Abstract
Cattle domestication from wild aurochsen was among the most important innovations during the Neolithic agricultural revolution. The available genetic and archaeological evidence points to at least two major sites of domestication in India and in the Near East, where zebu and the taurine breeds would have emerged independently. Under this hypothesis, all present-day European breeds would be descended from cattle domesticated in the Near East and subsequently spread during the diffusion of herding and farming lifestyles. We present here previously undescribed genetic evidence in contrast with this view, based on mtDNA sequences from five Italian aurochsen dated between 7,000 and 17,000 years B.P. and >1,000 modern cattle from 51 breeds. Our data are compatible with local domestication events in Europe and support at least some levels of introgression from the aurochs in Italy. The distribution of genetic variation in modern cattle suggest also that different south European breeds were affected by introductions from northern Africa. If so, the European cattle may represent a more variable and valuable genetic resource than previously realized, and previous simple hypotheses regarding the domestication process and the diffusion of selected breeds should be revised.
Keywords: domestication, Europe, mtDNA, aurochs
The domestication of cattle (Bos taurus and Bos indicus) from wild aurochsen (Bos primigenius) was an important step in human history, leading to extensive modifications of the diet, the behavior, and the socioeconomic structure of many populations (1). This process started ≈11,000 years ago (2, 3), and the deep genetic divergence between taurine (B. taurus) and zebu (B. indicus) cattle breeds points to at least two independent domestication events from two distinct aurochsen groups (4). Archaeological data suggest that the zebu domestication occurred probably in the Indus Valley (today’s Pakistan) (5), with a primary diffusion of these breeds in India and only a more recent (<3,000 years) secondary male introduction in Africa (6). Conversely, the most likely domestication site for the taurine breeds is considered a westernmost area in the Near East, the Fertile Crescent (FC), even though an independent domestication event may have occurred in Africa (7–9).
Approximately 480 cattle breeds are recognized today in Europe (10). All of them are of the taurine type, and all of them are considered to be in strict mitochondrial genetic continuity with the breeds selected in the FC (3, 6, 11, 12). The evidence for this hypothesis can be summarized as follows: (i) none of the British aurochs sequences typed so far was found in modern cattle, and the divergence between the two distinct clades (B. taurus and B. primigenius) predates by several thousand years the domestication event; (ii) European cattle belong almost exclusively to a single group of mtDNA sequences, haplogroup T3, which represents a subset of the variation observed in the Near East (where four major haplogroups, T, T1, T2, and T3, are present); and (iii) the shape and the age of the European network of sequences is compatible with a demographic expansion from a small population after domestication. Nuclear markers also seem to show a higher variability in the Near East than in other regions (13, 14), thus supporting the conclusions based on mitochondrial data.
The mtDNA and, possibly, the nuclear DNA of European cattle, therefore appear to descend from a group of aurochsen not typed yet (but very different from the British ones), and the evidence from modern breeds points to a Near Eastern ancestor. By following the south-east to north-west cultural and demographic diffusion of the new lifestyle of farming and herding (1, 15), cattle breeds would have dispersed in the continent with no genetic contact with local aurochsen. Interestingly, a crucial point of this hypothesis, that is, the assumption that all European aurochsen had DNA sequences similar to six British samples, already has been (and, as we will show, unsafely) used to define a genetic marker of cattle domestication. Accordingly, ancient remains, which are difficult to classify because morphological traits have overlapping distributions in cattle and aurochs and diagnostic features are identified only in horn and some cranial element, tend to be attributed to a cattle or to an aurochs depending on their mtDNA sequence (3, 16, 17).
