Abstract
Whether long interspersed element-1 (L1 or LINE-1) retrotransposition can occur in quiescent, nondividing, and/or terminally differentiated somatic cells has remained an unanswered fundamental question in human genetics. Here, we used a ubiquitously active phosphoglycerate kinase-1 promoter to drive the expression of a highly active human L1 element from an adenovirus-L1 hybrid vector. This vector system achieved retrotransposition in up to 91% of actively growing immortalized cells, and we demonstrated that L1 retrotransposition can be suppressed by the reverse transcriptase inhibitor 3′-azido-3′-deoxythymidine. This adenovirus vector enabled efficient delivery of the L1 element into differentiated primary human somatic cells and G1/S-arrested cells, resulting in retrotransposition in both cases; however, it was not detected in G0-arrested cells. Thus, these data indicate that L1 retrotransposition can occur in nondividing somatic cells.
Keywords: adenovirus, LINE-1, quiescent cell, hybrid vector
Retrotransposons are mobile elements that insert into new genomic locations by reverse transcription of an RNA intermediate. Human long interspersed element-1 (L1) elements (1, 2) are non-LTR retrotransposons that comprise >17% of the human genome. The vast majority (>99%) of L1s are inactive because of point mutations, truncations, and other rearrangements; however, it is estimated that the average human diploid genome contains ≈80–100 retrotransposition-competent (RC)- L1s (3). RC-L1s have played and continue to play a significant role in shaping the genome through insertional mutagenesis, nonallelic recombination, and by trans mobilization of non-L1 RNAs (2, 4, 5).
L1 retrotransposition requires transcription of L1 RNA, its transport to the cytoplasm, and translation of its two ORFs (ORF1 and ORF2). Both L1-encoded proteins (ORF1p and ORF2p) are thought to preferentially associate with their own encoding RNA (“cis preference”) to form a ribonucleoprotein particle (RNP) (6, 7), which is a proposed retrotransposition intermediate (8, 9). The L1 RNP must access the nucleus, where the L1 endonuclease cleaves genomic DNA at a degenerate consensus sequence (5′-TTTT/A and variant sequences) to liberate a 3′ hydroxyl residue that is subsequently used by the L1 reverse transcriptase (RT) as a primer to copy the L1 sequence in situ, a process termed “target-primed reverse transcription” (2, 10). The resultant L1 cDNA then is joined to target DNA, leading to typical L1 structural hallmarks [5′ truncations and/or internal inversions, 3′ poly (A) tail, and target-site duplications (TSDs)]. Although a putative nucleolar localization signal has been identified in ORF2p (11), it still remains unclear whether the L1 RNP crosses an intact nuclear membrane or whether its entry requires mitotic nuclear envelope breakdown.
We previously developed a hybrid vector system consisting of a high-capacity, helper-dependent adenovirus vector encoding a human RC-L1 element (L1.3) tagged with a neomycin-resistance (neoR) retrotransposition indicator cassette, and demonstrated that this L1 element could efficiently retrotranspose from the adenoviral backbone into HeLa genomic DNA (12). Here, we used the ubiquitously active heterologous promoter, mouse phosphoglycerate kinase-1 (pgk), to drive the expression of a more active human L1 (L1RP) tagged with an EGFP retrotransposition indicator cassette and inserted this engineered L1 into a second-generation adenovirus-retrotransposon (A/RT) hybrid virus (A/RT-pgk-L1RP-EGFP). This hybrid virus achieves retrotransposition frequencies of up to 91% in actively growing immortalized cell lines and can mediate L1 retrotransposition in certain differentiated human primary somatic cells, including dermal fibroblasts and hepatocytes. Furthermore, we have demonstrated that L1 can retrotranspose in nondividing cells arrested in the G1/S phase of the cell cycle.
Results
Design of a Second-Generation A/RT Vector.
An A/RT vector plasmid, pA/RT-pgk-L1RP-EGFP, was created by subcloning the pgk-RC-L1RP sequence tagged with an EGFP retrotransposition indicator cassette into an high-capacity, helper-dependent adenovirus vector plasmid encoding only the adenoviral ITRs and packaging signal (Fig. 1a). Additional stuffer sequences brought the overall A/RT vector construct contained in the plasmid above the lower size limit for packaging into adenovirus capsids. Once the pgk-L1RP-EGFP sequence is delivered into a cell, whether by chemical transfection of the original plasmid or by infection with helper-dependent adenovirus derived from encapsidation of the linearized A/RT construct, the retrotransposition indicator cassette becomes functional only after a cycle of transcription, reverse transcription, and integration (Fig. 1a; see also Fig. 2c) (13, 14).
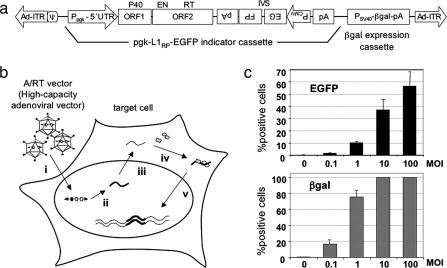
Fig. 1.
