Abstract
Considerable animal and human data have indicated that selenium is effective in reducing the incidence of several different types of cancer, including that of the prostate. However, the mechanism by which selenium inhibits carcinogenesis remains unknown. One possibility is that dietary selenium influences the levels of selenium-containing proteins, or selenoproteins. Selenoproteins contain selenium in the form of selenocysteine and perform a variety of cellular functions, including antioxidant defense. To determine whether the levels of selenoproteins can influence carcinogenesis independent of selenium intake, a unique mouse model was developed by breeding two transgenic animals: mice with reduced selenoprotein levels because of the expression of an altered selenocysteine-tRNA (i6A−) and mice that develop prostate cancer because of the targeted expression of the SV40 large T and small t oncogenes to that organ [C3(1)/Tag]. The resulting bigenic animals (i6A−/Tag) and control WT/Tag mice were assessed for the presence, degree, and progression of prostatic epithelial hyperplasia and nuclear atypia. The selenoprotein-deficient mice exhibited accelerated development of lesions associated with prostate cancer progression, implicating selenoproteins in cancer risk and development and raising the possibility that selenium prevents cancer by modulating the levels of these selenoproteins.
Keywords: cancer, selenium
Selenium is a chemopreventive agent for which there are considerable animal and human data indicating a protective role against cancer. Data reported from the Nutritional Prevention of Cancer (NPC) clinical trial indicated that dietary supplementation with 200 μg/day of selenium resulted in a 49% reduction in prostate cancer incidence (1). The link between selenium and prostate cancer risk has also been supported through prospective studies, which have shown an inverse relationship between prostate cancer risk and selenium levels in either serum or toenails (2, 3, 4). These data have provided impetus for additional human trials, including the SELECT trial, designed to assess the efficacy of selenium alone or in combination with vitamin E in preventing prostate cancer (5). The long latency period for prostate cancer development makes it a good candidate for early chemopreventive intervention through the use of nontoxic doses of nutritional supplements, such as selenium. However, to understand and maximize the effectiveness of these compounds, mechanisms by which they confer their benefits need to be understood.
The benefits of selenium may be mediated through its functions as a component of small selenium-containing metabolites or because of its role in the regulation and activity of selenium-containing proteins or selenoproteins. Selenium is incorporated into these selenoproteins as the 21st amino acid, selenocysteine, in response to the UGA codon in selenoprotein mRNAs (6). The insertion of selenocysteine in response to the UGA codon, instead of termination of translation, requires the presence of a recognition element within the 3′ UTR of the selenoprotein mRNA and is referred to as the selenocysteine insertion sequence (SECIS) element (7). Other factors required for selenoprotein synthesis include the selenocysteine tRNA (sec tRNA), the SECIS-binding protein 2 (SBP2) and the sec-specific elongation factor (EFsec) (6).
In humans, 25 selenoproteins have been identified, many of which have unknown functions (8). A link between selenoproteins and cancer risk is provided by genetic data that have implicated specific selenoproteins. Moscow et al. (9) provided the earliest evidence of loss of heterozygosity (LOH) of glutathione peroxidase-1 (GPx-1) in lung cancer, by using microsatellite markers that flank this gene at the 3p21 locus. LOH in the GPx-1 allele was subsequently observed in DNA from breast and head and neck tumors, implicating this protein in the development of these cancers (10, 11). Moscow et al. (9) also described a polymorphism at the 198 codon in GPx-1, which results in either a leucine or proline at that position. In a subsequent case control study, it was found that the allele with leucine at the 198 codon was associated with greater risk of lung cancer (12). In vitro studies in breast cancer cells engineered to specifically express either the leucine- or proline-containing GPx-1 allele indicate that the leucine-containing allele is less responsive to selenium supplementation than the proline allele, suggesting a possible functional consequence for the allelic identity at the 198 codon (10). Another selenoprotein, Sep15, is located on human chromosome 1p31, a region that is commonly deleted or mutated in cancer (13). Two polymorphisms within the 3′ UTR of the Sep15 gene have been reported, with a significant difference in the allelic frequency between African Americans and Caucasians (14). More recently, analysis in breast cancer DNA samples indicated a high frequency of LOH (28%) at the D1S2766 locus, which is tightly linked to Sep15, suggesting that allelic loss of this selenoprotein may be involved in breast cancer progression (15).
Despite the correlative genetic data, direct evidence of a role for selenoproteins in chemoprevention is lacking. A transgenic mouse model, referred to as i6A−, has been developed in which expression of selenoproteins is reduced because of the presence of a mutant sec tRNA (16). Sec tRNA contains relatively few modified residues, including isopentenyladenosine (i6A) at position 37. Expression of the i6A mutant Sec tRNA gene, lacking the modified residue at position 37 in transgenic mice (16), resulted in the reduction of most of the selenoproteins examined (i.e., GPx-1, GPx-3, D1, D2, and SelP) but not all (TR3). Importantly, these animals appear phenotypically normal and, therefore, provide a unique model to study the role of selenoproteins in chemoprevention (16).
