessary to produce an F2 generation to confirm germ-line transmission of the transgene, it generally takes 6–12 months after transfection to produce knockout mice from ES cells (36). Although the speed of gene targeting is comparable, the GS cell-based approach may confer an advantage in gene trapping, because gene trapping in GS cells permits the direct expression analysis of whole embryos in the F1 generation.
The most exciting application of our results will be the mutagenesis of other animal species. Given that >60% of genes have been disrupted by using ES cells (37), this technology may have little impact on mouse functional genomics. However, ES cells with germ-line potential are available only in mice. Even in mice, mutations are commonly made in the strain 129 background, and, owing to several genetic weaknesses in this strain, mutant animals are often backcrossed to another background (usually C57BL/6, B6), which takes 18–24 months even with speed congenics (38). Although targeted mutations have been produced recently in several animal species by using nuclear cloning, primary somatic cells have limited proliferative potential, which makes gene targeting more difficult in somatic cells than in ES cells (25). Furthermore, problems associated with nuclear cloning, such as low birth rate and abnormalities in the offspring, need to be resolved (39). In this sense, GS cells may provide the solutions to these problems. The ability to derive GS cells is now extended to different murine genetic backgrounds and to other species (29, 40). Extension of the methodology described here may lead to useful applications in diverse mammalian species.
Materials and Methods
Cell Culture and Genetic Modification.
GS cells were established from newborn DBA/2 (Japan SLC, Hamamatsu, Japan) or Green mice (gift from M. Okabe, Osaka University, Osaka, Japan) using a two-step enzymatic digestion, and the culture was initiated as described in ref. 10.
For the gene-trapping experiments, GS cells were infected with the ROSAβgeo retrovirus plasmid (gift from P. Soriano, Fred Hutchinson Cancer Research Center, Seattle) (17). Virus particles were produced by transfection of Plat-E cells and were concentrated as described in ref. 41. The virus titer was ≈108 colony forming units/ml. The infection of GS cells with the retrovirus was carried out as described in ref. 42. In brief, a single-cell suspension of 1.5 × 106 dissociated GS cells on mouse embryonic fibroblasts was mixed with the virus suspension, and the mixture was centrifuged at 1,750 × g for 1 h at 32°C.
For the gene-targeting experiments, we used a gene-targeting vector for the mouse occludin gene (gift from S. Tsukita, Kyoto University, Kyoto, Japan) (20). Five micrograms of the targeting vector was electroporated into GS cells by using Nucleofector (Amaxa Biosystems, Cologne, Germany). In brief, GS cells were suspended in 100 μl of Nucleofector Solution T, mixed with 5 μg of DNA, and subjected to electroporation. For gene targeting in mGS cells, the cells were suspended in 500 μl of PBS with 20 μg of DNA and subjected to a pulse of 250 V and 500 μF by using the Bio-Rad Gene Pulser II. The mutated cells were selected with 40–200 μg/ml G418 (Invitrogen). The colonies were screened individually by PCR using the following specific primers, P1, 5′-TTGTCACGTCCTGCACGACG-3′, and P2, 5′-CTTAGCTAGGAACCATCAGA-3′.
Transplantation.
For s.c. injection, 4 × 106 cells were injected into KSN mice (Japan SLC). For germ cell transplantation, ≈105 cells were microinjected into the seminiferous tubules of the testes of 5- to 10-day-old immunosuppressed W recipients (Japan SLC) (15).
Chimera Formation and Microinsemination.
Recipient blastocysts for chimera production were obtained by in vitro fertilization of (C57BL/6 × DBA/2)F1 (BDF1) oocytes with B6 sperm. Cells were microinjected into the blastocoels by using a Piezo-driven micromanipulator (43). Microinsemination was carried out by using BDF1 oocytes, as described in ref. 43. The embryos were transferred into the oviducts of ICR pseudopregnant females on the day after culture.
Histological Analysis.
The sample tissues or GS cells were fixed with 4% paraformaldehyde for 2 h and stained for LacZ activity (40). The tissues were embedded in paraffin, cut into sections, and stained with hematoxylin and eosin.
5′-RACE.
The 5′-RACE technique was performed by using a 5′-RACE system (Invitrogen) according to the manufacturer’s protocol, with a primer for cDNA synthesis (5′-TGGGATAGGTTACGTTGGTG-3′) and primers for the nested PCR (5′-CATTCGCCATTCAGGCTGCG-3′ and 5′-TGTTGGGAAGGGCGATCGGT-3′). The PCR products were sequenced directly by using the primer 5′-CTCTTCGCTATTACGCCAGC-3′.