Outline of the adenovirus/retrotransposon hybrid vector (A/RT vector) system. (a) Schematic structure of the A/RT vector. The A/RT vector is a high-capacity, helper-dependent adenovirus vector. The sequences composing the A/RT vector are, from left to right: the 5′ inverted terminal repeat (Ad-ITR) of human adenovirus type 5 including the packaging signal (ψ), the L1RP-EGFP cassette, a β-gal marker expression cassette, and the 3′ Ad-ITR. The pgk-L1RP-EGFP cassette consists of a mouse phosphoglycerate kinase promoter-1 (Ppgk), the 5′ UTR, and two ORFs [ORF1, P40; ORF2, endonuclease (EN)/RT] of L1RP, the EGFP transgene, and a polyadenylation signal (pA). The EGFP transgene cloned into the 3′ UTR of L1RP in the reverse orientation (PFGE) is flanked by the CMV promoter (PCMV) and inverted pA sequences also in the reverse orientation and is interrupted by a forward-splicing intron (IVS) (13, 14). The structure of the splice-interrupted EGFP indicator gene cassette before and after L1 retrotransposition is shown in Fig. 2c. This A/RT vector construct can be introduced into cells via direct transfection in the form of plasmid DNA or via infection after being packaged into an adenovirus. (b) Two-stage transduction with the A/RT virus. The A/RT virus infects target cells efficiently as an adenovirus (i). A full-length active L1 element is transcribed from the constitutively active pgk promoter to produce bicistronic mRNA (ii). The RNA undergoes processing and is exported from the nucleus (iii). In the cytoplasm, the ORF1 and ORF2 proteins are translated and specifically function on the RNA that transcribed them (cis preference) (6, 7). The L1 RNA molecule, ORF1, and ORF2 proteins assemble into a ribonucleoprotein complex that is an intermediate in retrotransposition (iv) and then imported back into the nucleus, where the L1 RNA is reverse transcribed and integrated into the host genome for stable gene expression (v) (2). (c) Retrotransposition after A/RT virus infection. Gli36 cells were infected with the A/RT virus at different mois. Seven days after infection, the cells were analyzed for expression of β-gal from the adenovirus backbone and EGFP from retrotransposition events. Data shown are averages and SDs from experiments performed in quadruplicate.
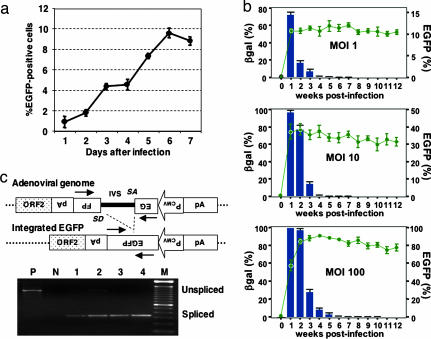
Fig. 2.
Time course of retrotransposition in cells infected with the A/RT virus. (a) Increased retrotransposition in cells infected with the A/RT virus. Gli36 cells were infected with A/RT virus at a moi of 10, harvested every day, and assayed for EGFP expression. The mean and SD from experiments performed in triplicate are shown. (b) Prolonged and stable EGFP expression in retrotransposed cells. Gli36 cells were infected with A/RT virus at different mois, passaged every week, and the expression of EGFP (green curves) and β-gal (blue bars) was assayed up to 12 weeks after infection. The mean and SD from experiments performed in quadruplicate are shown. (c) PCR assay for correct splicing of the artificial intron in the EGFP gene after retrotransposition from the A/RT virus. Genomic DNA was extracted from the infected cells at different mois and time points and used as the template for PCR. This strategy allows distinction of the spliced and reverse-transcribed form of the EGFP gene (343 bp) from the original unspliced form (1,244 bp) and confirmed integration via authentic retrotransposition (13, 14). Lane M, 100-bp DNA ladder (Stratagene); lane P, A/RT virion DNA; lane n, negative control genomic DNA from uninfected Gli36 cells. Lanes 1–4, genomic DNA extracted from the infected cells at a moi of 1 (lanes 1 and 3) or 10 (lanes 2 and 4) at 1 week (lanes 1 and 2) or 4 weeks (lanes 3 and 4) after infection.
The retrotransposition capacity of L1RP is roughly 3-fold that of L1.3 (13, 15), the element used in our first-generation A/RT system (12). Moreover, because the L1 promoter (residing in the L1 5′ UTR) is only active in certain cell types (14, 16, 17), we augmented the expression of L1RP with the mouse pgk promoter. This modification enhanced the retrotransposition efficiency of L1RP in HeLa and 143B osteosarcoma cell lines by roughly 3-fold (data not shown). Together, these modifications suggested a potential for 9-fold improvement in our second-generation A/RT system.
Confirmation of L1 Retrotransposition After A/RT Vector Plasmid Transfection.
After chemical transfection of the pA/RT-pgk-L1RP-EGFP plasmid into different cell lines, a low but significant percentage of cells positive for GFP fluorescence was observed after 2 weeks in culture (i.e., 2.7%, 1.0%, and 1.3% in 293T human embryonic kidney, Gli36 human glioma, and Hep3B human hepatoma cell lines, respectively). The number of EGFP-positive cells then remained at a similar level after 4 weeks in culture, indicating that L1 retrotransposition had resulted in stable integration of the EGFP gene. By comparison, cells transfected with the control plasmid pEGFP-N1, which does not contain any retrotransposon sequences, exhibited a steady decrease in EGFP expression over the course of the assay (from 97.2%, 58.8%, and 45.9%, respectively, in 293T, Gli36, and Hep3B cells on day 1 of the assay, to <0.5% in all cell lines by 3 weeks after transfection; data not shown). These data indicated that the A/RT hybrid sequence in the plasmid encoded a functional L1 that could undergo retrotransposition after cell transfection at efficiencies comparable with those reported previously for L1RP (13, 15).
