Abstract
The two-headed kinesin motor harnesses the energy of ATP hydrolysis to take 8-nm steps, walking processively along a microtubule, alternately stepping with each of its catalytic heads in a hand-over-hand fashion. Two persistent challenges for models of kinesin motility are to explain how the two heads are coordinated (“gated”) and when the translocation step occurs relative to other events in the mechanochemical reaction cycle. To investigate these questions, we used a precision optical trap to measure the single-molecule kinetics of kinesin in the presence of substrate analogs beryllium fluoride or adenylyl-imidodiphosphate. We found that normal stepping patterns were interspersed with long pauses induced by analog binding, and that these pauses were interrupted by short-lived backsteps. After a pause, processive stepping could only resume once the kinesin molecule took an obligatory, terminal backstep, exchanging the positions of its front and rear heads, presumably to allow release of the bound analog from the new front head. Preferential release from the front head implies that the kinetics of the two heads are differentially affected when both are bound to the microtubule, presumably by internal strain that is responsible for the gating. Furthermore, we found that ATP binding was required to reinitiate processive stepping after the terminal backstep. Together, our results support stepping models in which ATP binding triggers the mechanical step and the front head is gated by strain.
Keywords: motor coordination, optical tweezers, single-molecule biophysics, gating, processivity
Conventional kinesin, the founding member of the Kinesin-1 family, uses energy from ATP hydrolysis to transport cellular cargo, taking 8-nm steps along microtubules (MTs) (1). Kinesin molecules are formed from two identical heavy chains, whose N-terminal regions fold to form catalytic motor domains, or heads, which are joined by “neck-linker” regions to a common, coiled-coil stalk. With each step, the two kinesin heads exchange leading and trailing positions as they alternately hydrolyze ATP, generating “hand-over-hand” motion (2–7). This motion is processive under moderate loads for up to ≈100 steps in succession, a property that enables small numbers of motors to ferry cargo over long distances (8, 9). The mechanism responsible for the remarkable processivity of kinesin remains unresolved. Furthermore, the relative order of the translocation step with respect to other steps in the mechanochemical cycle is not established.
A corollary of kinesin processivity is that its two heads do not function independently, but in concert. Put another way, the relative phase between the heads must be synchronized by a gating mechanism that prevents both heads from dissociating simultaneously from the MT. Because of the 2-fold symmetry imparted by the kinesin structure, this mechanism must break symmetry to permit synchronization. The most attractive candidate for such a mechanism is the mechanical strain that develops when the two heads separate and bind to the MT, with one head leading the other (10–12). Evidence for strain-based gating has recently been reported for the processive stepping cycle of myosin V (13–15). Kinesin mutants with polypeptide insertions between the neck linker and coiled-coil region are less processive (16), presumably because strain between the bound heads is reduced. This interpretation is supported by evidence that external load modulates the affinity of kinesin for nucleotide and MT substrates (17).
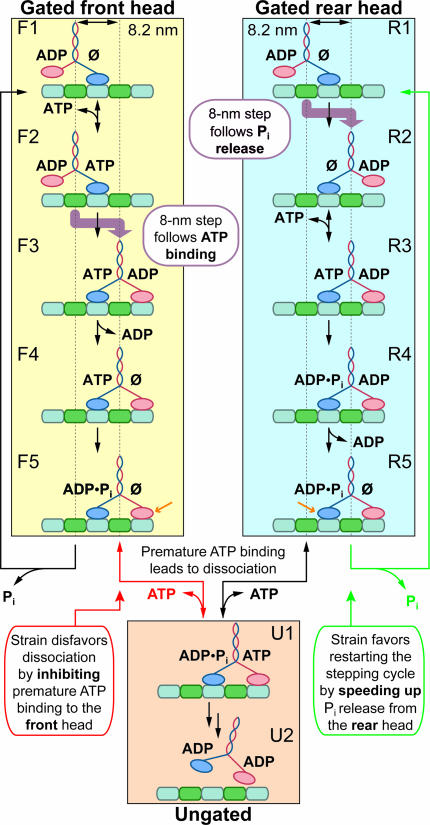
Broadly speaking, two distinct reaction schemes have been proposed for the kinesin cycle (Fig. 1). These competing models differ with respect to head gating, and each has several minor variants. In models where the front head is gated (see refs. 18 and 19 for details), internal strain developed between the two heads reduces the affinity of the leading head for ATP, preventing binding that might otherwise lead to dissociation of kinesin from the MT (i.e., slowing F5 → U1) (12, 18–20). Once the rear head releases Pi and detaches from the MT, strain is relieved and ATP may subsequently bind (F1 → F2). In models where the rear head is gated instead (see ref. 21 for details), strain between the heads weakens the MT affinity of the rear head, increasing the rate of trailing head detachment and the release of Pi (speeding R5 → R1) (21, 22). In both models, the effect of internal strain is to reduce the affinity of one of the heads for the substrate (either for ATP or the MT), possibly by destabilizing one or more structural elements. Without a gating mechanism, ATP could bind prematurely to the front head before the rear head released Pi, leading to an arbitrary phase between the catalytic cycles of the heads and inevitably to dissociation from the MT (U1 → U2). As with all mechanochemical processes, the relationship between mechanical strain and chemical substrate affinity underlying the gating mechanism is reciprocal: one may view the reduction of substrate affinity upon the simultaneous binding of both heads to the MT as a source of strain, or conversely, the presence of strain as a modulator of substrate affinity.
