Abstract
Precise characterization of the mutation histories of evolutionary lineages is crucial for understanding the evolutionary process, yet mutation identification has been constrained by traditional techniques. We sought to identify all accumulated mutations in an experimentally evolved lineage of the cooperative bacterium Myxococcus xanthus, which constructs fruiting bodies by a process of social multicellular development in response to starvation. This lineage had undergone two major transitions in social phenotype: from an ancestral cooperator to a socially defective cheater, and from the cheater to a competitively dominant cooperator that re-evolved social and developmental proficiency. The 9.14-Mb genome of the evolved, dominant cooperator (strain “PX”) was sequenced to ≈19-fold coverage by using recent “sequencing-by-synthesis” technology and partially sequenced (≈45%) by using capillary technology. The resulting data revealed 15 single-nucleotide mutations relative to the laboratory ancestor of PX after the two phases of experimental evolution but no evidence of duplications, transpositions, or multiple-base deletions. No mutations were identified by capillary sequencing beyond those found by pyrosequencing, resulting in a high probability that all mutations were discovered. Seven errors in the reference strain previously sequenced by the Sanger approach were revealed, as were five mutational differences between two distinct laboratory stocks of the reference strain. A single mutation responsible for the restoration of development in strain PX was identified, whereas 14 mutations occurred during the prior phase of experimental evolution. These results provide insight into the genetic basis of two large adaptive transitions in a social bacterium.
Keywords: cooperation, Myxococcus xanthus
Evolutionary biology seeks to understand how mutation, phenotypic variation, natural selection, chance, and history interact to shape the process of evolution. Because spontaneous mutation is the foundational biological source of phenotypic variation, the complete identification of mutational changes over defined evolutionary periods is necessary to fully understand processes of evolutionary change in particular lineages. This can be difficult when comparing extant natural organisms, because ancestral character states and generations since divergence can remain uncertain even when complete genome sequences and independent time-since-divergence data, such as fossil records, are available. Moreover, the selective histories of natural lineages are notoriously difficult to reconstruct.
Genotypes evolved under controlled selective conditions from a known ancestor over a defined period, however, are not subject to these ambiguities and provide a rich resource for understanding microevolutionary processes, particularly with the advent of new technologies for mutation identification. Because of their rapid growth, large population sizes, and the ability to freeze viable ancestors indefinitely, various microorganisms have been evolved in laboratory regimes to examine a wide variety of evolutionary questions (1). Although complete mutational characterizations have been made of experimentally evolved viruses (2) and silicon-based replicators (3), sequencing costs and traditional mutation identification techniques have previously hindered such comprehensive mutation analysis in free-living, carbon-based forms of life (4).
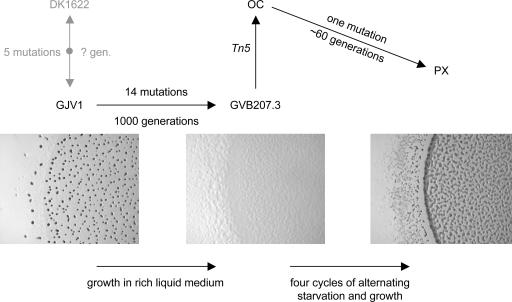
Myxococcus xanthus is a soil proteobacterium with several distinguishing social traits, including social motility, cooperative predation, and starvation-induced multicellular development in which a minority of cells within fruiting body populations differentiate into stress-resistant spores. M. xanthus has one of the largest known bacterial genomes (9.14 Mb), and a large proportion of its genes appear to be involved in its various social behaviors (W. C. Nierman, D. Kaiser, and B. S. Goldman, personal communication). A clone (GJV1) of the standard M. xanthus lab strain DK1622 (5) previously served as the ancestor of 12 experimental lineages that underwent 1,000 generations of evolution in nutrient-rich liquid medium (6) (Fig. 1). This selective regime placed no positive selection on M. xanthus social traits but rather on competitiveness under asocial growth conditions. The replicate lineages improved their maximum growth rates an average of 37% over the 1,000 generations, but all populations incurred partial or complete losses in their capacity for social motility and social development during this period of adaptation.
Fig. 1.
The previously sequenced strain DK1622 and its derivative clone GJV1 are separated by five mutations and an unknown number of generations of lab stock cultivation. The lineage from GJV1 to GVB207.3 incurred 14 mutations over 1,000 generations of growth in liquid medium, one or more of which eliminated the ability to undergo multicellular development (6). OC was generated by integration of a Tn5 transposon (which confers resistance to kanamycin) into the GVB207.3 genome (8). OC evolved into PX by regaining the ability to sporulate via social development during an extended developmental competition against a marked variant of GJV1 (9, 10). Only one mutation was found to distinguish PX from OC, and this mutation was subsequently shown to cause the restoration of developmental proficiency in PX (10).