High-Efficiency L1 Retrotransposition After Infection with A/RT Helper-Dependent Adenovirus.
We next generated a second-generation A/RT helper-dependent adenovirus by transfection of the linearized pA/RT-pgk-L1RP-EGFP vector plasmid into 293FLPe cells, followed by infection with FL helper adenovirus, which provides adenoviral proteins but itself cannot be packaged (18). The helper-derived adenoviral proteins transiently mediate packaging of the linearized A/RT vector sequence into virion particles as if it were an adenoviral genome, but because the encapsidated A/RT vector is devoid of any adenoviral coding sequences, the resultant A/RT adenovirus itself is replication-defective upon target cell infection in the absence of additional helper virus.
In addition to the RC-pgk-L1RP-EGFP cassette, the encapsidated A/RT vector sequence also contains a β-gal marker gene that is inserted into the stuffer sequences which replace the adenoviral backbone and whose expression does not require retrotransposition of the L1 element; β-gal expression therefore serves as an independent indicator of adenovirus infection efficiency (Fig. 1a). After serial propagation, the β-gal titer of the helper-dependent A/RT adenovirus on 293 cells was determined to be 4.1 × 109 to 1.8 × 1010 transducing units per ml (TU per ml). The helper-dependent adenovirus stock contained <0.1% helper virus contamination, as determined by Southern hybridization, by using a probe for the adenoviral packaging signal (data not shown).
Because the β-gal marker gene in the adenoviral backbone is independent of the retrotransposon cassette, it is possible to discriminate untransduced cells [β-gal(−)/EGFP(−)], A/RT virus-infected cells before retrotransposition [β-gal(+)/EGFP(-)], A/RT virus-infected cells after retrotransposition [β-gal(+)/EGFP(+)], and postretrotransposition cells from which the adenoviral backbone vector has disappeared [β-gal(−)/EGFP(+)]. After infection of Gli36 cells with the A/RT virus, L1 retrotransposition was observed to increase in a multiplicity of infection (moi)-dependent manner, with the percentage of EGFP-positive cells reaching 37% and 57% at a moi of 10 and 100, respectively (Fig. 1c). Thus, the retrotransposition efficiency from the second-generation A/RT virus was markedly greater than that achieved by direct plasmid transfection, or our first-generation A/RT virus (12). Similar results also were obtained by using various other cell lines, (i.e., 293T, Hep3B human hepatoma, HeLa human cervical carcinoma, and Hepa1–6 mouse hepatoma cells) (data not shown), indicating that the second-generation A/RT virus allowed retrotransposition in a variety of transformed cell types derived from different tissues and/or organisms.
Time Course of A/RT-Virus Infection-Mediated Retrotransposition.
To determine the time course of retrotransposition, Gli36 cells were infected with the A/RT-pgk-L1RP-EGFP virus at a moi of 10, and the infected cells were examined by flow cytometry once a day for 1 week. A very low percentage (0.8%) of EGFP-positive cells was detected 24 h after infection, but a >10-fold increase in the EGFP-positive population was seen over the next 6 days (Fig. 2a), suggesting the accumulation of ongoing retrotransposition events. Further increases in the percentage of EGFP-positive cells were detected during the following week after A/RT virus infection at mois of 1 and 10, and for at least 2 weeks after infection at a moi of 100, eventually reaching 91.5% EGFP (+) cells. At each moi, the level of retrotransposition then reached a plateau, which correlated with a marked decrease in β-gal expression (Fig. 2b). EGFP (+)/β-gal (−) cells were then observed for at least 12 weeks after infection (Fig. 2b), indicating stable expression from the retrotransposed EGFP gene after the loss of the original adenovirus vector. Finally, we also found that L1 retrotransposition was suppressed by >30-fold when cells were treated with 3′-azido-3′-deoxythymidine (from 55.7 ± 7.4% to 1.7 ± 0.4%, in triplicate; P < 0.01), which is comparable to the extent of suppression observed for lentiviral vectors (≈37-fold from 44.2 ± 1.5% to 1.2 ± 0.1%, in triplicate; P < 0.01).
Integration Events from A/RT Virus Infection Exhibit L1 Structural Hallmarks.
To verify that L1 retrotransposed into the target cell genome, genomic DNA was extracted from Gli36 cells infected with A/RT-pgk-L1RP-EGFP virus at mois of 1 and 10 both 1 week and 4 weeks after infection (Fig. 2c). PCR was then performed by using primers flanking the intron in the EGFP gene to determine whether the intron was spliced from the retrotransposition indicator cassette. At 1 week after infection, two bands were detected, a product of 343 bp, which represents the spliced EGFP gene, and a product of 1,244 bp, which is diagnostic for the unspliced EGFP gene in the A/RT-pgk-L1RP-EGFP adenovirus (Fig. 2c). By comparison, only the spliced PCR product was detected at 4 weeks after infection, which is consistent with the finding that β-gal expression is near baseline levels at this time point (<0.1% of control levels; Fig. 2 b and c).