We believe that contrasting emerging evidence calls now for a reevaluation of the single-origin hypothesis of the European cattle. In fact, (i) this hypothesis is based on limited DNA sampling of modern breeds from Southern Europe and Northern Africa and only six aurochsen from a single geographically restricted area in Northern Europe; (ii) the aurochs went extinct in Europe no more than 400 years ago (2, 18) and, hence, the cattle and local wild aurochsen coexisted for millennia during which interbreeding was possible; (iii) recently, a 4,000-year-old molar from Northern Spain, morphologically attributed to a cattle, was proved to have a mtDNA sequence of the British aurochs type (17), and the tooth could have belonged to a hunted aurochs morphologically misattributed to a cattle, but local domestication or cattle-aurochs hybridization cannot be excluded; (iv) the North African influence, at least on Iberian breeds, is well documented (14, 17, 19, 20), even if it is usually attributed to occasional historical (the Moorish occupation) or Bronze Age (exchanges via the Straits of Gibraltar) events; (v) archeological data suggest that farmers spread from the Near East to northwestern Europe by following continental routes but also westward through the Mediterranean Sea by following maritime routes, and, thus, a genetic influence from North African cattle is possible (21–23); finally, (vi) two recent analyses, based on the mtDNA variation observed in the pig (24), and the MHC variation expected when different domestication scenario are simulated for different mammals (25), seems to indicate that simple well accepted hypotheses regarding livestock domestication might be wrong.
Here we test explicitly the null hypothesis (H0) of a single origin of the European cattle by separately comparing it with two alternative, nonmutually exclusive, hypotheses of multiple origins: a genetic contribution of the European aurochsen, due to local domestication or introgression events (H1), and a significant genetic impact of cattle of North African origin introduced in Southern Europe (H2).
The H1 hypothesis will be tested by using previously undescribed ancient mtDNA sequences obtained from continental European aurochsen. The specimens we typed were recovered in Southern Italy and are dated between 7,000 and 17,000 years ago. Three of them are older than any previously sequenced aurochs. With this analysis, we also test the hypothesis that mtDNA sequences of British aurochsen are representative of the genetic variation of this species in Europe, which was assumed to be true in studies suggesting that European cattle have a single Near Eastern origin (11, 12).
The H2 hypothesis, introgression of African genetic lineages into Europe, does not refer to recent and occasional introductions of individuals in geographically restricted areas. It implies, on the contrary, a more widespread process that would have shaped the genetic composition of different breeds in different southern European regions. Being proved, such widespread contribution of North African cattle could be only explained by the seaborne dispersal of cattle and pastoralism across the Mediterranean Basin. We test this hypothesis by sequencing mtDNA control region in 520 modern individuals from 51 different breeds (and 17 countries; see Table 2, which is published as supporting information on the PNAS web site), and analyzing a joint data set of 1,197 published and unpublished sequences (ref. 11; see also refs. 10–22 in Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
We sequenced mtDNA because it has proved very useful for inferring the origins and phylogenetic history of many species including livestock and humans (26, 27) but also because it is the most reliable (and presently nearly the only) genetic locus used to study ancient samples (28). New techniques are opening also the possibility to study nuclear DNA from ancient specimens (e.g., ref. 29), but the hypotheses tested in our study need necessarily the comparison with previously published modern and ancient DNA sequences.
Results and Discussion
Testing the Hypothesis of a European Aurochsen Contribution in European Breeds (H1).
All of the Italian aurochs sequences fall within the range of variation observed in modern cattle, and specific relationships with the genetically distinct clade of aurochsen from the British Isles do not emerge (Fig. 1; see Data Set 1, which is published as supporting information on the PNAS web site, for the sequences of the clones). The nucleotide sites amplifiable from each sample, ranging from 151 to 384 bp, identified (i) three sequences (Au-It1, Au-It4, and Au-It5) identical to the typical modern European haplotype, T3; (ii) one sequence (Au-It2) differing from T3 by two substitutions but still belonging to the T3 clade; and (iii) one sequence (Au-It3) differing from T3 by two substitutions. Au-It3 has two of three typical substitutions that separate the African clade T1 from the European clade T3, but its attribution to either major clades observed in modern cattle is ambiguous. In fact, the two “African” substitutions in Au-It3 (a T in 16,050 and a C in 16,113) are affected probably by recurrent mutations (11), whereas the C in 16,255, which would strongly favor an African origin, is not observed in this specimen.
Fig. 1.
The polymorphic sites obtained by comparing the most frequent modern cattle sequences (T1–T4) with the Italian (this study; AU-It1–AU-It5) and British (refs. 11, 12; AU-Br1–AU-Br6) aurochs sequences. T1 is the most common African sequence, T2 is found almost only in Middle Eastern and Anatolian breeds, T3 is the most common European sequence, and T4 is found only in eastern Asiatic breeds. Haplotypes T1 and T3 are also the roots of the most common European and African clades, called T1 and T3 haplogroups, respectively. Note that a five-digit position number is shown vertically above the sequences.