Fig. 1.
Consensus models for processive kinesin stepping. In the gated front head pathway (yellow shaded box), ATP binding triggers the mechanical step, and gating results from strain reducing the ATP affinity of the front head (F5). In the gated rear head pathway (cyan shaded box), the mechanical step is triggered by Pi release, and gating results from strain reducing the MT affinity of the rear head (R5). The common dissociation pathway (orange shaded box) illustrates how premature ATP binding, in the absence of gating, leads to release of both heads from the MT and loss of processivity. The models differ with respect to the identity of the nucleotide on the rear head when ADP is bound to the front (compare F3 and R4). Gated heads are indicated (orange arrows; F5 and R5). Under the experimental conditions, ATP binding is assumed to be reversible and other steps are taken to be irreversible, as indicated (black arrows). For unstrained states, we display the ADP-bound “tethered head” in close proximity to the MT, but the true extent of interaction between this head and the MT is unknown (24, 54). The position of the centroid of the molecule, measured by the optical trap, is indicated (dotted vertical lines), along with the position of mechanical steps (thick purple arrows). MTs have their plus-ends on the right; forward motion is rightward. Kinesin heads not bound to nucleotide are indicated (Ø).
The two competing models (Fig. 1) also differ in the relative order of the 8-nm mechanical step with respect to other biochemical steps. This mechanical transition is sometimes referred to as a “working stroke” or “force-producing step.” In the scheme proposed by Rice et al. (23) and incorporated in models where the front head is gated (18, 19), ATP binding induces a conformational change that docks the neck-linker region alongside its head, advancing the common stalk (and thereby the partner head), eventually leading to an 8-nm motion of the molecule (24–26). The model where the rear head is gated relies on an alternative mechanism, whereby the concomitant release of Pi and the rear head lead to an 8-nm advance (21, 27).
Questions of mechanism may be addressed by probing intermediate biochemical states with nucleotide analogs, such as the complex formed when beryllium fluoride (BeFx) binds to an ADP-bound head, ADP·BeFx. Structural and kinetic studies of myosin have shown that ADP·BeFx acts as an ATP analog, much as does adenylyl-imidodiphosphate (AMP-PNP) (28–30). Kinesin associates tightly with MTs when bound to ADP·BeFx (31–33), exactly as for AMP-PNP or ATP (34–39). Moreover, complexes of kinesin with MTs generate identical EPR spectra in the presence of AMP-PNP or ADP·BeFx (40). The evidence indicates that the binding of AMP-PNP docks the neck linker (12, 23, 41–44), favoring a two-heads-bound configuration, with AMP-PNP present on the rear head and no nucleotide on the front head. We therefore anticipate that the binding of BeFx to kinesin induces a similar two-heads-bound state, but with ADP·BeFx bound to the rear head.
Here, we used single-molecule assays to investigate the load-dependent kinetics of BeFx binding and release during kinesin stepping, along with the dynamics of intermediate states. Our data imply that strain reduces the affinity of the front head for ATP with respect to the rear head and that one or more additional catalytic steps are also inhibited. The data also suggest that ATP binding, as opposed to Pi release, represents the trigger for the mechanical step. A model where strain coordinates stepping by gating the front head therefore best explains how the kinesin moves hand-over-hand.
Results
BeFx and AMP-PNP Induces Pauses with Backsteps.