Southern Blotting.
DNA was extracted from tail or brain tissue samples. For the gene trapping, a fragment that contained exon 3 of the occludin gene (≈630 bp) was amplified by PCR using the 5′-ATGTATGGCGGAGAGATGCAT-3′ and 5′-GGATCAACCACACAGTAGTGA-3′ primers and was used as a hybridization probe. For gene targeting, DNA samples were examined by both the exon 2 and intron 4 probes (20). The clones were further examined for single-site integration by hybridization with the neo gene (nucleotides 187–804).
RT-PCR and Western Blotting.
First-strand cDNA synthesis and subsequent PCR were carried out as described in refs. 15 and 20. For Western blotting, samples were separated by SDS/PAGE, transferred to poly(vinylidene difluoride) membranes (Hybond-P; Amersham Pharmacia Biosciences), and probed with rabbit polyclonal anti-mouse occludin antibody (F4) (gift from S. Tsukita).
Combined Bisulfite Restriction Analysis (COBRA).
The methylation statuses of the imprinted genes were assessed by COBRA using specific primers (14).
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Dr. Shoichiro Tsukita, who encouraged and stimulated the minds of many young people. We thank Dr. Y. Kaziro for encouragement and critical reading of the manuscript, Drs. M. Saitou and M. Furuse for discussion, and Ms. A. Wada for technical assistance. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; by grants from CREST (Japan Science and Technology Corporation) and the Human Science Foundation (Japan); and in part by MEXT Special Coordination Funds for Promoting Science and Technology.
Abbreviations
- EGFP
enhanced GFP
- ES
embryonic stem
- GS
germ-line stem
- neo
neomycin.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 7939.
References
- 1.Potten C. S. In: Oxford Textbook of Pathology. McGee J. O., Isaacson P. G., Wright N. A., editors. Oxford: Oxford Univ. Press; 1992. pp. 43–52. [Google Scholar]
- 2.Meistrich M. L., van Beek M. E. A. B. In: Cell and Molecular Biology of the Testis. Desjardins C., Ewing L. L., editors. New York: Oxford Univ. Press; 1993. pp. 266–295. [Google Scholar]
- 3.de Rooij D. G., Russell L. D. J. Androl. 2000;21:776–798. [PubMed] [Google Scholar]
- 4.Evans M. J., Kaufman M. H. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 5.Martin G. R. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinster R. L., Zimmermann J. W. Proc. Natl. Acad. Sci. USA. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinster R. L., Avarbock M. R. Proc. Natl. Acad. Sci. USA. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagano M., Brinster C. J., Orwig K. E., Ryu B.-Y., Avarbock M. R., Brinster R. L. Proc. Natl. Acad. Sci. USA. 2001;98:13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamra F. K., Gatlin J., Chapman K. M., GrellhesI D. M., Garcia J. V., Hammer R. E., Garbers D. L. Proc. Natl. Acad. Sci. USA. 2002;99:14931–14936. doi: 10.1073/pnas.222561399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S., Shinohara T. Biol. Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 11.Kubota H., Avarbock M. R., Brinster R. L. Proc. Natl. Acad. Sci. USA. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa T., Ohmura M., Tamura Y., Kita K., Ohbo K., Suda T., Kubota Y. Arch. Histol. Cytol. 2004;67:297–306. doi: 10.1679/aohc.67.297. [DOI] [PubMed] [Google Scholar]
- 13.Meng X., Lindahl M., Hyvönen M. E., Parvinen M., de Rooij D. G., Hess M. W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M., et al. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 14.Kanatsu-Shinohara M., Ogonuki N., Iwano T., Lee J., Kazuki Y., Inoue K., Miki H., Takehashi M., Toyokuni S., Shinkai Y., et al. Development (Cambridge, U.K.) 2005;132:4155–4163. doi: 10.1242/dev.02004. [DOI] [PubMed] [Google Scholar]
- 15.Kanatsu-Shinohara M., Miki H., Inoue K., Ogonuki N., Toyokuni S., Ogura A., Shinohara T. Biol. Reprod. 2005;72:985–991. doi: 10.1095/biolreprod.104.036400. [DOI] [PubMed] [Google Scholar]