After 1,000 generations, several evolved clones were isolated that are able to “cheat” on their ancestor during starvation-induced development in mixed populations (7). Although defective at developmental sporulation in clonal cultures, these cheats sporulate more efficiently than their socially proficient ancestor when present as a minority in mixed populations. One cheating clone, here termed “OC” (Obligate Cheater), is completely defective at social development during starvation and thus requires the presence of socially proficient cells to form stress-resistant spores (8). OC is marked with kanamycin-resistance (via chromosomal integration of a plasmid-borne Tn5 transposon) and was subsequently mixed as a 1% minority with a distinctly marked (rifampicin-resistant) variant of GJV1 and the chimeric population was subjected to six sequential cycles of alternating starvation and growth (9). After four cycles of this competition, OC had re-evolved the ability to undergo multicellular development, and the resulting genotype (PX, or Phoenix) shows a developmental phenotype that is distinct from, and competitively superior to, that of the GJV1 ancestor (10).
Recent advances in sequencing technology now make efficient identification of polymorphisms at the genomic scale possible (11, 12). Here, we report the first complete sequencing of an experimentally evolved cellular organism to identify all accumulated mutations relative to a known ancestor. To identify polymorphisms that distinguish PX, OC, and GJV1, we sequenced the PX genome using two independent approaches: PicoTiterPlate-based technology (454 Sequencing or “sequencing by synthesis”; 454 Life Sciences) (which generated 19-fold coverage of the 9.14-Mb genome) and traditional capillary-based sequencing (0.45-fold coverage). The resulting PX sequence from both approaches was assembled against the previously sequenced genome of DK1622 (the lab parent of GJV1; W. C. Nierman, D. Kaiser, and B. S. Goldman, personal communication), and true discrepancies were then identified by PCR-sequencing analysis of putative discrepancies.
Results
Sequencing by Synthesis.
Approximately 2.5 million reads with an average length of 96 bp were generated from twelve runs of PicoTiterPlates (12). Sequence reads were assembled into 104 contigs, and all but 16 of the remaining sequence gaps were closed by PCR-based sequencing in the laboratory of G.J.V. The remaining gaps are all in repeat sequence regions among which alignment software was unable to properly allocate reads. The total remaining unaligned sequence is 8,701 bp, or 0.095% of the genome.
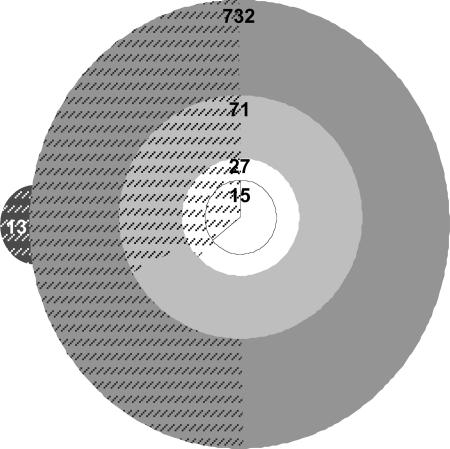
The 104 contig assembly showed 732 discrepancy units relative to DK1622 (Fig. 2). Single base deletion calls within long homopolymer runs and discrepancies very near contig ends are common false positive discrepancies generated by this technology and accounted for 661 of the differences. All 71 putative discrepancies outside of these categories (including double and triple base pair deletions within homopolymer runs and single base deletions within homogenous runs of only three bases and excluding putative discrepancies ≤5 bp from a contig terminus) were examined by PCR amplification and capillary sequencing. Using mutation identification software developed by 454 Life Sciences and corresponding parameters intended to conservatively exclude all false negatives, only 16 discrepancies were called as having a high probability of being true positives. All 16 were among the 71 potential discrepancies identified by assembly alignment against the DK1622 template, and all 16 calls were confirmed as true positives by PCR-sequencing analysis. However, 11 additional potential discrepancies were also confirmed as being true discrepancies, thus giving a total of 27 real discrepancies and 44 false positives among the 71 examined. Finding these additional discrepancies highlights the importance of testing all categories of putative discrepancies that are not known a priori to be predominantly or exclusively false positive artifacts of the 454 Sequencing technology.
Fig. 2.