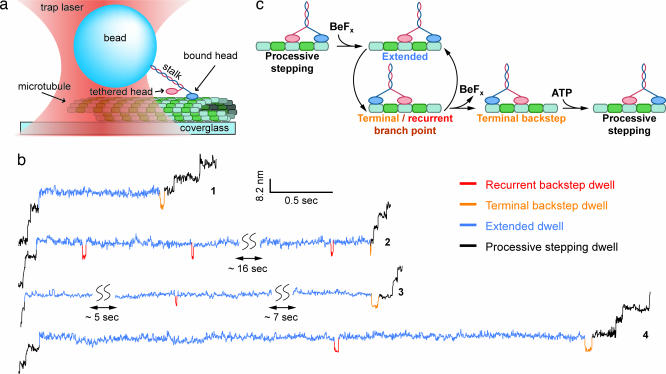
Polystyrene beads with small numbers of kinesin molecules attached to their surfaces were placed in the immediate vicinity of surface-bound MTs by using a specialized optical trapping apparatus, whereupon individual motors could bind and move. Motion was recorded with subnanometer precision under force-clamped conditions (Fig. 2a and see Materials and Methods) (45, 46). In the presence of ATP plus an analog (BeFx or AMP-PNP), the processive stepping of single kinesin molecules was frequently halted by lengthy pauses lasting from fractions of a second up to tens of seconds (Fig. 2b) (in the absence of analogs, such pauses were not present in records). Before the resumption of forward motion, long pauses were almost invariably terminated by a brief, backward step lasting a few tenths of a second (at low ATP levels). Rare instances of pauses that apparently ended without a terminal backstep could be accounted for by noise that occasionally obscured briefer motions (see Supporting Text, which is published as supporting information on the PNAS web site). In addition to this obligatory “terminal backstep,” records of most long pauses were interrupted by additional backsteps lasting only milliseconds (at all ATP levels), with statistically distinct properties from the terminal backstep. We termed these briefer events “recurrent backsteps.” The mean dwell times for forward steps immediately before the pause, and for steps immediately after a terminal backstep, were the same as the mean stepping times observed for normal, processive movement. All kinesin steps, forward or backward, involved abrupt displacements of ≈8 nm and were completed instantaneously on the time scale of our measurements. Long pauses induced by either AMP-PNP or BeFx appeared to be similar, so we focused on BeFx to facilitate rapid data collection (see Supporting Text). Although occasional backsteps are known to occur during normal stepping (47), particularly in response to high loads (25, 43), this report demonstrates backstepping induced by a chemical agent under moderate load.
Fig. 2.
Experimental geometry and detection of chemically induced backsteps. (a) Cartoon illustrating the optical trapping geometry used to apply load to single kinesin molecules moving on a MT (not drawn to scale). (b) Representative sections of stepping records showing long pauses induced in the presence of an admixture of nucleotide analogs and ATP. Experimental conditions for traces were: traces 1, 2, and 4, [BeFx] = 1 mM, [ATP] = 2 mM, F = 4.5 pN (hindering load); trace 3, [AMP-PNP] = 200 μM, [ATP] = 2 mM, F = 5.3 pN (hindering load). Each long pause consists of several phases: extended dwells (blue), zero or more recurrent backstep dwells (red), followed by a single, terminal backstep dwell (orange). Regions of processive stepping before and after each pause are indicated (black). (c) A minimal kinetic pathway for pauses induced by BeFx binding, consistent with both models shown in Fig. 1.
Our observations suggest a straightforward kinetic scheme (Fig. 2c). During processive stepping, BeFx binds to kinesin, arresting forward motion and initiating an extended dwell interval. With the analog bound, kinesin can only move backward, interrupting the extended dwell. Once in this rearward state, the motor may either reverse the mechanical process, stepping forward again to reinitiate the extended dwell while BeFx remains bound (a recurrent backstep), or it may release BeFx, after which it can resume processive forward motion (a terminal backstep). Because BeFx release occurs only after kinesin steps backward, exchanging the positions of its two heads, the nucleotide affinity of any particular head must depend on its position relative to the partner head.
Terminal Backstep and Extended Dwell Times Depend on [ATP].
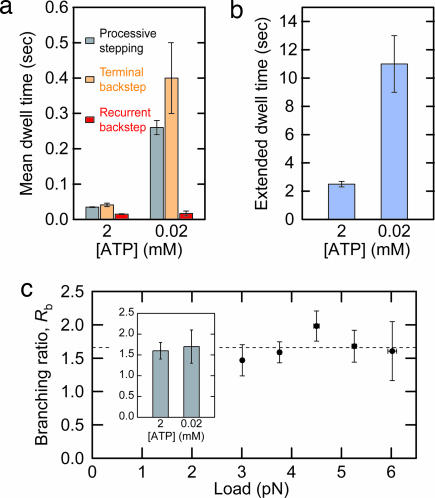
To ascertain whether ATP binding occurs during the period of a long pause, we measured the dependence of the times for backsteps and extended dwells on the ATP concentration. The mean recurrent backstep dwell time was unchanged at ATP levels of 2 and 0.02 mM. In contrast, the mean terminal backstep dwell time increased by roughly an order of magnitude in response to this same reduction in ATP level, similar to the increase in the mean dwell time for normal, processive stepping (Fig. 3a). This concentration dependence implies that ATP binding is required for the resumption of forward stepping. The same reduction in ATP levels also increased the mean extended dwell time by a factor of 4.4 (Fig. 3b). Extended dwells could be further prolonged when ATP was removed from solution by buffer exchange during a pause (data not shown). We therefore conclude that ATP also binds to kinesin during the extended dwell interval.