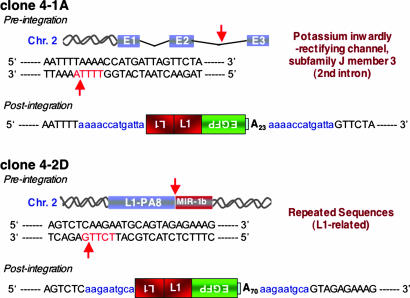
Analysis of the postintegration sites of two independent retrotransposition events revealed L1 structural hallmarks (Fig. 3). They inserted at preferred L1 endonuclease cleavage sites, contain a 5′ truncated, inversion/deletion structure, end in a 3′ poly(A) tail, and are flanked by short target-site duplications (9 and 13 bp, respectively). The first event (clone 4-1A) integrated into an annotated intron of the KCNJ3 gene, whereas the second event (clone 4-2D) integrated in a series of repeat sequences. Thus, these data demonstrated authentic retrotransposition from the A/RT virus.
Fig. 3.
A/RT virus-mediated retrotransposition events show structural hallmarks of LINE-1 retrotransposition. Two events derived from A/RT virus-infected Gli36 cells were characterized. The flanking genomic sequences were identified by using blat searches against the Human Genome Sequence (Assembly May 2004; http://genome.ucsc.edu). The cartoons show the relative positions of each insertion in chromosomal DNA and the sequences of the respective preintegration and postintegration sites. The blue letters in the postintegration sequences indicate the target site duplications. The vertical arrows and red lettering indicate the L1 endonuclease cleavage sites in the preintegration sequences. Both insertions analyzed (4-1A and 4-2D) show hallmarks of conventional target-primed reverse transcription-dependent retrotransposition (2, 19, 20).
Retrotransposition from A/RT Virus in Primary Human Cells.
Because adenoviruses can efficiently transduce nonimmortalized cells, we next tested whether primary cultures of human dermal fibroblasts and hepatocytes could accommodate L1 retrotransposition. Again, β-gal expression was used to determine the adenoviral transduction efficiency, and retrotransposition (i.e., EGFP expression) was analyzed 7 days after infection. Staining for β-gal demonstrated efficient infection of these primary cells by the A/RT virus (Table 1), and we observed a moi-dependent increase in L1 retrotransposition efficiency. Although the L1 retrotransposition efficiency was ≈10-fold lower in primary cells when compared with the previously tested immortalized cell lines (i.e., 1.8% at a moi of 10 in fibroblasts, and 2.7% at a moi of 10 in hepatocytes; Table 1), these data provide “proof of principle” that L1 retrotransposition can occur in certain primary cells when the expression of the retroelement does not depend on the 5′ UTR alone.
Table 1.
Retrotransposition in primary human fibroblasts and hepatocytes
| A/RT virus amount used for infection, moi | Hepatocyte |
Fibroblast |
||||
|---|---|---|---|---|---|---|
| Exp. 1 |
Exp. 2 |
|||||
| β-gal | EGFP | β-gal | EGFP | β-gal | EGFP | |
| 0 | 0 | 0.1 | 0 | 0.1 | 0 | 0.1 |
| 0.1 | 10 | 0.3 | 20 | 0.1 | 10 | 0.1 |
| 1 | 40 | 0.5 | 70 | 0.5 | 70 | 0.9 |
| 10 | 90 | 2.7 | 90 | 1.5 | 90 | 1.8 |
Cells (2 × 105) were infected with various amounts of A/RT virus and assayed for EGFP and β-gal expression 7 days after infection.
Retrotransposition from A/RT Virus Occurs in G1/S-Arrested Cells.
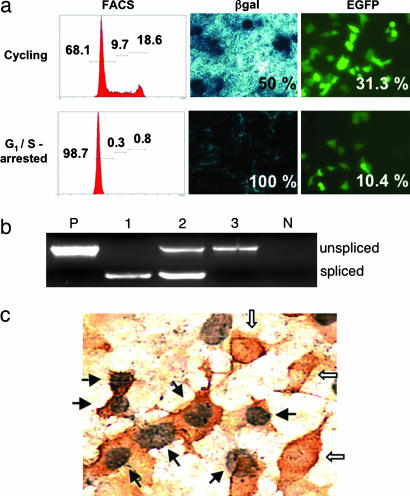
To investigate whether L1 retrotransposition could occur in nondividing cells, Gli36 cells were placed in G1/S arrest by serum reduction and aphidicolin treatment, and the cell cycle status of the population was confirmed by flow cytometry (Fig. 4a). One day after transduction with the A/RT-pgk-L1RP-EGFP adenovirus at a moi of 10, all of the arrested cells were β-gal-positive but EGFP-negative (data not shown). However, by 5 days after infection, EGFP fluorescence was detected in 10.4% of G1/S-arrested Gli36 cells and in 31.3% of a parallel culture of dividing Gli36 cells (Fig. 4a). The presence of the spliced EGFP gene in both cell populations was confirmed by PCR (Fig. 4b). Immunocytochemistry performed on confluent cultures of Gli36 cells incubated in the presence of BrdU identified EGFP(+)/BrdU(−) cells (Fig. 4c, open arrows), further verifying that L1 retrotransposition could take place in nondividing cells.