This result indicates that European aurochsen were structured geographically. British and Italian aurochsen were probably different populations, with a large average percentage of nucleotide divergence (6.2, SD = 1.5), even if the recent finding in northern Spain of a 4,000-year-old specimen with a mtDNA sequence belonging to the British aurochs haplogroup suggests that British haplotypes were not restricted to northern Europe. More data are needed to clarify the phylogeographic pattern of the aurochs, but the fact that all five Italian aurochsen had cattle-like mtDNA sequences is very informative. It implies that the mtDNA control region cannot be used as a genetic marker of domestication, and, more importantly, it also contradicts the view that British aurochs sequences can be regarded as the standard European aurochs sequences, leading to ruling out a genetic contribution of European aurochsen into European cattle. But can the sequences of the Italian aurochsen be taken as evidence in favor of this contribution? We present here two indirect pieces of evidence based on the comparison between the 116-bp highly variable region of maximum overlap between aurochs and 1,197 modern B. taurus sequences.
First, we estimate the frequency of the three Italian aurochsen haplotypes in modern breeds pooled in 12 major geographic regions (Table 1). The genetic affinity between Italian aurochsen and the cattle breeds from Europe, the FC area (Anatolia and the Near East), and America (recently introduced breeds) is confirmed by the fact that the haplotype observed in three of five aurochsen is largely the most common haplotype in all these modern groups. But the frequencies of this match are different. The highest value is observed in Italy (59.7%), followed closely by mainland Europe and northwestern Europe, with all of the other groups (including likely regions of initial domestication, such as Anatolia and Middle East) having a much smaller frequency of this sequence. Overall, the most frequent haplotype in the Italian aurochsen is observed in 44.3% of the European modern samples, but it is in 31.9% of Middle-Eastern and Anatolian cattle (P < 0.05 by using either χ2 or Fisher test). Interestingly, the frequency of this haplotype increases by an additional 10% in Italy when the only Italian breed with a well known northern European origin (Pezzata Rossa) is not considered. This pattern is not expected under the hypothesis H0 (single origin in the FC of European breeds). H0, in fact, predicts that European and Middle Eastern/Anatolian breeds should be, on the average, equally divergent from all European aurochsen (which have nothing to do with them under this hypothesis). On the other hand, if we exclude random convergence of allele frequencies, only the hypothesis H1, which assumes a genetic contribution of at least some European aurochsen into European breeds, could explain (i) a higher genetic proximity of these aurochsen with European breeds and (ii) that aurochsen from a specific area resemble mostly the breeds currently found in that area.
Table 1.
Percentage of individuals in modern breeds from different geographic areas identical to each of the three different sequences (116 bp) found in the Italian aurochs
| n | Au-It1, AU-It4, AU-It5, % | AU-It3, % | Au-It2, % | |
|---|---|---|---|---|
| Italy | 62 | 59.7* | 4.8 | 0.0 |
| Iberian Peninsula | 142 | 38.0* | 14.5 | 1.4 |
| Balkan Peninsula | 72 | 33.3* | 1.4 | 2.8 |
| Mainland Europe | 111 | 54.9* | 0.0 | 0.9 |
| Central-Eastern Europe | 30 | 23.3* | 0.0 | 0.0 |
| Britain | 81 | 37.0* | 0.0 | 1.2 |
| Northwestern Europe | 51 | 54.9* | 0.0 | 11.8 |
| Middle East | 37 | 29.7* | 0.0 | 0.0 |
| Anatolia | 60 | 33.3* | 1.6 | 0.0 |
| Eastern Asia | 59 | 16.9 | 0.0 | 0.0 |
| Africa | 250 | 0.8 | 38.0* | 0.0 |
| America | 242 | 37.6* | 6.2 | 0.4 |
Twelve groups were defined by following and updating the geographic scheme in ref. 11, Italy, Iberian Peninsula (Portugal and Spain), Balkan Peninsula (Greece, Albania, Serbia, Romania, Bulgaria, Serbia, and Slovenia), Mainland Europe (France and Germany), Central-Eastern Europe (Slovakia, Ukraine, and Hungary), Great Britain, Northwestern Europe (called Western-fringe Europe in ref. 11), Middle East, Anatolia, Eastern Asia (Tibet, Korea, and Japan), Africa, and America (Central and Southern breeds). n, sample size.