Assembly of pyrosequencing reads of strain PX resulted in 104 contigs bearing 732 discrepancy units relative to the DK1622 GenBank sequence (large gray circle). Seventy-one (71) of these were selected (by exclusion of putative single base pair deletions within homopolymer runs four bases or longer and discrepancies ≤5 bp from contig ends) and tested independently by PCR (light gray circle). Twenty-seven (27) of these discrepancies were real (open circle), with 15 of them being evolutionary mutations separating PX from GJV1 (bordered open circle) and the remaining 12 being differences between GJV1 and the DK1622 reference sequence. Of these latter 12 discrepancies, 5 are real mutational differences between GJV1 and the sequenced clone of DK1622, whereas the other 7 represented errors in the original GenBank sequence. Hatched regions indicate the proportion of putative discrepancies within each category that were covered by high-quality capillary sequence. Capillary sequencing generated 13 putative discrepancies (gray, hatched protrusion) not present among the 732 putative contig discrepancies. All 13 were shown to be false positives by subsequent PCR-sequencing analysis. Relative total circle areas are not proportionally accurate, whereas the proportions of hatched vs. unhatched areas within the four concentric circles are proportionally accurate.
Capillary Sequencing.
Independent testing of possible true positive discrepancies still left the possibility of unseen false negatives in the assembled contigs. To evaluate the accuracy of the assembled sequence from 454 Life Sciences, we sequenced 45.3% of the PX genome using traditional shotgun cloning and capillary sequencing (5,716 reads assembled, 4.14 Mb of genome covered, 4.44 Mb total high-quality sequence, and 712 bp average high-quality read-length). Of the 732 initial contig discrepancies, 368 (50.3%) were covered by capillary sequence of moderate to high quality (PHRED score ≥19). Ten (37%) of the 27 real discrepancies known from PCR-sequencing analysis were verified by high-quality capillary sequence.
Importantly, of the 368 initial contig discrepancies covered by high-quality shotgun reads (most of which were putative single base deletions in homopolymer runs) and not identified as real by PCR-sequencing analysis, all were shown to be false positives by the shotgun reads. This result indicates that the inclusion criteria used to select discrepancies between the sequencing-by-synthesis contigs and the DK1622 sequence template for independent PCR-sequencing analysis (described above) are likely to have resulted in zero or very few omissions of real differences.
Thirteen additional high-quality discrepancies were found between shotgun reads and the DK1622 reference that were not among the 732 initial contig discrepancies (Fig. 2). All of these 13, however, were shown to be false positives by independent PCR-sequencing analysis, indicating that these putative discrepancies were artifacts of the shotgun cloning approach that appear at a rate of ≈3 × 10−6. In shotgun cloning projects of low average coverage such as this one, such false positive discrepancies may not be corrected by multiple coverage and need to be tested independently.
Multibase Mutation Screen.
There is no evidence for genomic rearrangements, multiple-base deletions, duplications, or transpositions in the PX genome. All such mutation events would have resulted in at least one (in the case of proximate duplications), and in most cases two or more, new sequence junctures relative to the ancestral sequence. For example, a single duplication event would result in two new sequence junctures, one at each end of the duplicated region. Only one segment of shotgun sequence reads covering such novel junctures will align to the reference genome at any given location, whereas the remainder of the read will be high-quality but unaligned. Because the shotgun reads were long (712 bp on average) and cover ≈45% of the genome, nearly half of all such mutation events should have been detected in high-quality sequences that align only partially to the reference genome. Four such chimeric reads were initially identified, but the chimeras may have resulted either from real mutation events in the genome or as an artifact of shotgun cloning. One of the four chimeric reads covered one of the two junctures between the Tn5 marker and the pilS2 gene in which this marker was inserted.
For the remaining three chimeric junctures, genomic fragments extending ≈450 bp on either side of the putative mutation loci were amplified by PCR and sequenced. In all three cases, the fragment sizes were as predicted from the genomic sequence, and the PCR fragments revealed no sequence other than that expected from the genomic template. The chimeric reads, therefore, appear to be artifacts of the shotgun sequencing approach and occurred at a frequency of 0.0035 among the 5,716 shotgun reads. In addition, the chimeric shotgun reads were subjected to blast analysis against all 2.5 million sequencing-by-synthesis reads. Chimeric junctures (relative to the reference template) in shotgun reads that do not represent real corresponding junctures in the genome will not be covered by individual sequencing-by-synthesis reads that extend in both directions from the juncture point. Sequencing-by-synthesis reads that align to the three shotgun chimeras (other than the Tn5 insertion) show abrupt alignment stops at precisely the chimera junctures from both directions, indicating that these junctures do not occur in the PX genome. (In contrast, individual pyrosequencing reads do cover the Tn5–pilS2 juncture.) Thus, we infer that zero or extremely few mutations involving deletion or alteration of extended sequence segments occurred in this M. xanthus lineage. In contrast, deletion and transposition events involving insertion sequence elements were common among evolving laboratory populations of Escherichia coli after 1,000 or more generations (13, 14), suggesting that the dynamics of insertion sequence element-mediated mutation differ between E. coli and M. xanthus.
Gap Analysis.