Fig. 3.
Kinetic properties of pause events. (a) [ATP] dependence of processive stepping time (gray), terminal backstep duration (orange), and recurrent backstep duration (red). (b) [ATP] dependence of extended dwell time. F = 3.8 pN for data shown in a and b. (c) Load dependence of the branching ratio, Rb. Weighted average value is 1.7 ± 0.1 (dashed horizontal line). [ATP] = 2 mM. (Inset) [ATP] dependence of Rb at F = 3.8 pN.
The Branching Ratio Between Recurrent and Terminal Backsteps Is Load- and [ATP]-Independent.
To probe the nature of the transitions leading from the backstep dwell, we determined load and ATP dependence of the branching ratio, Rb, between the pathways leading to the reinitiation of an extended dwell and the commitment to pause termination (Fig. 2c). Values of Rb were computed by dividing the total number of recurrent backsteps by the total number of terminal backsteps observed. For 2 mM ATP, Rb displayed no particular load dependence over the full range of forces studied, with an average value of 1.7 ± 0.1 (Fig. 3c), suggesting that both branches must have a similar dependence on load (if any). Furthermore, Rb was independent of the ATP level (Fig. 3c), suggesting that ATP binding (or lack thereof) affects both branches similarly.
Discussion
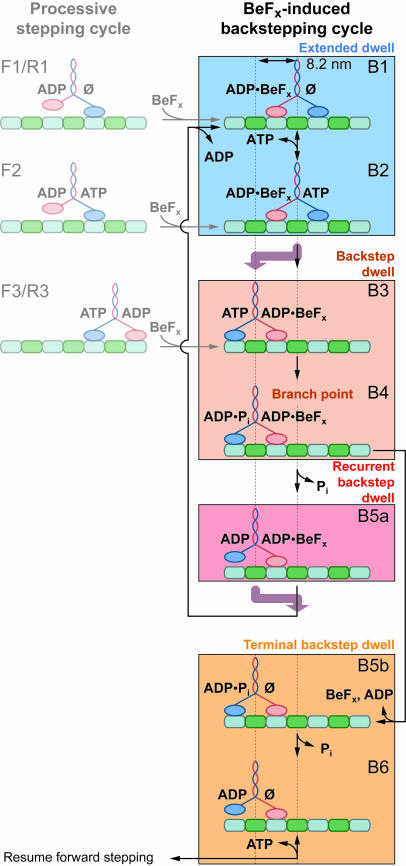
Taking the foregoing observations into account, we can now elaborate on the model for the BeFx-induced backstepping cycle (Fig. 4). BeFx association with an ADP-bound head induces kinesin to adopt a paused state with the rear head bound to ADP·BeFx and the front head free of nucleotide (B1). Because kinesin spends the overwhelming majority of time in an extended dwell state during a long pause, we surmise that ADP·BeFx remains bound to the rear head throughout extended dwells and to the front head during backstep dwells. With the rear head locked down, kinesin can only bind ATP (B2), step backward by 8 nm (B3), hydrolyze the ATP molecule (B4), and arrive at the branch point in the cycle. From this point, the release of Pi from the rear head commits the motor to a forward step (B5a), returning it to an extended dwell state and restarting the cycle, after ADP release from the front head. Conversely, release of BeFx along with ADP from the front head at this point terminates the pause (B5b), and after unbinding Pi, restores kinesin to a state where it is competent to resume processive stepping upon subsequent ATP binding (B6). Consistent with the data (Fig. 3a), recurrent backsteps are briefer, on average, than processive stepping dwells, because of kinetic partitioning from state B4 and the lack of an ADP release step. Because no exogenous ADP was added to our assay buffers, BeFx must bind to one or more states of kinesin where the head is bound to an ADP molecule retained from a previous ATP turnover. We were unable to exclude any of these alternative BeFx binding pathways (Fig. 4 Left) through measurements of the pausing frequency as a function of load and [ATP] (Fig. 5, which is published as supporting information on the PNAS web site). However, if there were multiple pause-initiation points, as shown by transitions from states F1, F2, and F3, then the first extended dwell should differ slightly in duration from subsequent extended dwell times. We found no systematic deviation between the first and subsequent extended dwells, likely because slower steps common to all extended dwells dominate the residence time.