To evaluate the role of selenoproteins in prostate cancer progression, i6A− mice were crossed with C3(1)/Tag mice, which develop prostate cancer because of the expression of the simian virus 40 (SV40) early-region large T and small t oncogenes (Tag). Tag expression is targeted to the prostate by the 5′-flanking region of the rat C3(1) gene. The biology of prostatic neoplasia in the C3(1)/Tag mouse closely resembles that observed in humans (17, 18). Because the course of prostate cancer development in this model is highly predictable and progresses over a relatively long time period, it provides a unique opportunity to investigate the histopathogenesis and molecular alterations that arise during tumor progression (19). In this study, a role for selenoproteins in prostate cancer etiology was investigated by using the transgenic mouse models described.
Results
GPx-1 Levels in Prostate Tissue from i6A−/Tag and WT/Tag Mice.
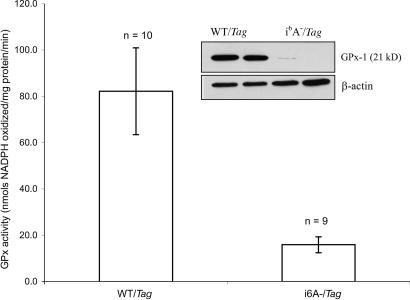
Because previous studies have not reported data on selenoprotein levels in the prostates of the i6A−/Tag mice, the levels of a representative selenoprotein, GPx-1, were determined in prostates obtained from 20-week-old mice by Western blot analyses using GPx-1-specific antibodies and direct enzyme assay. GPx-1 levels were dramatically reduced in prostates from the i6A−/Tag mice compared with the WT/Tag controls (Fig. 1Inset), bands representing GPx-1 in i6A−/Tag mice being apparent only at longer exposures (data not shown). This observation is corroborated by the significant decrease in GPx-1 enzyme activity in the prostates obtained from the i6A−/Tag mice as compared with the WT/Tag mice (Fig. 1). The observed decrease in GPx protein and enzyme activity in the prostate of the i6A− mice is consistent with data reported for other tissues in these mice (16).
Fig. 1.
GPx activity (nmol of NADPH oxidized per mg of protein per min) in ventral prostate tissues from 32-week-old WT/Tag and i6A−/Tag mice. Tissues were homogenized in 0.1M Na2HPO4 buffer (pH 7.5) and enzyme activity measured by using a standard coupled spectrophotometric method. Values are expressed as mean ± SEM. As compared with control WT/Tag mice, GPx-1 activity was 5-fold lower in ventral prostates and 7-fold lower in the dorsal prostates (data not shown) of i6A−/Tag mice (P < 0.05; two-tailed Student's t test). (Inset) A representative Western blot of GPx-1 protein levels in 20-week prostate tissues. A significant reduction in GPx-1 expression was observed in prostate tissues from i6A−/Tag mice relative to the WT/Tag animals, confirming the selenoprotein-deficient genotype of the i6A−/Tag animals.
Prostate Histopathology of Tissue Obtained from the i6A−/Tag and WT/Tag Mice.
To compare the presence, degree, and progression of prostatic epithelial hyperplasia and nuclear atypia in i6A−/Tag and WT/Tag mice, prostates were histologically analyzed at 12, 20, 32, and 42 weeks in a manner blinded for both time and genotype. Progressive hyperplasia, low-grade prostatic intraepithelial neoplasia (LGPIN), high-grade prostatic intraepithelial neoplasia (HGPIN), and microinvasion were observed with a positive linear trend over time in the Tag mice (Peto test, P < 0.001). Specific examples of each classified lesion are shown in Fig. 2. Epithelial hyperplasia was common from week 12 onward, and no differences were observed in incidence between the genotypes. In both genotypes, the incidence and severity of prostatic intraepithelial neoplasia (PIN) lesions and microinvasion were lower in the dorsolateral prostate as compared with the ventral lobe. In 39% of samples, the dorsolateral lobe did not exhibit nuclear atypia when PIN lesions were observed in the ventral prostate. In contrast, there were no instances when the ventral lobe lacked atypia and the dorsolateral prostate possessed PIN lesions. A reduced incidence of PIN in the dorsal prostates of the C3(1)/Tag mouse model has been reported (20) and is likely related to the promoter used for prostate targeting. C3(1), a subunit of prostate binding protein, is the major secretory product of the rodent ventral prostate and is normally expressed at minimal to undetectable levels in the dorsolateral prostate (20). Thus, further analysis of prostatic lesions focused on events occurring in the ventral lobe.
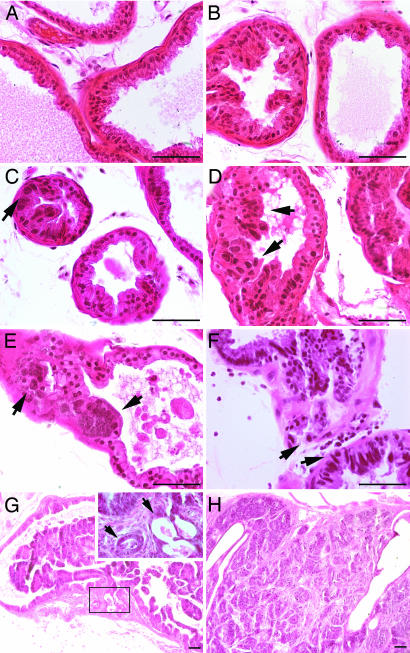
Fig. 2.