Eighty-eight of the 104 gaps in the 454 Sequencing contig assembly were closed by either PCR-based sequencing or shotgun reads (and several of the remaining 16 gaps were partially covered by shotgun sequence), leaving a total of 8,701 bp of unaligned sequence. If the genome were composed entirely of unique sequence regions, the level of coverage obtained should have left no gaps, and in fact all of the gap sequences were completely covered by pyrosequencing reads as revealed by blast analysis. However, gaps in the assembly result from the inability of assembly software to correctly localize short reads that align within repeat sequence regions. Thus, the majority of gaps lie within genes encoding insertion element transposases (of which there are several dozen) or multiple copy ribosomal RNA genes. All unclosed gaps fall within sequences repeated elsewhere in the genome. No additional discrepancies were found within the sequenced gap regions. Thus, no mutations were identified by the capillary-based sequences of the PX genome beyond those identified from the PicoTiterPlate data, indicating that this analysis is likely to have captured all mutational differences between the genomes.
Discrepancy Histories.
Three additional strains were examined for the presence or absence of the 27 discrepancies found between PX and the original (November 17, 2004) DK1622 GenBank template sequence. These strains were GJV1 (a putatively identical clone of DK1622), GVB207.3 (the unmarked, immediate parent of OC that was isolated from a 1,000-generation culture evolved from GJV1), and OC (the proximate evolutionary ancestor of PX). Twenty-six (26) of the discrepancies between PX and DK1622 were also present in GVB207.3 and OC. This left only a single discrepancy unique to PX (position 1258238; Table 1), and this mutation was subsequently shown to have caused the phenotypic transition from OC to PX (10). Twelve of the discrepancies relative to DK1622 are also present in GJV1 and could have resulted either from errors in the original DK1622 sequence or from real mutations accumulated in one of the lab-stock lineages leading to both clones since their last common ancestor in the laboratory of D. Kaiser in 1990. We checked these 12 discrepancies by sequencing PCR products amplified from the original genomic DNA preparation used by Monsanto/TIGR (The Institute for Genomic Research) to sequence strain DK1622. Seven of these discrepancies represented errors in the original TIGR sequence, whereas the remaining five reflected real mutational differences between GJV1 and DK1622 (Fig. 1 and Table 2). This result sheds light on the rate of sequence evolution that can occur during propagation of laboratory stocks between periods of frozen storage (see Materials and Methods for strain histories).
Table 1.
Experimental evolution mutations
| Mutation position | Gene name/position | TIGR annotation | Nucleotide, codon change | Amino acid change (no.) | Frequency |
|---|---|---|---|---|---|
| 1162101 | 1000 ← (1161497..1162060) | c.h.p. | C→A (−41 bp) | Noncoding | 0.07 |
| 1258238* | 1079 → (1258366..1259202) | GNAT family acetyltransferase | C→A (−128 bp) | Noncoding | n.a. |
| 3033111 | 2606 → (3032327..3035239) | Histidine kinase/response regulator | GAG→GGG | Glu→Gly (262) | 1.00 |
| 3091652 | 2647 → (3089922..3091961) | ftsI, cell division protein FtsI | GAG→GAT | Glu→Asp (577) | 1.00 |
| 3103359 | 2659 ← (3102167..3103702) | h.p. | GAC→GGC | Asp→Gly (115) | 0.07 |
| 4429794 | 3713 → (4428765..4430204) | Peptidase, M1 6 (pitrilysin) family | GCG→ACG | Ala→Thr (344) | 0.04 |
| 5310523 | 4310 ← (5310256..5311341) | h.p. | TCC→TCT | Ser  Ser (273) Ser (273) |
1.00 |
| 7036274 | 5673 ← (7035787..7036197) | h.p. | G→−(−77 bp) | Noncoding | 1.00 |
| 7147834 | 5772 ← (7146368..7149073) | pilQ, type IV pilus biogenesis protein | AAG→TAG | Lys→Stop (414) | 0.87 |
| 7354908 | 5932 ← (7354354..7355226) | Formamidopyrimidine-DNA glycosylase | GAT→AAT | Asp→Asn (107) | 1.00 |
| 7533448 | 6087 → (7533016..7534686) | coxA, cytochrome c oxidase, subunit I | GGC→AGC | Gly→Ser (145) | 0.04 |
| 8238547 | 6698 → (8238412..8239941) | c.h.p. | AAA→GAA | Lys→Glu (46) | 0.04 |
| 8685509 | 7114 → (8685277..8686311) | Periplasmic ABC transporter | GTG→GGG | Val→Gly (78) | 0.04 |
| 8870771 | 7265 ← (8870110..8870802) | c.h.p. | GCG→GTG | Ala→Val (11) | 1.00 |
| 8966897 | 7345 ← (8965180..8966719) | rrsD, 16S ribosomal RNA | A→C (−178 bp) | Noncoding | 1.00 |
c.h.p., conserved hypothetical protein; h.p., hypothetical protein; n.a., not applicable. Arrows next to gene numbers indicate the direction of transcription relative to the DK1622 genome sequence direction; numbers below amino acid transitions indicate the amino acid/codon position in the protein/gene; distances upstream from the start if the nearest gene are given as negative numbers for mutations in noncoding sequence.