Fig. 4.
Kinetic model for the BeFx-induced backstepping cycle. Each pause begins with an extended dwell, which is followed by one or more backsteps. From the branch point B4, the backstepped enzyme may release Pi and enter the recurrent backstep state B5a or release BeFx along with ADP, and then enter the terminal backstep state B5b. Recurrent backsteps are followed by a forward step (thick purple arrow) and reentry into the extended dwell (B1). Terminal backsteps are followed by a resumption of processive forward stepping. States from which BeFx binding may directly induce a pause (reproduced from Fig. 1) are shown (Left; grayed out). The color shading of boxes corresponds to the scheme used for coloring the corresponding dwell intervals in Fig. 2.
Two key features of the backstepping scheme follow directly from our data and favor models for processive stepping with a gated front head, rather than models with a gated rear head. First, the resumption of stepping only occurs subsequent to the obligatory terminal backstep and the resulting exchange in position of the two kinesin heads to permit BeFx and ADP release from the new front head. From this, we conclude that the affinity of the front head for the analog is significantly lower than that of the rear head when kinesin is in a strained, two-heads-bound configuration. This scenario supports stepping models where gating by internal strain prevents or slows the front head from tightly binding ATP. We can estimate a lower bound for the relative difference in binding affinity by comparing the mean extended dwell time at 20 μM ATP (representing a lower limit on how long BeFx remains bound to the rear head) with the mean recurrent backstep dwell time, multiplied by (1 + Rb) (representing an upper limit on how long BeFx remains bound to the front head during all backstep dwells for a given pause). The computed ratio is ≈200, a value sufficient to account fully for kinesin processivity without a need to invoke additional gating of the rear head.
Second, we found that the terminal backstep dwell time depended on the ATP concentration (Fig. 3a). This finding implies that ATP binding to state B6 is required for the step forward after BeFx release, consistent with models where the front head is gated, but inconsistent with models where the rear head is gated and movement takes place before ATP binding. To be consistent with the latter, the terminal backstep dwell time would need to be independent of ATP levels. This conclusion is supported by recent results showing that the tethered head of kinesin is poised behind the nucleotide-free head as the dimer is waiting for ATP to bind (24).
Although the strained front head has a lower affinity for ATP than the rear, it does eventually bind ATP productively (B2). An observation of longstanding has been that ATP binding can promote the release of an ATP analog (AMP-PNP) from an arrested kinesin–MT complex (39). Our results confirm and extend this idea. ATP does promote release of the bound analog (here, ADP·BeFx); however, this rate of release is not fully governed by ATP availability. Lowering the ATP concentration by 100-fold increased the time spent in the extended dwell state, but only by a factor of 4.4 (Fig. 3b), suggesting that another step influences the extended dwell time. Internal strain may therefore continue to inhibit the kinetic properties of the front head for at least one biochemical step beyond ATP binding. This interpretation is consistent with recent results suggesting that premature ATP binding can speed up the fast head of a mutant kinesin heterodimer (18) and can be directly observed as a slow phase in the binding transient of a fluorescent nucleotide (34).
Candidates for this additional step include hydrolysis, neck linker docking, or possibly some other conformational change, such as the rapid transition after ATP binding noted in presteady-state kinetics measurements (48–50). With this step inhibited, the strained kinesin head is unable to maintain a tight affinity for the MT upon binding ATP, leading to a backstep. A similar mechanism may be invoked to explain previous reports of kinesin backsteps induced by external load. Load seems likely to place the front head under strain, resulting in weak, rather than strong, affinity for the MT upon ATP binding. This explanation for load-induced backsteps is supported by the observation that ATP binding is required for load-induced backsteps (25, 43), just as it is for the analog-induced backsteps described here.