Representative prostate histopathology observed in the ventral lobes of WT/Tag and i6A−/Tag mice across time. (A) Normal ventral prostate epithelium (WNL) from a week-12 WT/Tag animal. (B) Epithelial hyperplasia without evidence of nuclear atypia (left acini) in a week-20 WT/Tag specimen. (C) Low-grade prostatic intraepithelial neoplasia (LGPIN) characterized by slight nuclear enlargement (arrow) and cellular crowding in a week-20 WT/Tag prostate. (D and E) HGPIN in week-20 i6A−/Tag prostate, as characterized by markedly enlarged or elongated nuclei, irregular nuclear membranes, increased nuclear/cytoplasmic ratios, hyperchromasia, and loss of polarity. (F) Microinvasion in week-32 i6A−/Tag prostate showing basement membrane breakdown and invasion of epithelium into the acinar wall (arrows) of associated HGPIN epithelium. (G) Focal carcinoma in week-42 i6A−/Tag prostate. Low-power image shows excessive intralumenal proliferation, PIN, basement membrane breakdown, and locally invasive carcinoma (boxed region). Higher magnification reveals discreet small glands (arrows). (H) Carcinoma in 42-week i6A−/Tag prostate with extensive invasion into stromal and loose periprostatic connective tissue and small-gland formation of variable size and shape with moderately differentiated epithelium. The magnification is indicated by the bar in each image, which is 50 μm.
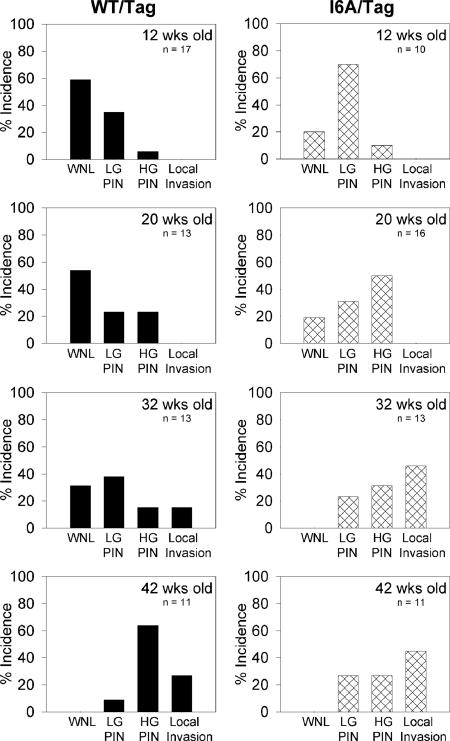
At every time point examined, there was clear evidence that the decrease in selenoproteins in the prostate gland resulted in acceleration of prostate pathology in the Tag animal model (Fig. 3). Statistical analysis confirmed that there was a significantly higher proportion of “within normal limits” (WNL) classification in the WT/Tag group across time as compared with the i6A−/Tag animals (Cochran–Mantel–Haenszel common relative risk test, P < 0.05), and there was a positive linear trend for the pathological event (LGPIN plus HGPIN plus microinvasion) between the two groups adjusted by time (Peto test, P < 0.05). At 12 weeks of age, 59% of WT/Tag ventral prostates were classified as WNL for nuclear morphology, whereas only 20% were WNL in the i6A−/Tag mice (P < 0.05). The incidence of LGPIN and HGPIN in the WT/Tag mice was 35% and 6%, respectively, at 12 weeks, whereas, for the i6A−/Tag mice, it was 70% and 10%, respectively. Similar findings were observed at 20 weeks, with a WNL classification of 54% for WT/Tag mice as compared with 19% of i6A−/Tag mice (P < 0.05). At this time, a shift was observed in the incidence of LGPIN and HGPIN, with 23% and 23%, respectively in the WT/Tag mice and 31% and 50%, respectively, in the i6A−/Tag mice, suggesting that pathologic progression were slowly taking place. As time elapsed, the i6A−/Tag mice displayed accelerated formation of higher-grade lesions compared with the WT/Tag controls. By 32 weeks of age, 31% of the ventral lobes in the WT/Tag mice were classified as WNL for nuclear morphology, whereas all i6A−/Tag mice had atypical nuclear lesions at this point in time (P < 0.03). Of added significance was the observation of focal microinvasions with associated PIN lesions in 46% of the i6A−/Tag ventral prostates, whereas only 15% of the WT/Tag mice had progressed to this invasive stage (P < 0.02). By 42 weeks of age, the WT/Tag prostates progressed to the same pathologic status as the i6A−/Tag mice, with no normal prostates and a high incidence (64%) of HGPIN lesions. Although not statistically significant (P < 0.10), the incidence of microinvasive and local carcinoma was higher in the i6A−/Tag prostates (45%) in contrast to the WT/Tag mice (27%). In addition, the development of aggressive disease as a function of time was found to be higher in the i6A− mice compared with the WT mice, assessed by estimating the proportion of prostates with HGPIN and microinvasive lesions at each time point, indicated as p̂/time; the higher the proportion of aggressive lesions at a given time point (p̂/time) the faster the development of the event (Table 1). Compared with the i6A− mice, the WT mice harbored fewer aggressive lesions at both the 12- and 20-week time points; however, the observed difference between the two groups was not statistically significant (Table 1). This difference was found to increase with elapsing time, and, at the 32-week time point, the difference reached statistical significance (P = 0.0182). The smaller proportion of HGPIN plus microinvasive lesions in the prostates from the WT mice suggested a delay of aggressive events in this group of mice.