Fig. 4.
Functional analysis of retrotransposition in G1/S-arrested nondividing cells. (a) A/RT retrotransposition occurs in G1/S-arrested cells. G1/S-arrested cells were infected initially with the A/RT virus at a moi of 10 and maintained in the nondividing state without passaging (G1/S-arrested) for 5 days or passaged the day after infection and allowed to resume cell division (Cycling). At 5 days after infection, all sets were analyzed for cell cycle status by propidium iodide staining (FACS; inset numbers from the left indicate percentages of cells in G1/G0, S, and G2/M, respectively) and for expression of β-gal from the adenovirus backbone and EGFP from retrotransposition events (inset numbers indicate percentages of positive cells). (b) PCR assay for retrotransposition in Gli36 cells after A/RT infection. G1/S-arrested and G0 cells were infected with A/RT virus (moi 10). On day 5 after infection, total DNA was extracted from the infected cells and used as the template for PCR. Lane P, original A/RT vector plasmid (pA/RT-pgk-L1RP-EGFP); lane 1, growing cells; lane 2, G1/S-arrested cells; lane 3, G0 cells; lane N, noninfected cells as a negative control. (c) BrdU incorporation and EGFP expression in confluent Gli36 cells after A/RT infection. Confluent cells were infected with A/RT virus at a moi of 10 and incubated in the presence of BrdU. Five days later, cells were stained for BrdU incorporation (nuclear staining, black) and EGFP expression (cytoplasmic staining, red). The solid arrows point to cells that are BrdU-positive (indicating proliferation) and EGFP-positive (indicating retrotransposition), and the open arrows point to cells that are BrdU-negative (nonproliferating) yet EGFP-positive (indicating retrotransposition) (×400).
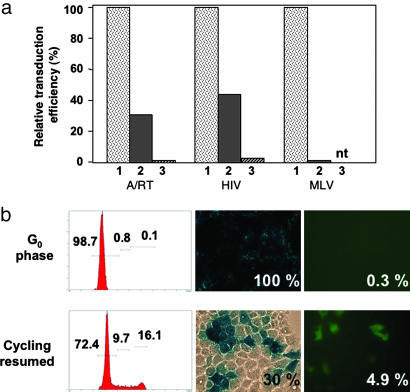
The nuclear accessibility of the G1/S arrested population also was confirmed functionally by challenging the cells with either an EGFP-containing HIV-based replication-defective lentiviral vector (HIV; sinSKcmv-EGFP vector) or an EGFP-containing amphotropic Moloney murine leukemia virus (MLV)-based replication-competent retroviral vector (MLV; ACEemd vector) (21). As predicted, the HIV-based lentiviral vector still was able to transduce these cells under G1/S-arrest conditions, consistent with its ability to undergo active nuclear uptake in the absence of active mitosis (Fig. 5a, HIV 2). In contrast, no transduction of G1/S-arrested cells could be observed with the MLV-based retroviral vector (Fig. 5a, MLV 2), even though this MLV vector was a replication-competent virus designed to be fully capable of efficient horizontal transmission with concomitant amplification of the GFP signal if there had been any significant level of residual cell division (21). Thus, A/RT adenovirus-mediated delivery to these nondividing cells allowed demonstration of L1 retrotransposition under conditions permissive for lentivirus but not oncoretrovirus infection (Fig. 5a, A/RT 2).
Fig. 5.
Retrotransposition occurs in G1/S-arrested but not in G0 cells. (a) Retrotransposition in stationary cells infected with A/RT virus. Actively dividing (1), G1/S-arrested (2), and G0 phase (3) Gli36 cells infected with A/RT virus at a moi of 10 were prepared as described in Materials and Methods, and after confirmation of cell cycle status by FACS and adenovirus infection efficiency by β-gal, the cells were analyzed for expression of EGFP. In parallel control experiments, actively dividing, G1/S-arrested, and G0 phase cells also were infected with either HIV-based self-inactivating lentivirus vector (HIV) or replication-competent retrovirus (MLV) and also were analyzed for EGFP expression. nt, not tested. Data are representative of three independent experiments, all yielding similar results. (b) A/RT retrotransposition does not occur in G0 phase cells. G0 phase cells were infected with the A/RT virus at a moi of 10 and maintained in the nondividing state without passaging (G0 phase) for 5 days. At 5 days after infection, cell cycle status and expression of β-gal and EGFP were examined as above. A parallel set of cells infected as above were then passaged and allowed to resume cell division (cycling resumed) and analyzed 7 days later.
Lack of Retrotransposition from A/RT Virus in G0-Phase Cells.
To determine whether L1 retrotransposition also could occur under fully quiescent (G0) conditions, we prepared both G1/S-arrested and G0-arrested Gli36 cells (see Materials and Methods). Flow cytometry and Western analysis together confirmed that these cell populations were arrested in, respectively, the G1/S phase [Ki67(+/−)/cyclin D1(+)] or the G0 phase [Ki67(−)/cyclin D1(+/−)] of the cell cycle (Fig. 5b and data not shown). The cell cycle status under each condition was confirmed again functionally by challenge with the HIV-based lentiviral vector or the MLV-based retroviral vector, and as predicted, the MLV-based vector was unable to transduce either G1/S-arrested or G0-phase cells, whereas the HIV-based vector was capable of transducing G1/S-arrested cells (Fig. 5a). However, the lentiviral vector could not transduce G0-phase cells (Fig. 5a, HIV 3), presumably because of the lack of adequate free nucleotide pools to support reverse transcription (22–27).