*The corresponding allele is also the most frequent in that area. In Eastern Asia, the most frequent alleles is T4 (see Fig. 1) with 20.3%.
Second, we focus only on the modern Italian breeds and their potential ancestors, which were aurochsen from the FC area under H0, or, with unknown proportions, both aurochsen from Italy and FC under H1. Call D(IC-IA) the genetic distance between Italian cattle and Italian aurochsen and D(IC-FCA) the genetic distance between Italian cattle and aurochsen from the FC area. If Italian and FC aurochsen were not genetic differentiated, we expect that d = D(IC-FCA) − D(IC-IA) = 0, both under H0 and H1. However, if Italian and FC aurochsen were genetically differentiated, we expect d < 0 [D(IC-FCA) < D(IC-IA)] under H0 and d > 0 [D(IC-FCA) > D(IC-IA)] under H1. We tested formally this expectation by using a resampling approach (see Fig. 3, which is published as supporting information on the PNAS web site) to find the empirical confidence intervals around d. The average nucleotide divergence was used as a measure of genetic distance between populations, because it is less affected than random exact matches by recurrent mutations or single-base errors in ancient DNA typing. Present-day Middle Eastern and Anatolian breeds were used to estimate the unknown genetic variation of their likely ancestors, i.e., the FC aurochsen. This kind of approximation is based on the assumption that, when different evidence suggest a certain degree of chronological continuity, the best available proxy for a past population is the modern population dwelling in the same area (see e.g., refs. 30–32). This approximation seems justified for the FC aurochsen, given the lack of evidence for major displacement processes since the Neolithic period and the large demographic increase that reduced possible drift effects. The 90% confidence interval (C.I.) around d excludes 0 and the 95% C.I. is (–0.103, 1.195). In 95.4% of the resampled data set, d > 0. This finding, which suggests that Italian cattle are more similar to Italian than FC aurochsen ancestors, is clearly unexpected under H0 but compatible with H1.
Small to moderate levels of local gene flow from wild B. primigenius females in cattle breeds are consistent with the reasonable idea that, at least initially, cattle herds were free-ranging (and contacts with aurochsen not infrequent), and/or that cattle breeders might have favored the introgression of wild animal genomes adapted to the local environment. British and Italian aurochs were different, possibly monophyletic, populations, and the open question is why the large fraction of aurochs mtDNA variation represented by the Northern groups was apparently lost during domestication. It is possible that pastoralist societies in southern and northern Europe used different breeding techniques, with the latter more concerned with herd guarding. This hypothesis might also represent an alternative explanation for the different patterns of microsatellite variation observed between Mediterranean and northern European breeds, which is currently interpreted as evidence for the distinct Mediterranean and Danubian routes of migration from the Near East (14). The analysis of more breeds in northern Europe and northwestern Asia possibly could clarify whether all of the genetic variation referred to Northern aurochsen really was lost forever.
Testing the Hypothesis of an African Cattle Contribution in Southern European Breeds (H2).