Fig. 3.
Incidence of PIN lesions and local invasive carcinoma in the ventral prostate lobes of WT/Tag (Left) and i6A−/Tag (Right) mice at 12, 20, 32, and 42 weeks of age. Prostate specimens were examined histologically for prostatic lesions in a double-blinded manner and scored for the highest grade PIN or carcinoma lesion observed across multiple sections. For all Tag mice, there was a decreasing incidence of WNL and an increasing incidence of a lesion-associated event (LGPIN plus HGPIN plus microinvasion) over time, indicating appropriate progression in this transgenic model.
Table 1.
Proportion of aggressive disease (HGPIN plus microinvasion) as a function of time
| Group/time | 12 weeks | 20 weeks | 32 weeks | 42 weeks |
|---|---|---|---|---|
| WT/Tag | p̂/time = 0.0049 | p̂/time = 0.0115 | p̂/time = 0.0096 | p̂/time = 0.0261 |
| i6A−/Tag | p̂/time = 0.0083 | p̂/time = 0.0250 | p̂/time = 0.0240 | p̂/time = 0.0173 |
| P = 0.6930 | P = 0.1374 | P = 0.0182 | P = 0.2692 |
p̂/time, proportion of prostates with HGPIN and microinvasion at each time point within each genotype.
Comparisons for the presence of aggressive disease classified as HGPIN plus microinvasion were made between WT/Tag and i6A−/Tag at each time point. P ≤ 0.05 was considered significant.
In each genotype, there was a single case of macroscopic tumor histologically classified as carcinoma at 42 weeks (Fig. 2H). No evidence of lymph node involvement or metastasis was observed in this Tag model at 42 weeks of age. Collectively, the data indicate that, compared with the WT/Tag mice, selenoprotein-deficient mice exhibited accelerated development of lesions associated with prostate cancer progression.
To determine whether accelerated carcinogenesis in the selenoprotein-deficient mice may be a function of altered proliferation rates, Ki-67 expression was examined by immunohistochemistry. Week 20 was chosen for analysis because carcinogenesis was underway in prostates of both genotypes, with a significant difference in progression rates in the ventral lobe at this time point. For each separate lobe, overall proliferation throughout the tissue was calculated from representative regions. Because the ventral lobe exhibited a high incidence of HGPIN at 20 weeks, proliferation rates were additionally calculated for regions with no nuclear atypia and for regions with HGPIN lesions. For all three lobes, there was no difference in the proliferation index between the WT/Tag and the i6A−/Tag transgenic mice (Table 2). Although there was a trend for reduced proliferation within the ventral lobes of i6A−/Tag as compared with WT/Tag mice (P = 0.6), separation into normal versus HGPIN regions revealed that areas characterized as WNL had identical proliferation rates between the genotypes. Progression to HGPIN in the ventral lobe was associated with a significant increase in the epithelial proliferation index within each genotype; however, this difference was not significantly different between animals of different genotypes. These findings suggest that the protective role of selenoproteins is likely not attributed to lower rates of cell proliferation in the WT mice but, rather, may involve intracellular protective pathways that are reduced in the selenoprotein-deficient animals.
Table 2.
Epithelial proliferation index in prostate lobes from week-20 WT/Tag and i6A−/Tag transgenic mice
| Lobe | WT/Tag | i6A−/Tag |
|---|---|---|
| VP | ||
| All regions | 2.98 ± 0.66 | 1.29 ± 0.24 |
| Normal | 1.06 ± 0.28* | 1.06 ± 0.19* |
| HGPIN | 4.52 ± 0.69 | 2.43 ± 0.55 |
| LP | 1.10 ± 0.23 | 1.12 ± 0.34 |
| DP | 0.64 ± 0.15 | 0.53 ± 0.28 |
VP, ventral prostate; LP, lateral prostate; DP, dorsal prostate. *, P < 0.001 versus HGPIN region within each genotype (ANOVA with Bonferroni post hoc analysis).
Discussion
A genetic approach was used to examine the role of selenoproteins in prostate cancer progression by mating two transgenic mouse strains, one that expressed reduced selenoproteins but was otherwise asymptomatic to another strain that develops prostate pathology representing early events in cancer. This experimental approach permitted the examination of the role of selenoproteins independent of selenium intake, and these studies are consistent with the conclusion that animals expressing reduced selenoprotein levels exhibit accelerated development of PIN lesions with microinvasive carcinoma. At all time points examined, prostates obtained from i6A−/Tag mice demonstrated accelerated pathology associated with prostate cancer development as compared with WT/Tag controls. This accelerated pathology was apparent as selenoprotein-deficient animals exhibited PIN lesions at 12, 20, and 32 weeks and an earlier onset of microinvasive carcinoma as compared with the wild-type animals. By 42 weeks of age, all animals of both genotypes had developed progressive PIN lesions, and there were no statistical differences between the groups, although there was a trend (P < 0.10) for more microinvasive carcinoma in the i6A− mice.