After infection with the A/RT virus, retrotransposition once again was observed in G1/S-arrested cells but was not observed in G0 phase cells (0.3%) (Fig. 5). The inability of G0 cells to support retrotransposition was confirmed by PCR analysis, which demonstrated only the presence of the unspliced EGFP sequence (Fig. 4b, lane 3). Thus, although the A/RT adenovirus successfully entered the G0-arrested cells, the cells could not support L1 retrotransposition. However, when those cells were replated and were allowed to resume proliferation, EGFP-positive cells were observed readily after 7 days of incubation (4.9% of the cells were EGFP-positive; Fig. 5b).
Discussion
To date, L1 retrotransposition has been detected only in germ cells (16, 28–32), transformed or immortalized cells (13, 33–35), and, most recently, in select rodent somatic cells (16), and it remained unclear whether cell division is required for L1 retrotransposition. Unfortunately, the inability to efficiently introduce engineered RC-L1s expressed from EBNA/ori P-based plasmids into quiescent cells has made it difficult to answer this question unambiguously. The pgk-driven L1RP element delivered via helper-dependent adenovirus achieved a significant enhancement in retrotransposition efficiencies when compared with direct plasmid transfection of the same element or our first generation A/RT virus containing the L1.3 element. This technical improvement was instrumental in allowing the detection of retrotransposition in G1/S-arrested cells and in a cohort of primary human cell lines.
Why is the second-generation A/RT virus vector system so efficient for L1 retrotransposition? First, it is possible that the adenoviral nucleoprotein structure shields the encapsidated viral sequences from the action of cellular nucleases, allowing more-efficient delivery and prolonged expression of the encoded L1 genome, compared with previously used plasmid-based transient transfection protocols. For example, it has been reported that adenoviral terminal-binding protein enables the linear adenoviral genome to resist degradation by exonucleases (36). Second, the inclusion of a ubiquitous promoter (pgk) in addition to the L1 5′ UTR driving L1 transcription likely allows L1 expression in cell types that usually do not support the expression of endogenous L1s. It should be noted that, although our previous A/RT vector also contained a ubiquitous (CMV) promoter driving the L1.3 element, quantitative head-to-head comparison of efficiencies is difficult because of differences in the experimental conditions [e.g., prep-to-prep variation in virion particle: infectious unit ratio, differences in indicator gene detection sensitivity (neoR vs. GFP), and in target cell types, and time points examined]. Furthermore, the genomic structure of the first-generation and second-generation A/RT viruses are different, because the former contains two copies of the L1.3 cassette forming a “tail-to-tail” concatemer (12), which is potentially recombinogenic and could incur antisense inhibitory effects, whereas the current vector is large enough that it does not require concatemerization for efficient viral packaging and contains only a single copy of the L1RP cassette. It is also noteworthy that the retrotransposition efficiency observed in primary cells was significantly less than that observed in G1/S-arrested immortalized cell lines, suggesting that other factors act to limit retrotransposition in primary cells.
Nonetheless, the ability of this engineered L1 to retrotranspose in primary cells and nondividing cells not only suggests the potential for use of modified retroelements as a gene delivery vehicle, but more importantly, made it feasible for us to address a fundamental unanswered question in L1 biology; i.e., whether L1 elements can be active in quiescent cells in the absence of nuclear membrane breakdown. We have demonstrated that, in fact, L1 retrotransposition can occur in G1/S-arrested cells, under conditions in which MLV retrovirus cannot gain entry because of its lack of nuclear localization signals, whereas lentiviruses, which do display such signals, can be actively transported into the nucleus.
In G0-phase cells, however, despite successful infection by the A/RT adenovirus, L1 retrotransposition did not occur but proceeded only when the cells were allowed to resume proliferation. This phenomenon was strikingly reminiscent of the inability of lentiviruses such as HIV to infect cells in G0 phase, which has been attributed to insufficient availability of free nucleotide pools to sustain efficient reverse transcription (22–27). This mechanism also might be invoked to explain the ability of L1 to retrotranspose in G1/S but not in G0 phase.
Previous velocity sedimentation analyses have shown that the ORF1p and L1 RNA colocalize in an RNP complex that cosediments with ribosomes and polyribosomes (7–9, 37, 38). If this complex also contains ORF2p and is indeed a retrotransposition intermediate, it most probably is too large to pass through the nuclear pore complex by simple diffusion. Instead it must gain nuclear access by an energy-dependent, active transport mechanism. The ability of L1 to retrotranspose in G1/S-arrested cells suggests that the L1 RNP is transported across the nuclear membrane, which is reminiscent of the situation observed for the Tad-1 non-LTR retrotransposon from Neurospora crassa (39). Future studies should focus on the specific nuclear import pathways used by L1 elements.
Materials and Methods
Cells.
Cell lines [293 (40), 293T (41), Gli36 human glioma (42), and Hep3B human hepatocellular carcinoma] were cultured under standard conditions as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Primary human dermal fibroblasts (NHDF; CC-2509), hepatocytes (CC-2591), and their specific medium were purchased from Clonetics.