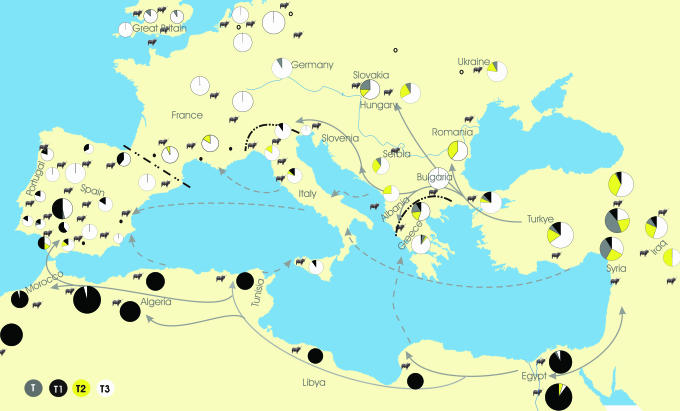
Our extensive sampling across North Africa reveals that the T1 haplogroup is almost fixed across this region (Fig. 2). Nevertheless, 63 different sequences with the T1 motif are observed, producing a total nucleotide diversity in North Africa (1.76%, SD = 0.15) slightly higher than observed in the Middle East (1.65%, SD = 0.14) or in Anatolia (1.48%, SD = 0.13), where all four major haplogroups are found. These observations, together with the fact that T1 haplotypes are very rare in the Middle East and Anatolia, appear consistent with the previously suggested hypothesis (7, 11) that African cattle were independently domesticated. This hypothesis, however, also would imply that Northern African and Near Eastern aurochsen were genetically differentiated even without major barriers limiting their dispersion (with the former being mainly T1-like and the latter being non-T1-like) or that the African and Near Eastern domestication processes were very different (with the former producing a much more intense bottleneck than the latter). As far as genetic data are concerned, the simpler hypothesis of an introduction in Africa of few T1-like cattle domesticated in the Near East, and their subsequent demographic expansion and genetic diversification appears more parsimonious.
Fig. 2.
European spread of agropastoralism. Each black cattle figure represents a population sample point. Different hypothesized maritime routes (dashed line with arrow) and continental route (solid line with arrow) are indicated. Dash-dot lines are suggestive of the geographic limits of African cattle influence in Europe. Pie chart represents the frequencies of the four major mtDNA haplogroups, with circle sizes proportional to sample sizes. The figure is adapted from ref. 33.
Regardless of the origin of the African breeds, T1 mtDNA sequences are clearly a distinctive feature of their genetic composition. The distribution of the T1 haplogroup outside Africa thus can be used to understand the relationships between cattle breeds across the Mediterranean, and an interesting pattern seems to emerge in Europe (Fig. 2): T1 sequences are relatively common (with frequencies ranging from 5% to 30%) in different breeds from Portugal, Spain, Italy, and Greece.
The presence of T1 mainly along the Mediterranean shores of Europe (near Africa), but not in central and northern Europe, is suggestive of the occasional introduction of cattle by boat from North Africa into southern Europe and is difficult to reconcile with any gene flow process unrelated with the sea. But when did this process occur? The presence of T1 haplotypes previously observed in Portugal was attributed to historical migration due to North African, possibly Moorish, conquerors (19). However, even if 63 and 11 different T1 haplotypes are observed in Africa and Europe, respectively, only two of them are present in both regions. In addition, (i) T1 haplotypes can be found well beyond the area of maximum Moorish expansion, (ii) recent introductions of exotic cattle are usually male mediated (not affecting mtDNA) (34), and (iii) one T1 haplotype has been recently observed in a sample of 16 Bronze Age cattle remains from Spain. So, the hypothesis of a recent and geographically restricted introduction of African cattle does not seem sufficient to explain the T1 distribution in Europe. On the contrary, DNA data are compatible with earlier gene flow into several Mediterranean regions. There is evidence of early diffusion of cattle pastoralism by people crossing arms of sea (21–23), and, hence, the same process may have led to the dispersal in Europe of breeds carrying the T1 haplotype.
Conclusions
The modern and ancient mtDNA sequences we present here do not support the currently accepted hypothesis of a single Neolithic origin in the Near East. The processes of livestock domestication and diffusion were certainly more complex than previously suggested, and our data provide some evidence in favor of the hypothesis that the origin of European cattle is multiple. Breeds domesticated in the Near East and introduced in Europe during the Neolithic diffusion probably intermixed, at least in some regions, with local wild animals and with African cattle introduced by maritime routes. As a consequence, European breeds should represent a more diverse and important genetic resource than previously recognized, especially in the Southern regions.
Materials and Methods
Ancient B. primigenius Samples.