* Sole mutation that occurred during the second evolutionary phase of a chimeric developmental competition experiment and that caused the OC→PX phenotypic transition (10).
Table 2.
Mutational differences between two laboratory stocks of M. xanthus, DK1622 and GJV1
| Mutation position | Gene name/position | TIGR annotation | Nucleotide, codon change | Amino acid change (no.) |
|---|---|---|---|---|
| 1717943 | 1458 ← (1716791..1718092) | Putative RNA methylase family | CGC vs. CGA | Arg  Arg (50) Arg (50) |
| 2304201 | 1970 → (2303987..2304883) | Transcriptional regulator, ArsR family | TTG G vs. TGG | Frameshift (72) |
| 5893144 | 4700 ← (5888844..5891669) | Serine/threonine protein kinase | A vs. C | Noncoding (−1475 bp) |
| 6287570 | 5030 → (6287648..6289036) | Outer membrane efflux protein family | C vs. A | Noncoding (−78 bp) |
| 7101833 | 5735 → (7101624..7103015) | ftsY, cell division protein FtsY | ACT vs. ACC | Thr  Thr (70) Thr (70) |
Experimental Evolution Mutations.
A total of 15 mutations were found to have accumulated in the evolutionary lineage from GJV1 to PX, all of which were single base pair discrepancies that included six transversions, eight transitions, and one deletion. Fourteen single base mutations accumulated in the lineage from GJV1 to GVB207.3 (the unmarked parent of OC), whereas only one mutation (1258238) appeared in the 60 generation lineage from OC to PX (10) (Fig. 1 and Table 1).
We sought to determine whether the 14 mutations that accumulated in GVB207.3 were also present in other clones from the same 1,000-generation population from which GVB207.3 was isolated. To estimate the frequency of these mutations in the general population, 15 randomly selected clones were genotyped for each mutation locus. An additional 12 random clones were genotyped for any mutation that was not present in a majority of the initial 15 clones (Table 1). Eight mutations appear to have been fixed or nearly fixed in the 1,000-generation population, whereas six loci were polymorphic with the mutations at frequencies of 0.07 or lower.
As a whole, the population in which GVB207.3 evolved was limited to ≈6.6 generations per day by a daily dilution factor of 100 (6). However, each newly arisen mutant with a fitness advantage (and not lost from the population by chance) would have undergone more generations per day than the population as a whole until the new genotype reached fixation, whereupon the number of replications per day would have become limited by the dilution factor. Thus, the exact number of generations undergone by the ancestral lineage of any particular clone isolated from the general population will be greater than an estimate calculated for the population as a whole as a function of the number of distinct selective sweeps in the clone’s ancestral lineage. For an adaptive mutant to rise from a single cell to fixation in a population bottlenecked daily at ≈2 × 108 individuals (6) requires the sweeping clone to undergo ≈28 more cell divisions than the general population. If we assume that at least 1 mutation, and a maximum of 14, rose to fixation (or near fixation) through independent successive adaptive sweeps, then the total number of genome replications undergone by the GJV1 → GVB207.3 lineage ranges between 1,028 and 1,392.
Nine mutations occurred in or near genes with annotated functions (Table 1). The most straightforward mutation to interpret occurred in the pilQ gene (7147834), which encodes the PilQ secretin homolog required for biosynthesis of the type IV pili that drive social motility in M. xanthus (15). Strains GVB207.3 (the immediate, unmarked parent of OC) (Fig. 1), OC, and PX are all defective in type IV pilus-mediated social motility, and it was previously shown that transfer of the ancestral pil region surrounding the pilQ gene from DK1622 restores social motility in GVB207.3 (8). This restoration of social motility function in the evolved clone caused a 4% decrease in evolutionary fitness (in the liquid evolution regime), showing that the previous mutational loss of social motility was adaptive under asocial growth conditions. The pilQ mutation prematurely terminates translation, resulting in a 414-residue peptide rather than the full-length 906-residue PilQ protein. This mutation is the only alteration found within the pil region that rescued social motility in GVB207.3 (8), thus implicating it as the cause of social motility loss in that lineage. The pilQ mutation, however, does not appear to mediate cheating behavior in OC, because rescue of S-motility in GVB207.3 with the ancestral pilQ region did not diminish the cheating phenotype of the resulting S-motility-proficient transformants relative to OC (8).