Vectors.
The plasmid pA/RT-pgk-L1RP-EGFP contains two independent cassettes, the marker gene cassette SV40-β-gal and the RC-L1 retrotransposon-reporter cassette pgk-L1RP-EGFP, both subcloned into the pSTK68 high-capacity, helper-dependent adenovirus vector backbone (43). Direct plasmid transfection experiments were performed by lipofection of pA/RT-pgk-L1RP-EGFP or control plasmid pEGFP-N1 (Clontech), followed by flow cytometry to examine EGFP expression at serial time points. For infection experiments, A/RT-pgk-L1RP-EGFP virus was prepared by using the FLPe/FRT helper virus system as described in ref. 18, and the β-gal titer of the virus in transducing units per ml was determined by X-Gal staining of infected 293 cells. Helper virus contamination levels were determined by Southern blot by using a probe for the adenoviral packaging signal (43).
Lentivirus preparations were produced by cotransfection of 293T cells with the lentiviral vector construct psinSKcmv-EGFP [a variant of pRRL-sin.hCMV-EGFP-pre (44)], pMDLg/p (encoding gag-pol), pRSV-Rev (encoding rev), and pMD.G (encoding VSV-G env) as described in ref. 45.
Retrovirus preparations were produced by transfection of 293T cells with retroviral vector construct pACEemd (21), which encodes a fully replication-competent amphotropic MLV vector carrying an internal ribosome entry site-EGFP marker gene cassette inserted between the env gene and 3′ UTR, as described in ref. 21. Further details are provided in Supporting Materials and Methods.
Retrotransposition Analysis.
After infection with the A/RT-pgk-L1RP-EGFP virus at various mois, retrotransposition was examined at serial time points by FACS analysis for EGFP and X-Gal staining for β-gal expression. Genomic DNA from infected cells was used for PCR analysis of retrotransposon integration as described in refs. 13 and 14.
To test the effect of RT inhibitors on retrotransposition, cells infected with either A/RT virus (moi 10) or control lentivirus vector (sinSKcmv-EGFP) were maintained with or without 5 μM 3′-azido-3′-deoxythymidine (Sigma), followed by FACS analysis as above.
For integration site analysis, XbaI- or SspI-digested genomic DNA from individual A/RT virus-infected Gli36 cell subclones was self-ligated, and amplification products of inverse PCR by using nested primers hybridizing to the EGFP cassette (7, 35) were cloned and sequenced. Additional details are provided in Supporting Materials and Methods.
Cell Cycle Arrest.
G1/S-arrest was induced by exposing Gli36 cells to aphidicolin (10 μg/ml) with reduced serum (5%) 24 h before and after infection. G0 phase was induced by maintaining cells at confluency with reduced serum (2%) for 1 week. Cell cycle status was confirmed by propidium iodide/FACS and Ki67/cyclin D1 immunoblots. After infection with replication-competent retrovirus (ACEemd, MLV) (21), self-inactivating lentiviral vector (sinSKcmv-EGFP, HIV), or A/RT-pgk-L1RP-EGFP virus (moi 10), EGFP and β-gal expression were examined as above. Immunocytochemistry of confluent Gli36 cells infected with A/RT virus also was performed to compare BrdU incorporation and EGFP expression. Further details are provided in Supporting Materials and Methods.
Statistical Analysis.
The results are presented as mean ± SD. Student’s t test was used to evaluate differences, and P < 0.01 was considered significant.
Supplementary Material
Acknowledgments
We thank Pedro Lowenstein (Cedars-Sinai Medical Center, Los Angeles) for kindly providing the FLPe/FRT helper virus system, Stefan Kochanek (University of Ulm, Ulm, Germany) for the pSTK plasmid, Didier Trono (University of Geneva, Geneva) for lentiviral constructs, and Christopher Logg (University of California, Los Angeles) for the ACEemd vector and for suggesting the use of 3′-azido-3′-deoxythymidine as a potential inhibitor of retrotransposition. This work was supported by National Institutes of Health Grants R01 CA93709 (to N.K.), R01 GM45398 (to H.H.K.), and R01 GM60518 (to J.V.M.). H.S.S. is supported by a Beckman Fellowship from the Arnold and Mabel Beckman Foundation, and J.L.G.P. was supported by Ministerio de Educación y Cultura/Fulbright Postdoctoral Grant EX-2003-0881 (Spain).