DNA was extracted from the teeth of five Italian B. primigenius samples and the hypervariable control region of the mitochondrial genome was typed. The remains were recovered from three different caves (Paglicci and Grotta delle Mura in Puglia and Termini Imerese in Sicily) in southern Italy. Low levels of humidity typical of caves are considered an important factor that retards DNA degradation (35). Radiocarbon determination dated three of them at 17,100 ± 300 (Au-It1), 16,260 ± 160 (Au-It2), and 15.860 ± 80 (Au-It3) years ago, respectively (36). The fourth sample (Au-It4) was recovered in the stratigraphic unit 130. This unit is not dated, but it is included between unit 135 (radiocarbon date: 11,420 ± 100 BP) and unit 62 (radiocarbon date: 15,860 ± 80 B.P.). The fifth sample (Au-It5) does not have a radiocarbon reference but was morphologically attributed to B. primigenius, and, on the basis of the layers information, it was dated at ≈7,000 years ago. Genetic typing of ancient samples is technically challenging because DNA is degraded and present in small amounts, and stringent standards for the authentication of ancient DNA are required (37, 38).
In particular, for all samples (a) DNA was extracted in a laboratory room in Florence exclusively dedicated to ancient DNA analyses; (ii) for each sample, at least two independent DNA extractions were performed, and PCR controls produced negative results; (iii) amplification of long DNA fragments, unusual in ancient DNA analyses, was not observed, and the final consensus sequences make phylogenetic sense; (iv) different primer pairs were used to amplify different overlapping fragment; (v) several clones (total = 102, see Data Set 1) were analyzed for each fragment; the average rate of Taq misincorporation across fragments was low (5.1 for every 1,000 bp), and at least two-thirds of the clones showed the consensus nucleotide at each DNA fragment; (vi) DNA extraction, amplification, cloning, and sequencing of four fragments were independently repeated in a different laboratory (Barcelona) for Au-It1 by using sets of primers different from the primers used in Florence, and the sequences were consistent across laboratories; (vii) an independent set of extraction, amplification, and cloning was performed for Au-It3 by using the uracil-N-glycosylase treatment, and no evidence of sequence artifacts due to postmortem damages was found; and (viii) the degree of racemization for three amino acids was low in all samples, suggesting that DNA preservation was good. The results obtained by applying these criteria suggest, therefore, that the aurochs sequences we present can be confidently considered authentic. Additional details of the methods used to type the ancient samples are given in Supporting Materials and Methods.
Conclusions on the Authentication of Ancient DNA.
The authentication of DNA sequences from ancient remains with absolute certainty is virtually impossible when identical or very similar sequences are also found in modern samples. However, we are confident that the sequences we present are endogenous, because (i) contamination with external DNA in teeth specimens, which are protected by a layer of enamel and inserted in the dental alveolus, is uncommon (39); (ii) before their arrival in the molecular laboratory, the remains were manipulated only by archaezoologists; (iii) one of the laboratory (Barcelona) involved in this study never was involved in the genetic analysis of cattle samples; the other laboratory (Florence) produced the sequences for 21 Italian cattle presented in this study, but ancient and modern DNA were extracted, amplified, and cloned in different isolated rooms; and (iv) stringent criteria and protocols for DNA validation were applied. In addition, the contamination of all five specimens seems very unlikely, and the major conclusion of our study would not change even considering only one or two samples.
Modern B. taurus Samples.
Sampling procedures.
We sampled a total of 520 native cattle individuals from across the Mediterranean Basin and Eastern Europe, representing 51 breeds representing 17 countries (see Table 2). To avoid exotic crosses with native cattle (which are, in general, unlikely to affect the maternal lineages), we only sampled small countryside villages, taking also great care to exclude related individuals. After local anesthesia, small (3 mm3) skin biopsies were collected and stored in 95% ethanol before DNA extraction.
DNA extraction, PCR amplification, and sequencing.
DNA was extracted from ethanol preserved tissue by using standard commercial kits (tissue and blood kits; Qiagen, Chatsworth, CA). A 320-bp fragment from mtDNA hypervariable region I control region by using the primers as suggested in ref. 11, and PCR products were purified by using the QIAquick PCR columns (Qiagen). All sequences were obtained for both DNA strands by using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) in a 20-μl volume containing 40–50 ng of purified genomic DNA and 3.2 pmol of primer and were electrophoresed in ABI3100 sequencer (Applied Biosystems). Raw sequences were edited and aligned by using seqscape (Applied Biosystems). The resulting sequences then were aligned with other published sequences (ref. 11 and refs. 10–22 in Supporting Materials and Methods), and a database of 1,197 sequences thus was available.