Another mutation (3091652) caused an amino acid substitution in FtsI, a cell division transpeptidase necessary for peptidoglycan synthesis that is localized to the division septum ring (16). This mutation may have improved the efficiency of peptidoglycan synthesis under conditions where nutrient level does not limit growth rate. A further mutation at position 7533448 caused an amino acid change within a transmembrane helix of cytochrome c oxidase, a catalyst of molecular oxygen reduction into water as the terminal step of ATP production in the electron transport pathway (17).
Formamidopyrimidine-DNA glycosylase repairs oxidative damage to DNA that occurs during chromosome replication (18). The mutM gene encoding this enzyme incurred an amino acid change in strain OC (mutation 7354908), possibly altering the function of MutM. Inactivation of mutM in E. coli causes a mutator (i.e., high mutation rate) phenotype that specifically generates a large excess of G·C to T·A transversions. However, only one of the 14 GJV1 → GVB207.3/OC mutations is such a transversion, suggesting that the PX mutM mutation is unlikely to cause a mutator phenotype.
Additional amino acid changes occurred in pitrilysin (E. coli protease III, mutation 4429794), a peptidase localized to the periplasm (19), an undefined histidine kinase/response regulator (mutation 3033111), and a periplasmic ATP binding cassette transporter (mutation 8685509). The latter two protein classes are highly represented in the M. xanthus genome and are involved in a wide variety of social function pathways (W. C. Nierman, D. Kaiser, and B. S. Goldman, personal communication). The histidine kinase mutation falls within the enzyme’s structural domain (a coiled-coil region upstream of the phosphorylation site), suggesting an effect on enzymatic rate or specificity. One noncoding mutation (8966897) occurs 178 bp above one copy of the 16S rRNA gene. The remaining mutations occurred in or near uncharacterized hypothetical proteins.
Mutation Rate Estimation.
Four mutations that accumulated in the GVB207.3/OC genome do not affect the amino acid sequence of a protein, including one synonymous change in a coding region and three others that lie in noncoding sequence. The single synonymous mutation in a coding region (mutation 5310523) is likely to be selectively neutral but nonetheless appears to have reached fixation in the population from which GVB207.3 was isolated (Table 1). This mutation may have risen to high frequency by “hitchhiking” in the same genomic background as a distinct, selectively advantageous mutation, or by random genetic drift.
The substitution rate of neutral mutations is determined directly by the steady-state mutation rate independent of population size (20). Thus, the total number of synonymous fixation events expected is the product of the background genome-wide mutation rate, the number of neutral sites in the evolving genome, and the number of generations. To make an initial estimate of the M. xanthus per base pair mutation rate, we assume that only the single silent coding substitution is neutral, although we cannot exclude the possibility that additional mutations might be neutral as well. Given one synonymous substitution event and 2.2 − 3.0 × 109 base pair generations at potentially silent sites in the experiment [one population × no. of generations (1,028 − 1,392) × (2.2 × 106 coding-region bp susceptible to silent mutations; see Materials and Methods], the steady-state mutation rate most likely to result in one substitution is 3.3 − 4.5 × 10−10 per bp per generation. This estimate for the GJV1 → GVB207.3/OC lineage of M. xanthus is intermediate between two previous estimates for the mutation rates of nonmutator strains of E. coli [1.44 × 10−10 (4) and 5.40 × 10−10 (21) per bp per generation], further suggesting that OC is not a mutator. However, because this estimate is based on only a single substitution (presumably drawn from a Poisson distribution), its confidence interval is large and it may be severalfold inaccurate in either direction, a caveat shared by a previous estimate for E. coli that was also based on mutations accumulated during a laboratory evolution experiment (4).
Discussion
Two lines of evidence support the view that natural selection was a major force in driving mutations accumulated in OC to fixation or detectable frequency. First, if the mutation rate of the OC lineage was similar to previous estimates of (nonmutator) mutation rates for other Gram-negative bacteria such as E. coli (as we have provisionally inferred above), only one or very few neutral mutations should have risen to high frequency within 1,000 generations. Because (i) none of the accumulated mutations appear to cause a mutator phenotype, (ii) 14, rather than 1 or 2, mutations rose to detectable frequency, and (iii) the population from which GVB207.3 was sampled underwent a large increase in fitness (6), it is likely that a large proportion of these mutations increased to high frequency by the power of natural selection (either by direct selection or by hitchhiking of one mutation in the genomic background of a distinct mutation favored by selection). Second, only 1 of the 11 mutations found within predicted coding sequence is synonymous, whereas three synonymous changes would have been expected had there been no bias toward the accumulation of nonsynonymous mutations. Although this discrepancy has a 14% probability of having occurred by chance with a sample size of eleven (binomial test), it is nonetheless suggestive of a bias among coding-region mutations toward changes that altered proteins, which are much more likely to have affected fitness significantly than are synonymous mutations.