The backstepping model (Fig. 4) also shows a load-independent step (Pi release) competing with BeFx along with ADP release from the front head at the branch point (B4). Release of BeFx and ADP cannot occur during the state that immediately precedes the mechanical step (B5a), because the front head, being unstrained, tightly binds ADP·BeFx. The experimentally observed load independence of Rb (Fig. 3c), which requires that both branches have identical load behavior, supports this interpretation. Because the commitment step for the terminal backstep (B4 → B5b) involves no longitudinal motion along the MT and is therefore load-independent, the commitment step for the recurrent backstep (B4 → B5a) must be equally load-independent. Similarly, the invariance of Rb with ATP level is consistent with the commitment step being something other than ATP binding. Any other transition not generating motion along the MT, such as ATP hydrolysis or Pi release, could therefore represent a candidate commitment step. However, Pi release (with concomitant rear-head detachment) represents the most attractive candidate, because head detachment directly relieves strain and thereby restores the affinity of the front head for ADP·BeFx. Other possibilities for the commitment step, such as hydrolysis, do not directly relieve strain, leading to more complex models where BeFx may be released from multiple states. Furthermore, placement of the branch point before the forward mechanical step argues against an alternative mechanism where backsteps occur after the front head hydrolyzes ATP, releases Pi, and is left with ADP in its active site. Thereafter, once the heads exchange positions in a backstep, only ADP would be bound to the rear head, precluding the possibility of Pi release or hydrolysis as commitment steps for reentry into the pause.
We have used nucleotide analogs to probe the mechanochemistry of the kinesin stepping cycle. The data support a model where internal strain gates the front head by inhibiting premature ATP binding and subsequent catalytic steps (which would otherwise lead to loss of processivity), and the 8-nm mechanical step is triggered by ATP binding to the front head. We anticipate that additional studies of nucleotide analogs and other small-molecule inhibitors, in combination with single-molecule biophysics, have the potential to reveal further mechanistic details of kinesin function.
Materials and Methods
Assays.
Polystyrene beads (0.5-μm diameter) functionalized with sulfate groups (Interfacial Dynamics, Portland, OR) were incubated in the presence of 2 mM ATP for at least 2 h with native kinesin purified from the squid, Loligo pealei. For all incubations and experiments, buffers contained 80 mM Pipes (pH 6.9), 50 mM potassium acetate, 4 mM MgCl2, 2 mM DTT, 1 mM EGTA, and 10 μM taxol. After incubation, the ATP level was adjusted to the final concentration, and BSA blocking proteins were added to 10 mg/ml along with an oxygen-scavenging system (235 μg/ml glucose oxidase, 42 μg/ml catalase, and 4.6 mg/ml glucose). Finally, analogs were added to final concentrations of either 200 μM AMP-PNP or 1 mM BeFx (5 mM [NaF] and 1 mM [BeSO4]). MTs were polymerized from bovine brain tubulin (Cytoskeleton, Denver), taxol-stabilized, and immobilized on cleaned coverglasses coated with poly-l-lysine. To ensure that all beads measured were bound only to one molecule of kinesin, data were analyzed only from assays where fewer than half the beads tested moved on average (9). All chemicals were purchased from Sigma, except glucose oxidase (Calbiochem), catalase (Roche Applied Science), and DTT (Invitrogen). Details of the motility assays have been described (51).
Instrument.
Constant loads along the MT axis were applied to the moving beads by using an optical force clamp apparatus as described (45). The apparatus is based on an inverted microscope (Nikon) modified for mechanical stability and illuminated by a high-power, 1,064-nm trapping laser (Spectra-Physics) and a low-power, 830-nm position-detector laser (Point Source, Southampton, U.K.). Bead position was measured by focusing the low-power laser onto an optically trapped bead and monitoring the scattered light with a position-sensitive detector (Pacific Silicon Sensor, Westlake Village, CA). Software, written in labview 7 (National Instruments, Austin, TX), controlled the trap position and intensity via acousto-optical deflectors (IntraAction, Bellwood, IL) and the specimen position via a piezo stage (Physik Instruments, Karlsruhe, Germany). The position detector was calibrated for each bead recorded. Trap stiffness was checked regularly; stiffness was determined by an average of variance and Lorentzian roll-off methods (46, 52). Force clamping was activated by rapidly moving the stage to bring the bead a preset distance from the trap center whenever the kinesin molecule bound to a MT. After clamp activation, bead position was sampled at 2 kHz (and filtered at 1 kHz) while trap position was maintained at a fixed distance of 70 nm from the bead, updated at 200 Hz. At low [ATP], when BeFx binding was more frequent, kinesin would often pause on the MT before force clamping could be activated. In such cases, a brief application of high force was used to pull kinesin off the MT and release BeFx, restarting motion in the active clamp zone. About 20% of kinesin molecules yielded data that were too noisy for further analysis, leaving n = 82 that were measured. All loads applied were hindering loads, specified with a positive sign convention. Room temperature was regulated at 18.3°C ± 0.1°C. Kinesin velocity is known to depend moderately on the temperature (53), but the effect of temperature on individual kinetic rates remains largely unexplored.
Data Analysis.