These data provide evidence for a role of selenoproteins in cancer risk and development. Numerous studies have reported an inverse association between selenium status and cancer risk, including cancer of the prostate (2–4, 21–23), and it is possible that low selenium status is associated with risk because of the consequential reduction in the levels of one or more selenoproteins. In this regard, it is interesting that the results of the Nutritional Prevention of Cancer trial indicated that the protective effect of selenium supplementation was significant to only those individuals in the lowest baseline plasma selenium levels (24). This observation indicates the possibility that selenium supplementation may serve to return reduced levels of selenoproteins to baseline levels required to maximize their benefits. Furthermore, in vitro data have indicated that polymorphisms in the genes of two selenoproteins, Sep15 (14) and GPx-1 (10), may result in a requirement for higher selenium levels to achieve baseline levels of the corresponding proteins, and these genetic variations may affect cancer susceptibility (25).
The human genome encodes 25 selenoproteins, approximately half of which have known functions or enzyme activities (8). Several have antioxidant roles, including four members of the GPx family, three thioredoxin reductases, and possibly others (26). There is significant data implicating antioxidants in the prevention of human prostate cancer (27), and it is possible that the reduction in this class of selenoproteins accounts for the accelerated prostate pathology reported in this article.
Reduced levels of several individual selenoproteins may also account for an increased risk of prostate cancer. Perhaps the best candidate is GPx-1, because an association with polymorphisms and the risk of lung cancer has been reported, as has loss of heterozygosity at this locus in several cancer types (9–12). In fact, an association between a polyalanine polymorphism in exon 1 and the risk of young-onset prostate cancer has been reported (28). Reduced levels of GPx-1 could conceivably influence cancer risk through its role as an antioxidant or possibly through the modulation of DNA repair and cell-survival molecules (15, 29). Direct indication of a role for GPx genes in carcinogenesis was provided by a study in which double-knockout mice for GPx-1 and GPx-2 genes developed ileal tumors associated with bacteria-induced intestinal inflammation. This antitumorigenic involvement of GPx in intestinal cancers was attributed to their antioxidant ability to quench inflammation-related increases in hydroperoxide concentrations in the gut (30).
Among the selenoproteins, a role for Sep15 in prostate cancer has been suggested, based on the findings that both humans and mouse prostate express high levels of this protein (31). Differences in Sep15 allele frequency between tumors from breast and head and neck cancer subjects compared with cancer-free individuals (14) as well as observations indicating the growth arrest and the induction of apoptosis of mesothelioma cells by selenium depends on the Sep15 genotype (32) provide additional support for the role of Sep15 in cancer incidence. It is noteworthy that the increased risk observed for prostate cancer in African Americans (33) may be associated with a particular Sep15 allele that is 5-fold more frequent in this population and was found to be less responsive to selenium supplementation in an in vitro assay (14). Another possible candidate protective selenoprotein is thioredoxin reductase, for which there is considerable evidence indicating a possible role in carcinogenesis (34). Interestingly, thioredoxin reductase levels have been shown to be responsive to both selenium deficiency and supplementation in the rat (35).
The data presented in this article support the notion that selenoprotein levels can influence prostate cancer development. Because low dietary selenium intake can result in reduced levels of selenoproteins and increased cancer risk, these data also suggest that the benefits of selenium are mediated, at least in part, by this class of proteins. Future studies aimed at resolving this hypothesis include the effect of selenium supplementation in the i6A− background and the use of mice null for specific selenoproteins.
Materials and Methods
Animal Breeding and Genotyping.
All animals were handled in accordance with the principles and procedures of the 1996 Guide for the Care and Use of Laboratory Animals by the Institute of Laboratory Animal Resources, National Academy Press, Washington, DC, and the experiments were approved by the Institutional Animal Care and Use Committee. The C3(1)/Tag mice were obtained from the National Institutes of Health (NIH) Mouse Models of Human Cancers Consortium. The i6A−/i6A− mice were obtained from D. Hatfield (Section of the Molecular Biology of Selenium, NIH). Animals were maintained on a basal diet of corn oil, 20%; vitamin-free casein, 23.5%; dextrose, 44.7%; AIN-76 mineral mix, 4.1%; vitamin mix with 1980 modification, 1.2%; alpacel, 5.9%; DL methionine, 0.3%; and chlorine bitartrate. The AIN-76 mineral and vitamin mixtures provide 0.1 mg of selenium per kg of diet. To generate bigenic mice, homozygous C3(1)/Tag males were crossed with homozygous i6A− females to produce mice that were heterozygous for both transgenes, referred to as i6A−/Tag. WT/Tag mice were produced by mating homozygous C3(1)/Tag males with WT FVB/N females. Both i6A− and C3(1)/Tag mice were developed on a FVB/N background. Therefore, strain issues were not a confounding factor in any of the presented studies.