Supplementary Material
Acknowledgments
We thank Dan Bradley for his critical reading of an earlier version of this paper. This research was supported by funds from the University of Florence; the University of Ferrara; the Italian Ministry of Education, University, and Research; the Azienda Regionale Sviluppo Innovazione Settrore Agro Forestale Toscana; and the Departament d’Universitats, Recerca i Societat de la Informaciò de la Generalitat de Catalunya. A.B.-P. is supported by Fundação para a Ciência e Tecnologia, Portugal, Research Grant SFRH/BPD/17822/2004.
Abbreviation
- FC
Fertile Crescent.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bellwood P. First Farmers. Oxford: Blackwell; 2004. [Google Scholar]
- 2.Clutton-Brock J. A Natural History of Domesticated Mammals. 2nd Ed. Cambridge, U.K: Cambridge Univ. Press; 1999. [Google Scholar]
- 3.Edwards C. J., MacHugh D. E., Dobney K. M., Martin L., Russell N., Horwitz L. K., McIntosh S. K., Mac Donald K. C., Helmer D., Tresset A., et al. J. Archaeol. Sci. 2004;31:695–710. [Google Scholar]
- 4.Loftus R. T., MacHugh D. E., Bradley D. G., Sharp P. M., Cunningham P. Proc. Natl. Acad. Sci. USA. 1994;91:2757–2761. doi: 10.1073/pnas.91.7.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meadow R. H. In: Harappan Civilisation. Possehl G., editor. New Delhi: Oxford Univ. Press and India Book House; 1993. pp. 295–320. [Google Scholar]
- 6.Bradley D. G., Loftus R. T., Cunningham P., MacHugh D. E. Evol. Anthropol. 1998;6:79–86. [Google Scholar]
- 7.Bradley D. G., MacHugh D. E., Cunningham P., Loftus R. T. Proc. Natl. Acad. Sci. USA. 1996;93:5131–5135. doi: 10.1073/pnas.93.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall F., Hildebrand E. J. World Prehistory. 2002;2:99–102. [Google Scholar]
- 9.Wendorf F., Schild R., Close E. The Palaeoecology of Africa and the Surrounding Islands. Vol. 18. The Netherlands: Balkema, Rotterdam; 1987. pp. 441–448. [Google Scholar]
- 10.Food and Agriculture Organization . In: World Watch List for Domestic Animal Diversity. Scherf B. D., editor. Rome: Food Agric. Org.-U.N. Environ. Programme; 2000. [Google Scholar]
- 11.Troy C. S., MacHugh D. E., Bailey J. F., Magee D. A., Loftus R. T., Cunningham P., Chamberlain A. T., Sykes B. C., Bradley D. G. Nature. 2001;410:1088–1091. doi: 10.1038/35074088. [DOI] [PubMed] [Google Scholar]
- 12.Bailey J. F., Richards M. B., Macaulay V. A., Colson I. B., James I. T., Bradley D. G., Hedges R. E., Sykes B. C. Proc. Biol. Sci.; 1996. pp. 1467–1473. [DOI] [PubMed] [Google Scholar]
- 13.Loftus R. T., Ertugrul O., Harba A. H., El-Baroyd M. A. A., MacHugh D. E., Park S. D. E., Bradley D. G. Mol. Ecol. 1999;8:2015–2022. doi: 10.1046/j.1365-294x.1999.00805.x. [DOI] [PubMed] [Google Scholar]
- 14.Cymbron T., Freeman A. R., Malheiro M. I., Vigne J.-D., Bradley D. G. Proc. Biol. Sci.; 2005. pp. 1837–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ammerman A. J., Cavalli-Sforza L. L. The Neolithic Transition and Genetics of Populations in Europe. Princeton: Princeton Univ. Press; 1984. [Google Scholar]
- 16.Kühn R., Ludt C., Manhart H., Peters J., Neumair E., Rottmann O. J. Anim. Breed. Genet. 2005;122:36–44. doi: 10.1111/j.1439-0388.2005.00507.x. [DOI] [PubMed] [Google Scholar]