Data analysis software was written with the igor pro 5 analysis package (WaveMetrics, Lake Oswego, OR). All data were median-filtered with a 3.5- to 5.5-ms window. Velocities were determined by taking the arithmetic means of the slopes of linefits to regions of processive stepping containing a minimum of eight steps. Slopes were weighted by the run length of the record for computations of arithmetic mean and standard error. A pause region was defined for any period when the kinesin molecule halted for an interval in excess of twice its mean stepping dwell time (for normal forward stepping) before taking a backstep. We estimate that in the worst-case scenario, when processive stepping and extended dwell times differ by only a factor of 10, this definition ensures that <5% of pauses scored might be caused by natural backstepping events.
Backsteps were identified by an algorithm that compared the median-filtered displacement during the pause to a baseline computed by filtering data with a 500-ms boxcar filter, or, for pauses <500 ms, with the mean value for displacement over the entire pause region. Backsteps were operationally defined whenever the displacement signal dipped >5 nm below the baseline. Backstep and extended dwell times appeared to be exponentially distributed (Fig. 6, which is published as supporting information on the PNAS web site). The uncertainty in applied force was estimated from the known displacement and the error in stiffness estimated from measurements of (n ≥ 5) beads. Errors in dwell durations were estimated by bootstrapping.
Supplementary Material
Acknowledgments
We thank current and former members of S.M.B.'s laboratory for helpful discussions and reading the manuscript, Susan Gilbert and Joe Howard for suggestions, Josh Shaevitz for guidance during the initial phase of this work, and Arthur La Porta for discussions on simulations. N.R.G. was supported by a National Science Foundation Fellowship. This work was supported by National Institutes of Health Grant R01-GM51453 (to S.M.B.).
Abbreviations
- BeFx
beryllium fluoride
- MT
microtubule
- AMP-PNP
adenylyl-imidodiphosphate
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Svoboda K., Schmidt C. F., Schnapp B. J., Block S. M. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 2.Hackney D. D. Proc. Natl. Acad. Sci. USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnitzer M. J., Block S. M. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 4.Hua W., Young E. C., Fleming M. L., Gelles J. Nature. 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- 5.Kaseda K., Higuchi H., Hirose K. Nat. Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- 6.Asbury C. L., Fehr A. N., Block S. M. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yildiz A., Tomishige M., Vale R. D., Selvin P. R. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 8.Howard J., Hudspeth A. J., Vale R. D. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 9.Block S. M., Goldstein L. S., Schnapp B. J. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 10.Block S. Cell. 1998;93:5–8. doi: 10.1016/s0092-8674(00)81138-1. [DOI] [PubMed] [Google Scholar]
- 11.Tomishige M., Vale R. D. J. Cell Biol. 2000;151:1081–1092. doi: 10.1083/jcb.151.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skiniotis G., Surrey T., Altmann S., Gross H., Song Y.-H., Mandelkow E., Hoenger A. EMBO J. 2003;22:1518–1528. doi: 10.1093/emboj/cdg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell T. J., Sweeney H. L., Spudich J. A. Proc. Natl. Acad. Sci. USA. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veigel C., Schmitz S., Wang F., Sellers J. R. Nat. Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 15.Rosenfeld S. S., Houdusse A., Sweeney H. L. J. Biol. Chem. 2005;280:6072–6079. doi: 10.1074/jbc.M412717200. [DOI] [PubMed] [Google Scholar]
- 16.Hackney D. D., Stock M. F., Moore J., Patterson R. A. Biochemistry. 2003;42:12011–12018. doi: 10.1021/bi0349118. [DOI] [PubMed] [Google Scholar]
- 17.Uemura S., Kawaguchi K., Yajima J., Edamatsu M., Toyoshima Y. Y., Ishiwata S. Proc. Natl. Acad. Sci. USA. 2002;99:5977–5981. doi: 10.1073/pnas.092546199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenfeld S. S., Fordyce P. M., Jefferson G. M., King P. H., Block S. M. J. Biol. Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klumpp L. M., Hoenger A., Gilbert S. P. Proc. Natl. Acad. Sci. USA. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auerbach S. D., Johnson K. A. J. Biol. Chem. 2005;280:37048–37060. doi: 10.1074/jbc.M502984200. [DOI] [PubMed] [Google Scholar]