Litters were produced, and genotyping of all offspring indicated the expected i6A−/Tag genotype. To genotype the i6A− mice, we took advantage of the fact that the mutation that renders the selenocysteine tRNA incapable of being modified at position 37 lies in the recognition sequence for Ecor571 (MBI Fermentas). DNA was prepared from either ear punches or tail snips by using the DNA Purification kit (Promega) following the vendor's instructions. The DNA was used as a template for PCR using an oligonucleotide (5′-CAGTGGTCTGGGGTGCA-3′) as the forward primer and (5′-GAAAGGTGGAATTGAACCAC-3′) as the reverse primer. Cleavage of the PCR product with Ecor571 was indicative of the presence of the transgene. The i6A− genotype was also verified by the direct measurement of GPx enzyme activity in prostate tissues (see below).
WT/Tag (n = 54) and i6A−/Tag mice (n = 50) were killed on weeks 12, 20, 32, and 42 to evaluate prostate disease progression over time as a function of genotype. Mice were killed by cervical dislocation and the urogenital tissues quickly removed and placed in ice-cold PBS. The prostatic complex was dissected and either fixed en masse or further microdissected into the ventral and dorsolateral lobes and snap-frozen in liquid nitrogen.
Histopathology.
The prostatic complex was fixed overnight in 10% buffered formalin (Fisher Diagnostics, Middletown, VA) and stored in 70% ethanol until paraffin embedding. Fixed tissues were processed, paraffin embedded, and sectioned along the longitudinal axis at three levels in the paraffin block to provide a minimum of 50 sections per prostate. The animal genotypes were coded at this point to avoid observer bias in the pathological diagnosis. Two independent evaluators, a prostate biologist (G.S.P.) and a board-certified pathologist (V.H.R.), separately analyzed all specimens. Observations were made on the presence and degree of epithelial hyperplasia and the presence of nuclear atypia in the separate ventral and dorsolateral lobe regions. Samples with no nuclear atypia were classified as WNL, whereas specimens with nuclear atypia were further classified as LGPIN or HGPIN lesions according to accepted criteria of the College of American Pathologists and further characterized for mouse models (36). In addition, both evaluators looked for evidence of basement membrane breakdown, invasion, and the presence of tumors. Microinvasion was positively scored only when basement membrane breakdown and epithelial cells within the adjacent stroma were observed in the immediate vicinity of PIN lesions, which would suggest appropriate progression. Discrepancies between evaluators were resolved by mutual consent after reading the slides together. After the diagnoses were recorded, the code for genotype was broken and the data tabulated.
Immunohistochemistry.
To assess the prostatic epithelial proliferation rates, week-20 specimens (n = 10 from each genotype) were immunohistochemically labeled for Ki-67. Paraffin sections (4 μm) were mounted on Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA), deparaffinized in xylene, and rehydrated in water. Antigen retrieval was performed by heat treatment in a Decloaker Chamber (Biocare Medical, Walnut Creek, CA) for 30 min in 0.01 M citrate buffer (pH 6.0). Endogenous peroxidase was removed with 3% H2O2, and nonspecific binding was controlled by a 30-min treatment in Superblock (Pierce). The slides were incubated overnight at 4°C with a polyclonal Ki-67 primary antibody (NovoCastra, Newcastle-upon-Tyne, U.K.) at a 1:2,500 dilution, whereas adjacent sections were incubated in normal rabbit IgG (0.5 μm/ml) as a negative control. All sections were next incubated with biotinylated goat anti-rabbit secondary antibody (Vector Laboratories) at 1:200 dilution for 30 min and detected with an avidin–biotin peroxidase kit (Vector Laboratories). Diaminobenzidine (0.07%) was used as a chromagen (Sigma), and a 1:4 dilution of Gill's hematoxylin #3 was used as a counterstain (Fisher Diagnostics). Slides were dehydrated in ethanol, cleared in xylene, and coverslipped with Permount (Fisher Scientific). Multiple representative areas of each ventral, lateral, and dorsal lobe region were selected and digitally captured with an Axioskop microscope and a color digital AxioCam camera (Zeiss). Positive and negative Ki-67-stained epithelial cells were counted by using Zeiss image version 3.0 (Zeiss), with a minimum of 500 cells analyzed per slide.
GPx Activity.
GPx activity was measured by a standard coupled spectrophotometric method (37) in tissue samples from the ventral and dorsolateral prostate lobes. Briefly, tissues were homogenized on ice in sodium phosphate buffer (0.1 M Na2HPO4, pH 7.5), centrifuged, and disrupted by sonication (Ultrasonic homogenizer 4710 series; Cole–Palmer). Protein levels were quantified in the supernatant by using the Dc Protein Assay kit (Bio-Rad). GPx activity was determined by using 100–400 μl of the supernatant (130–328 μg of proteins), assayed in a 1-ml reaction volume containing NADPH, reduced glutathione reductase, sodium azide, and H2O2. The oxidation of NADPH to NADP was monitored at 339 nm and expressed as nmol of NADPH oxidized per mg of protein per min.
Western Blot Analysis.