A new mutant genotype present as a single cell in the population immediately after the first daily dilution transfer (≈2 × 108 individuals) would have needed a relative fitness advantage of ≈2% to achieve fixation by selection alone over 1,000 generations (for the population as a whole). [Relative fitness is defined as the ratio of two competitors’ actualized Malthusian parameters (22).] Alternatively, if all 13 mutations in GVB207.3 other than the synonymous codon substitution rose to high frequency in nonoverlapping selective sweeps that together spanned the entire 1,000 generations, then the average sweep time would have been ≈77 generations. The fitness advantage required for each mutation to sweep so rapidly under such a model, however, is very high (≈29% advantage for fixation in 77 generation for mutants first appearing immediately after a dilution transfer). The population from which GVB207.3 was isolated, however, had only increased its maximum growth rate a total of ≈28% in 1,000 generations (6), indicating a more complex history of mutation-frequency dynamics.
Selective sweeps may have overlapped such that any given adaptive mutation may have occurred in a genomic background already carrying a prior adaptive mutation but before the first mutation had risen to high frequency. Under another scenario, mutations of relatively small (or zero) fitness effect may have hitchhiked to high frequency in the same genomic background as an alternative mutation of large positive effect. Horizontal gene transfer between cells of laboratory M. xanthus cultures has not been reported, despite attempts to detect it (e.g., ref. 23). Thus, genomic evolution in the GJV1 → GVB207.3/OC lineage may have been primarily clonal, and accumulated mutations are likely to have occurred successively within a single lineage rather than having been recombined into the same chromosome from distinct mutant lineages. Assuming clonal reproduction and average mutation fitness benefits well below 29%, we expect some mutation sweeps to have been overlapping or to have involved hitchhiking, or both.
Only one mutation distinguishes the PX genotype from its immediate ancestor OC, and this mutation was subsequently shown to be the sole cause of the evolutionary transition from a socially incompetent cheater to a superior cooperator (10). This mutation altered the central position of a seven-base cytosine run located 128 bases above the start codon of a predicted GNAT-family acetyltransferase (Table 1) (24). The role of this acetyltransferase in M. xanthus physiology and development has not been characterized. Real-time PCR and microarray data show that the developmental expression patterns of this acetyltransferase and many other genes in the PX genome are significantly different from that of either GJV1 or OC (10) (S. V. Kadam and G.J.V., unpublished results), suggesting that the compensatory PX mutation generated a functionally novel transcriptome phenotype associated with development.
Materials and Methods
Strain DK1622 from the laboratory of D. Kaiser at Stanford University served as the type strain for a sequencing project that was initiated by Monsanto and completed by TIGR (W. C. Nierman, D. Kaiser, and B. S. Goldman, personal communication). This project was carried out by whole-genome random shotgun cloning and Sanger-type sequencing of plasmid libraries using capillary DNA sequencers. The initial sequence obtained was 7,139,763 bp and was deposited at TIGR in GenBank format November 17, 2004. This initial annotation was subsequently updated (February 28, 2006; NCBI accession CP000113). The NCBI CP000113 sequence and annotation will be made publicly available upon publication of the DK1622 genome manuscript. The DK1622 genome sequence (November 17, 2004, version) was obtained from TIGR and used for assembly of PX sequences (from both 454 Sequencing and capillary reads). Strain GJV1 is a derived clone of DK1622. GJV1 was obtained from a frozen stock of DK1622 in the laboratory of L. Kroos (Michigan State University, East Lansing) in March 1995. The stock of L. Kroos from which GJV1 was obtained had been received from D. Kaiser in January 1990 in a postal shipment on nutrient agar. Upon receipt, the culture was streaked for isolation of a single colony, which was grown to high density and frozen. From 1990 until the DK1622 sequencing project began, the DK1622 stock of D. Kaiser was kept frozen except for periodic growth phases in 5 ml of liquid medium to replenish the freezer stock [approximately once every 2 years (D. Kaiser, personal communication)] and a final growth phase in 1 liter of liquid to prepare genomic DNA for the Monsanto/TIGR sequencing project initiated in 1999.
Strain GJV1 is referred to as strain “S” in refs. 6 and 8, as “DK1622” in ref. 7, and “W1” in ref. 9. GVB207.3 is one of three clones initially isolated from a 1,000-generation population of lineage S2 in ref. 6 for analyses reported in ref. 8. Strain OC was created by integration of Tn5 (which encodes resistance to kanamycin) into the GVB207.3 genome after transformation with the plasmid pDW79 (8). Strain PX evolved directly from OC during a competition experiment described in refs. 9 and 10. The estimate of 60 generations (or fewer) separating PX from OC includes the period of preconditioning before mixing of OC with its competitor GJV2 (a spontaneous rifampicin-resistant mutant of GJV1).