- 21.Hancock W. O., Howard J. Proc. Natl. Acad. Sci. USA. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schief W. R., Clark R. H., Crevenna A. H., Howard J. Proc. Natl. Acad. Sci. USA. 2004;101:1183–1188. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., et al. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 24.Hackney D. D. Proc. Natl. Acad. Sci. USA. 2005;102:18338–18343. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter N. J., Cross R. A. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 26.Peterman E. J., Sosa H., Goldstein L. S., Moerner W. E. Biophys. J. 2001;81:2851–2863. doi: 10.1016/S0006-3495(01)75926-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandelkow E., Johnson K. A. Trends Biochem. Sci. 1998;23:429–433. doi: 10.1016/s0968-0004(98)01278-x. [DOI] [PubMed] [Google Scholar]
- 28.Coureux P. D., Sweeney H. L., Houdusse A. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher A. J., Smith C. A., Thoden J. B., Smith R., Sutoh K., Holden H. M., Rayment I. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 30.Gulick A. M., Bauer C. B., Thoden J. B., Rayment I. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- 31.Rosenfeld S. S., Rener B., Correia J. J., Mayo M. S., Cheung H. C. J. Biol. Chem. 1996;271:9473–9482. doi: 10.1074/jbc.271.16.9473. [DOI] [PubMed] [Google Scholar]
- 32.Ma Y. Z., Taylor E. W. J. Biol. Chem. 1997;272:717–723. doi: 10.1074/jbc.272.2.717. [DOI] [PubMed] [Google Scholar]
- 33.Shibuya H., Kondo K., Kimura N., Maruta S. J. Biochem. (Tokyo) 2002;132:573–579. doi: 10.1093/oxfordjournals.jbchem.a003259. [DOI] [PubMed] [Google Scholar]
- 34.Vale R. D., Reese T. S., Sheetz M. P. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romberg L., Vale R. D. Nature. 1993;361:168–170. doi: 10.1038/361168a0. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert S. P., Webb M. R., Brune M., Johnson K. A. Nature. 1995;373:671–676. doi: 10.1038/373671a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitta R., Kikkawa M., Okada Y., Hirokawa N. Science. 2004;305:678–683. doi: 10.1126/science.1096621. [DOI] [PubMed] [Google Scholar]
- 38.Lasek R. J., Brady S. T. Nature. 1985;316:645–647. doi: 10.1038/316645a0. [DOI] [PubMed] [Google Scholar]
- 39.Schnapp B. J., Crise B., Sheetz M. P., Reese T. S., Khan S. Proc. Natl. Acad. Sci. USA. 1990;87:10053–10057. doi: 10.1073/pnas.87.24.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naber N., Minehardt T. J., Rice S., Chen X., Grammer J., Matuska M., Vale R. D., Kollman P. A., Car R., Yount R. G., et al. Science. 2003;300:798–801. doi: 10.1126/science.1082374. [DOI] [PubMed] [Google Scholar]
- 41.Ma Y. Z., Taylor E. W. J. Biol. Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 42.Kawaguchi K., Ishiwata S. Science. 2001;291:667–669. doi: 10.1126/science.291.5504.667. [DOI] [PubMed] [Google Scholar]
- 43.Nishiyama M., Higuchi H., Yanagida T. Nat. Cell Biol. 2002;4:790–797. doi: 10.1038/ncb857. [DOI] [PubMed] [Google Scholar]
- 44.Asenjo A. B., Krohn N., Sosa H. Nat. Struct. Biol. 2003;10:836–842. doi: 10.1038/nsb984. [DOI] [PubMed] [Google Scholar]
- 45.Lang M. J., Asbury C. L., Shaevitz J. W., Block S. M. Biophys. J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuman K. C., Block S. M. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svoboda K., Block S. M. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 48.Gilbert S. P., Johnson K. A. Biochemistry. 1994;33:1951–1960. doi: 10.1021/bi00173a044. [DOI] [PubMed] [Google Scholar]
- 49.Ma Y. Z., Taylor E. W. Biochemistry. 1995;34:13242–13251. doi: 10.1021/bi00040a040. [DOI] [PubMed] [Google Scholar]
- 50.Rosenfeld S. S., Jefferson G. M., King P. H. J. Biol. Chem. 2001;276:40167–40174. doi: 10.1074/jbc.M103899200. [DOI] [PubMed] [Google Scholar]
- 51.Block S. M., Asbury C. L., Shaevitz J. W., Lang M. J. Proc. Natl. Acad. Sci. USA. 2003;100:2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visscher K., Block S. Methods Enzymol. 1998;298:460–489. doi: 10.1016/s0076-6879(98)98040-5. [DOI] [PubMed] [Google Scholar]
- 53.Kawaguchi K., Ishiwata S. Cell Motil. Cytoskeleton. 2001;49:41–47. doi: 10.1002/cm.1019. [DOI] [PubMed] [Google Scholar]
- 54.Carter N. J., Cross R. A. Curr. Opin. Cell Biol. 2006;18:61–67. doi: 10.1016/j.ceb.2005.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.