Protein extracts (100 μg) were electrophoresed on a 14% SDS/polyacrylamide gel, and transferred onto polyvinylidine fluoride (PVDF) membranes (Immobilon-P; Millipore). To evaluate GPx-1 protein levels, membranes were blocked with 5% nonfat dry milk in TBST (Tris buffered saline with 0.1% Tween 20) overnight at 4°C and then incubated with the primary antibody for GPx-1 (diluted 1:1,000 in blocking solution) for 1 h at room temperature (RT). Antibodies were affinity-purified from eggs of hens immunized with a KLH-conjugated peptide representing amino acid residues 83–100 of the human GPx-1 gene, CZHQENAKNEEILNSLKYVR (Aves Labs, Tigard, OR). Membranes were washed three times with TBST and incubated with a rabbit anti-chicken horseradish peroxidase-conjugated secondary antibody (diluted 1:5,000 in 5% nonfat dry milk in TBST) for 1 h at RT and washed three times with TBST.
Statistics.
The differences in PIN incidence at fixed time points between the WT/Tag and i6A−/Tag mice were assessed by the χ2 and Fisher exact tests. The Peto test was used to evaluate the trend in the occurrence of increasing PIN grades between the two groups. The Cochran–Mantel–Haenszel common relative risk test was performed to compare the trend in event occurrence across time between the two groups. The delay of tumor progression in i6A−/Tag mice versus WT/Tag mice was determined by using a combination of Student's t test and the Peto test. All statistical tests were performed by using the program sas (SAS Institute, Cary, NC). Differences were considered significant at P ≤ 0.05.
Acknowledgments
This work was supported by National Institutes of Health Grant R01 CA101053 (to A.M.D., G.S.P., and S.M.S.) and Department of Defense Postdoctoral Fellowship W81XWH-05-1-0009 (to V.D.-N.).
Abbreviations
- GPx-1
glutathione peroxidase-1
- i6A
isopentenyladnosine
- HGPIN
high-grade PIN
- LGPIN
low-grade PIN
- PIN
prostatic intraepithelial neoplasia
- WNL
within normal limits.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Clark L. C., Combs G. F., Turnbull B. W., Slate E. H., Chalker E. H., Chow J., Davis L. S., Glover R. A., Graham G. F., Gross E. G., et al. J. Am. Med. Assoc. 1996;276:1957–1963. [PubMed] [Google Scholar]
- 2.Yoshizawa K., Willett W. C., Morris S. J., Stampfer M. J., Speigelman D., Rimm E. B., Giovannucci E. J. Natl. Cancer Inst. 1998;90:1219–1224. doi: 10.1093/jnci/90.16.1219. [DOI] [PubMed] [Google Scholar]
- 3.Li H., Stampfer M. J., Giovannucci E. L., Morris J. S., Willett W. C., Gaziona M., Ma J. J. Natl. Cancer Inst. 2004;96:696–703. doi: 10.1093/jnci/djh125. [DOI] [PubMed] [Google Scholar]
- 4.van den Brandt P. A., Zeegers M. P. A., Bode P., Goldbohm R. A. Cancer Epidemiol. Biomarkers Prevention. 2003;12:866–871. [PubMed] [Google Scholar]
- 5.Lippman S. M., Goodman P. J., Klein E. A., Parnes H. L., Thompson I. M., Jr., Kristal A. R., Santella R. M., Probstfield J. L., Moinpour C. M., Albanes D., et al. J. Natl. Cancer Inst. 2005;97:94–102. doi: 10.1093/jnci/dji009. [DOI] [PubMed] [Google Scholar]
- 6.Hatfield D. L., Gladyshev V. N. Mol. Cell. Biol. 2002;11:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry M. J., Banu L., Chen Y. Y., Mandel S. J., Kieffer J. D., Harney J. W., Larsen P. R. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 8.Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigo R., Gladyshev V. N. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 9.Moscow J. A., Schmidt L., Ingram D. T., Gnarra J., Johnson B., Cowan K. H. Carcinogenesis. 1994;15:2769–2773. doi: 10.1093/carcin/15.12.2769. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y. J., Diamond A. M. Cancer Res. 2003;63:3347–3351. [PubMed] [Google Scholar]
- 11.Hu Y. J., Dolan E., Bae R., Yee H., Roy M., Glickman R., Kiremidjian-Schumacher L., Diamond A. M. Biol. Trace Elem. Res. 2004;101:97–106. doi: 10.1385/BTER:101:2:097. [DOI] [PubMed] [Google Scholar]
- 12.Ratnasinghe D., Tangrea J. A., Andersen M. R., Barrett M. J., Virtamo J., Taylor P. R., Albanes D. Cancer Res. 2000;60:6381–6383. [PubMed] [Google Scholar]