- 16.Kanatsu-Shinohara M., Toyokuni S., Shinohara T. Biol. Reprod. 2005;72:236–240. doi: 10.1095/biolreprod.104.035659. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich G., Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 18.Capecchi M. R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 19.Koller B. H., Smithies O. Annu. Rev. Immunol. 1992;10:705–730. doi: 10.1146/annurev.iy.10.040192.003421. [DOI] [PubMed] [Google Scholar]
- 20.Saitou M., Furuse M., Sasaki H., Schulzke J.-D., Fromm M., Takano H., Noda T., Tsukita S. Mol. Biol. Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanatsu-Shinohara M., Inoue K., Lee J., Yoshimoto M., Ogonuki N., Miki H., Baba S., Kato T., Kazuki Y., Toyokuni S., et al. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Ohta H., Tohda A., Nishimune Y. Biol. Reprod. 2003;69:1815–1821. doi: 10.1095/biolreprod.103.019323. [DOI] [PubMed] [Google Scholar]
- 23.Hatada S., Nikkuni K., Bentley S. A., Kirby S., Smithies O. Proc. Natl. Acad. Sci. USA. 2000;97:13807–13811. doi: 10.1073/pnas.240462897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy A., Gertsenstein M., Vintersten K., Behringer R., editors. Manipulating the Mouse Embryo. Woodbury, NY: Cold Spring Harbor Lab. Press; 2003. pp. 431–451. [Google Scholar]
- 25.Denning C., Priddle H. Reproduction. 2003;126:1–11. doi: 10.1530/rep.0.1260001. [DOI] [PubMed] [Google Scholar]
- 26.Sonoda E., Sasaki M. S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S. Mol. Cell. Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palitti F., Tanzarella C., Cozzi R., Ricordy R., Vitagliano E., Fiore E. Mutat. Res. 1982;103:191–195. doi: 10.1016/0165-7992(82)90028-8. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Schimenti J., Magnuson T. Mamm. Genome. 2000;11:598–602. doi: 10.1007/s003350010114. [DOI] [PubMed] [Google Scholar]
- 29.Hamra F. K., Chapman K. M., Nguyen D. M., Williams-Stephens A. A., Hammer R. E., Garbers D. L. Proc. Natl. Acad. Sci. USA. 2005;102:17430–17435. doi: 10.1073/pnas.0508780102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.te Riele H., Maandag E. R., Berns A. Proc. Natl. Acad. Sci. USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Wu H., Loring J., Hormuzdi S., Disteche C. M., Bornstein P., Jaenisch R. Dev. Dyn. 1997;209:85–91. doi: 10.1002/(SICI)1097-0177(199705)209:1<85::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 32.Longo L., Bygrave A., Grosveld F. G., Pandolfi P. P. Transgenic Res. 1997;6:321–328. doi: 10.1023/a:1018418914106. [DOI] [PubMed] [Google Scholar]
- 33.Dean W., Bowden L., Aitchison A., Klose J., Moore T., Menesses J. J., Reik W., Feil R. Development (Cambridge, U.K.) 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 34.Matsui Y., Zsebo K., Hogan B. L. M. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 35.Resnick J. L., Bixler L. S., Cheng L., Donovan P. J. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 36.Papaioannou V. E., Behringer R. R., editors. Mouse Phenotypes. Woodbury, NY: Cold Spring Harbor Lab. Press; 2005. pp. 1–11. [Google Scholar]
- 37.Austin C. P., Battey J. F., Bradley A., Bucan M., Capecchi M. R., Collins F. S., Dove W. F., Duyk G., Dymecki S., Eppig J. T., et al. Nat. Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seong E., Saunders T. L., Stewart C. L., Burmeister M. Trends Genet. 2004;20:59–62. doi: 10.1016/j.tig.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Yanagimachi R. Adv. Exp. Med. Biol. 2003;518:247–252. doi: 10.1007/978-1-4419-9190-4_22. [DOI] [PubMed] [Google Scholar]
- 40.Ryu B.-Y., Kubota H., Avarbock M. R., Brinster R. L. Proc. Natl. Acad. Sci. USA. 2005;102:14302–14307. doi: 10.1073/pnas.0506970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanatsu-Shinohara M., Toyokuni S., Shinohara T. Biol. Reprod. 2004;71:1202–1207. doi: 10.1095/biolreprod.104.031294. [DOI] [PubMed] [Google Scholar]
- 42.Ye S.-K., Maki K., Kitamura T., Sunaga S., Akashi K., Domen J., Weissman I. L., Honjo T., Ikuta K. Immunity. 1999;11:213–223. doi: 10.1016/s1074-7613(00)80096-5. [DOI] [PubMed] [Google Scholar]
- 43.Kimura Y., Yanagimachi R. Development (Cambridge, U.K.) 1995;121:2397–2405. doi: 10.1242/dev.121.8.2397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.