- 17.Anderung C., Bouwman A., Persson P., Carretero J. M., Ortega A. I., Elburg R., Smith C., Arsuaga J. L., Ellegren H., Götherström A. Proc. Natl. Acad. Sci. USA. 2005;102:8431–8435. doi: 10.1073/pnas.0503396102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein H. The Origin of Domestic Animals of Africa. New York: Africana; 1971. [Google Scholar]
- 19.Cymbron T., Loftus R. T., Malheiro M. I., Bradley D. G. Proc. R. Soc. London. 1999;266:597–603. doi: 10.1098/rspb.1999.0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miretti M. M., Dunner S., Naves M., Contel E. P., Ferro J. A. J. Hered. 2004;95:450–453. doi: 10.1093/jhered/esh070. [DOI] [PubMed] [Google Scholar]
- 21.Price D. T. Europe’s First Farmers. Cambridge, U.K.: Cambridge Univ. Press; 2000. [Google Scholar]
- 22.Bellwood P. First Farmers. Oxford: Blackwell; 2004. [Google Scholar]
- 23.Zilhão J. Proc. Natl. Acad. Sci. USA. 2001;98:14180–14185. doi: 10.1073/pnas.241522898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson G., Dobney K., Albarella U., Fang M., Matisoo-Smith E., Robins J., Lowden S., Finlayson H., Brand T., Willerslev E., et al. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 25.Vilà C., Seddon J., Ellegren H. Trends Genet. 2005;21:214–218. doi: 10.1016/j.tig.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Bruford M. W., Bradley D. G., Luikart G. Nat. Rev. Genet. 2003;4:900–910. doi: 10.1038/nrg1203. [DOI] [PubMed] [Google Scholar]
- 27.Cavalli-Sforza L., Feldman M. Nat. Genet. 2003;33:266–275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- 28.Paabo S., Poinar H., Serre D., Jaenicke-Despres V., Hebler J., Rohland N., Kuch M., Krause J., Vigilant L., Hofreiter M. Annu. Rev. Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 29.Noonan J. P, Hofreiter M., Smith D., Priest J. R., Rohland N., Rabeder G., Krause J., Detter J. C., Paabo S., Rubin E. M. Science. 2005;309:597–599. doi: 10.1126/science.1113485. [DOI] [PubMed] [Google Scholar]
- 30.Cavalli-Sforza L. L., Menozzi P., Piazza A. Science. 1993;259:639–646. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- 31.Wilson J. F., Weiss D. A., Richards M., Thomas M. G., Bradman N., Goldstein D. B. Proc. Natl. Acad. Sci. USA. 2001;98:5078–5083. doi: 10.1073/pnas.071036898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupanloup I., Bertorelle G., Chikhi L., Barbujani G. Mol. Biol. Evol. 2004;21:1361–1372. doi: 10.1093/molbev/msh135. [DOI] [PubMed] [Google Scholar]
- 33.Zilhão J. J. Mediterranean Archaeol. 1993;6:5–53. [Google Scholar]
- 34.Hanotte O., Tawah C. L., Bradley D. G., Okomo M., Verjee Y., Ochieng J., Rege J. E. Mol. Ecol. 2000;9:387–396. doi: 10.1046/j.1365-294x.2000.00858.x. [DOI] [PubMed] [Google Scholar]
- 35.Poinar H. N., Kuch M., McDonald G., Partin P., Pääbo S. Curr. Biol. 2003;13:1150–1152. doi: 10.1016/s0960-9822(03)00450-0. [DOI] [PubMed] [Google Scholar]
- 36.Palma di Cesnola A. Il Paleolitico Superiore in Italia. Firenze, Italy: Garlatti e Razzai Editori; 1993. [Google Scholar]
- 37.Cooper A., Poinar H. N. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 38.Hofreiter M., Serre D., Poinar H. N., Kuch M., Paäbo S. Nat. Rev. Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- 39.Herrmann B., Hummel S. Ancient DNA. New York: Spinger; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.