Abbreviations
- A/RT
adenovirus-retrotransposon hybrid
- L1
long interspersed element-1
- MLV
murine leukemia virus
- moi
multiplicity of infection
- pgk
phosphoglycerate kinase-1
- RC
retrotransposition-competent
- RNP
ribonucleoprotein particle
- RT
reverse transcriptase
- TU
transducing units.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Singer M. F., Krek V., McMillan J. P., Swergold G. D., Thayer R. E. Gene. 1993;135:183–188. doi: 10.1016/0378-1119(93)90064-a. [DOI] [PubMed] [Google Scholar]
- 2.Ostertag E. M., Kazazian H. H., Jr. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 3.Sassaman D. M., Dombroski B. A., Moran J. V., Kimberland M. L., Naas T. P., DeBerardinis R. J., Gabriel A., Swergold G. D., Kazazian H. H., Jr. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 4.Deininger P. L., Moran J. V., Batzer M. A., Kazazian H. H., Jr. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Kazazian H. H., Jr. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 6.Esnault C., Maestre J., Heidmann T. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 7.Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M., Kazazian H. H., Boeke J. D., Moran J. V. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohjoh H., Singer M. F. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 9.Kulpa D. A., Moran J. V. Hum. Mol. Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 10.Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 11.Goodier J. L., Ostertag E. M., Engleka K. A., Seleme M. C., Kazazian H. H., Jr. Hum. Mol. Genet. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 12.Soifer H., Higo C., Kazazian H. H., Jr., Moran J. V., Mitani K., Kasahara N. Hum. Gene Ther. 2001;12:1417–1428. doi: 10.1089/104303401750298571. [DOI] [PubMed] [Google Scholar]
- 13.Ostertag E. M., Prak E. T., DeBerardinis R. J., Moran J. V., Kazazian H. H., Jr. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostertag E. M., DeBerardinis R. J., Goodier J. L., Zhang Y., Yang N., Gerton G. L., Kazazian H. H., Jr. Nat. Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 15.Kimberland M. L., Divoky V., Prchal J., Schwahn U., Berger W., Kazazian H. H., Jr. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 16.Muotri A. R., Chu V. T., Marchetto M. C., Deng W., Moran J. V., Gage F. H. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 17.Swergold G. D. Mol. Cell. Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umana P., Gerdes C. A., Stone D., Davis J. R., Ward D., Castro M. G., Lowenstein P. R. Nat. Biotechnol. 2001;19:582–585. doi: 10.1038/89349. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert N., Lutz-Prigge S., Moran J. V. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert N., Lutz S., Morrish T. A., Moran J. V. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logg C. R., Logg A., Matusik R. J., Bochner B. H., Kasahara N. J. Virol. 2002;76:12783–12791. doi: 10.1128/JVI.76.24.12783-12791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick P. J., Danhauser L. L., Rustum Y. M., Bertram J. S. Biochim. Biophys. Acta. 1983;756:36–40. doi: 10.1016/0304-4165(83)90021-1. [DOI] [PubMed] [Google Scholar]
- 23.Meyerhans A., Vartanian J. P., Hultgren C., Plikat U., Karlsson A., Wang L., Eriksson S., Wain-Hobson S. J. Virol. 1994;68:535–540. doi: 10.1128/jvi.68.1.535-540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korin Y. D., Zack J. A. J. Virol. 1998;72:3161–3168. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 26.Schuitemaker H., Kootstra N. A., Fouchier R. A., Hooibrink B., Miedema F. EMBO J. 1994;13:5929–5936. doi: 10.1002/j.1460-2075.1994.tb06938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen A., Barankiewicz J., Lederman H. M., Gelfand E. W. J. Biol. Chem. 1983;258:12334–12340. [PubMed] [Google Scholar]
- 28.Kazazian H. H., Jr., Wong C., Youssoufian H., Scott A. F., Phillips D. G., Antonarakis S. E. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 29.Holmes S. E., Dombroski B. A., Krebs C. M., Boehm C. D., Kazazian H. H., Jr. Nat. Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 30.Narita N., Nishio H., Kitoh Y., Ishikawa Y., Minami R., Nakamura H., Matsuo M. J. Clin. Invest. 1993;91:1862–1867. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Divoky V., Indra K., Mrug M., Brabec V., Huisman T. H. J., Prchal J. T. Blood. 1996;88:148a. [Google Scholar]
- 32.Schwahn U., Lenzner S., Dong J., Feil S., Hinzmann B., van Duijnhoven G., Kirschner R., Hemberger M., Bergen A. A., Rosenberg T., et al. Nat. Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- 33.Miki Y., Nishisho I., Horii A., Miyoshi Y., Utsunomiya J., Kinzler K. W., Vogelstein B., Nakamura Y. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 34.Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., Kazazian H. H., Jr. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 35.Morrish T. A., Gilbert N., Myers J. S., Vincent B. J., Stamato T. D., Taccioli G. E., Batzer M. A., Moran J. V. Nat. Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 36.Lieber A., He C. Y., Kay M. A. Nat. Biotechnol. 1997;15:1383–1387. doi: 10.1038/nbt1297-1383. [DOI] [PubMed] [Google Scholar]
- 37.Martin S. L., Bushman F. D. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolosha V. O., Martin S. L. Proc. Natl. Acad. Sci. USA. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinsey J. A. Proc. Natl. Acad. Sci. USA. 1993;90:9384–9387. doi: 10.1073/pnas.90.20.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham F. L., Smiley J., Russell W. C., Nairn R. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 41.DuBridge R. B., Tang P., Hsia H. C., Leong P. M., Miller J. H., Calos M. P. Mol. Cell. Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sena-Esteves M., Hampl J. A., Camp S. M., Breakefield X. O. J. Gene Med. 2002;4:229–239. doi: 10.1002/jgm.276. [DOI] [PubMed] [Google Scholar]
- 43.Kubo S., Mitani K. J. Virol. 2003;77:2964–2971. doi: 10.1128/JVI.77.5.2964-2971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Follenzi A., Ailles L. E., Bakovic S., Geuna M., Naldini L. Nat. Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 45.Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., Naldini L. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.