- 13.Kumaraswamy E., Malykh A., Korotkov K. V., Kozyavkin S., Hu Y. J., Kwon S. Y., Moustafa M. E., Carlson B. A., Berry M. J., Lee B. J., et al. J. Biol. Chem. 2000;275:35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y. J., Korotkov K. V., Mehta R., Hatfield D. L., Rotimi C. N., Luke A., Prewitt E., Cooper R. S., Stock W., Vokes E. E., et al. Cancer Res. 2001;61:2307–2310. [PubMed] [Google Scholar]
- 15.Nasr M., Fedele M. J., Esser K. A., Diamond A. M. Free Radical Biol. Med. 2004;37:187–195. doi: 10.1016/j.freeradbiomed.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 16.Moustafa M. E., Carlson B. A., El-Saadani M. A., Kryukov G. V., Sun Q.-A., Harney J. W., Hill K. E., Combs G. F., Feigenbaum L., Mansur D. B., et al. Mol. Cell. Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata M. A., Ward J. M., Devor D. E., Liu M. L., Green J. E. Cancer Res. 1996;56:4894–4903. [PubMed] [Google Scholar]
- 18.Bostwick D. G., Ramnani D., Qian J. Prostate. 2000;43:286–294. doi: 10.1002/1097-0045(20000601)43:4<286::aid-pros8>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 19.Shibata M. A., Jorcyk C. L., Liu M. L., Yoshidome K., Gold L. G., Green J. E. Toxicol. Pathol. 1998;26:177–182. doi: 10.1177/019262339802600121. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y. L., Parker M. G., Bakker O. Mol. Endocrinol. 1990;4:1219–1225. doi: 10.1210/mend-4-8-1219. [DOI] [PubMed] [Google Scholar]
- 21.Zeegers M. P. A., Goldbohm R. A., Bode P., van den Brandt P. A. Cancer Epidemiol. Biomarkers Prevention. 2002;11:1292–1297. [PubMed] [Google Scholar]
- 22.Hartmann T. J., Taylor P. R., Alfthan G., Fagerstrom R., Virtamo J., Mark S. D., Virtanen M., Barrett M. J., Albanes D. Cancer Causes Control. 2002;13:923–928. doi: 10.1023/a:1021912117067. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs E. T., Jiang R., Alberts D. S., Greenberg E. R., Gunter E. W., Karagas M. R., Lanza E., Ratnasinghe L., Reid M. E., Schatzkin A., et al. J. Natl. Cancer Inst. 2004;96:1669–1675. doi: 10.1093/jnci/djh310. [DOI] [PubMed] [Google Scholar]
- 24.Duffield-Lillico A. J., Dalkin B. L., Reid M. E., Turnbull B. W., Slate E. H., Jacobs E. T., Marshall J. R., Clark L. C. Br. J. Urol. Inter. 2003;91:608–612. doi: 10.1046/j.1464-410x.2003.04167.x. [DOI] [PubMed] [Google Scholar]
- 25.Diwadkar-Navsariwala V., Diamond A. M. J. Nutr. 2004;134:2899–2902. doi: 10.1093/jn/134.11.2899. [DOI] [PubMed] [Google Scholar]
- 26.Gladyshev V. N., Hatfield D. L. J. Biomed. Sci. 1999;6:151–160. doi: 10.1007/BF02255899. [DOI] [PubMed] [Google Scholar]
- 27.Klein E. A., Thomas I. M. Curr. Opin. Urol. 2004;14:143–149. doi: 10.1097/00042307-200405000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Kote-Jarai Z., Durocher F., Edwards S. M., Hamoudi R., Jackson R. A., Ardern-Jones A., Murkin A., Dearnaley D. P., Kirby R., Houlston R., et al. Prostate Cancer Prostatic Dis. 2002;5:189–192. doi: 10.1038/sj.pcan.4500569. [DOI] [PubMed] [Google Scholar]
- 29.Cheng W.-H., Quimby F. W., Lei X. G. Free Radical Biol. Med. 2003;34:918–927. doi: 10.1016/s0891-5849(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 30.Chu F.-F., Esworthy R. S., Chu P. G., Longmate J. A., Huycke M. M., Wilczynski S., Doroshow J. H. Cancer Res. 2004;64:962–968. doi: 10.1158/0008-5472.can-03-2272. [DOI] [PubMed] [Google Scholar]
- 31.Gladyshev V. N., Jeang K.-T., Wootton J. C., Hatfield D. L. J. Biol. Chem. 1998;273:8910–8915. doi: 10.1074/jbc.273.15.8910. [DOI] [PubMed] [Google Scholar]
- 32.Apostolou S., Klein J. O., Mitsuuchi Y., Shetler J. N., Poulikakos P. I., Jhanwar S. C., Kruger W. D., Testa J. R. Oncogene. 2004;23:5032–5040. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- 33.Crawford D. E. Urology. 2003;62:3–12. [Google Scholar]
- 34.Mustacich D., Powis G. Biochem. J. 2000;15:1–8. [PMC free article] [PubMed] [Google Scholar]
- 35.Berggren M. M., Mangin J. F., Gasdaka J. R., Powis G. Biochem. Pharmacol. 1999;57:187–193. doi: 10.1016/s0006-2952(98)00283-4. [DOI] [PubMed] [Google Scholar]
- 36.Shappell S. B., Thomas G. V., Roberts R. L., Herbert R., Ittmann M. M., Rubin M. A., Humphrey P. A., Sundberg J. P., Rozengurt N., Barrios R., et al. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- 37.Samuels B. A., Murray J. L., Cohen M. B., Safa A. R., Sinha B. K., Townsend A. J., Beckett M. A., Weichselbaum R. R. Cancer Res. 1991;51:521–527. [PubMed] [Google Scholar]