Genomic DNA of strain PX was prepared by using a Qiagen Genomic-tip kit. Primer sequences and PCR-sequencing conditions are available upon request. Possible mutation effects on protein structure were evaluated by using the Simple Modular Architecture Research Tool (SMART) available at http://smart.embl-heidelberg.de (25). Mutation positions and gene start/end positions >6727645 in Tables 1 and 2 are increased by one relative to their positions in the February 28 annotation sequence of DK1622 due to identification of a base pair missing at that position in the annotation sequence present in the actual sequence.
To estimate the percentage of single base pair substitutions in the DK1622 genome likely to be neutral (or near neutral) with respect to fitness, we excluded noncoding regions (9.5% of the genome) from our analysis because of ignorance about the likely fitness effects of most noncoding mutations. We calculated that 26.3% of all possible mutations within coding regions would lead to silent mutations. To do this, the codon usage table was calculated for each gene by using the program cusp from the emboss package (26). The output of this program contains the number of occurrences of each codon type in a gene. For every codon “c” with a number of occurrences “#c” in a gene, the number of mutations “s_c” that leave the respectively coded amino acid unchanged is known. Thus, the total number of silent mutations for an individual gene could be computed by multiplying #c*s_c and summing up these numbers over all possible codons. This procedure was repeated for all genes in the genome. The number of possible silent mutations over all genes divided by the number of all possible mutations in coding regions leads to the fraction of coding-region mutations that are silent.
Acknowledgments
We thank E. Holmes, R. Lenski, and the anonymous reviewers for helpful comments on the manuscript.
Abbreviation
- TIGR
The Institute for Genomic Research.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Elena S. F., Lenski R. E. Nat. Rev. Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 2.Sachs J. L., Bull J. J. Proc. Natl. Acad. Sci. USA. 2005;102:390–395. doi: 10.1073/pnas.0405738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenski R. E., Ofria C., Pennock R. T., Adami C. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- 4.Lenski R. E., Winkworth C. L., Riley M. A. J. Mol. Evol. 2003;56:498–508. doi: 10.1007/s00239-002-2423-0. [DOI] [PubMed] [Google Scholar]
- 5.Kaiser D. Proc. Natl. Acad. Sci. USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velicer G. J., Kroos L., Lenski R. E. Proc. Natl. Acad. Sci. USA. 1998;95:12376–12380. doi: 10.1073/pnas.95.21.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velicer G. J., Kroos L., Lenski R. E. Nature. 2000;404:598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 8.Velicer G. J., Lenski R. E., Kroos L. J. Bacteriol. 2002;184:2719–2727. doi: 10.1128/JB.184.10.2719-2727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiegna F., Velicer G. J. Proc. R. Soc. London Ser. B; 2003. pp. 1527–1534. [Google Scholar]
- 10.Fiegna F., Yu Y.-T. N., Kadam S. V., Velicer G. J. Nature. 2006;441:310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
- 11.Shendure J., Porreca G. J., Reppas N. B., Lin X. X., McCutcheon J. P., Rosenbaum A. M., Wang M. D., Zhang K., Mitra R. D., Church G. M. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 12.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z. T., et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider D., Lenski R. E. Res. Microbiol. 2004;155:319–327. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Cooper V. S., Schneider D., Blot M., Lenski R. E. J. Bacteriol. 2001;183:2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S. S., Kaiser D. Mol. Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 16.Wissel M. C., Wendt J. L., Mitchell C. J., Weiss D. S. J. Bacteriol. 2005;187:320–328. doi: 10.1128/JB.187.1.320-328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabel C., Maier R. J. Nucleic Acids Res. 1990;18:6143. doi: 10.1093/nar/18.20.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaels M. L., Pham L., Cruz C., Miller J. H. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claverie-Martin F., Diaz-Torres M. R., Kushner S. R. Gene. 1987;54:185–195. doi: 10.1016/0378-1119(87)90486-0. [DOI] [PubMed] [Google Scholar]
- 20.Kimura M. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 21.Drake J. W. Proc. Natl. Acad. Sci. USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C. Am. Nat. 1991;138:1315–1341. [Google Scholar]
- 23.Caberoy N. B., Welch R. D., Jakobsen J. S., Slater S. C., Garza A. G. J. Bacteriol. 2003;185:6083–6094. doi: 10.1128/JB.185.20.6083-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vetting M. W., LP S. d. C., Yu M., Hegde S. S., Magnet S., Roderick S. L., Blanchard J. S. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Letunic I., Copley R. R., Schmidt S., Ciccarelli F. D., Doerks T., Schultz J., Ponting C. P., Bork P. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice P., Longden I., Bleasby A. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]