Abstract
Cell division is a carefully orchestrated procedure. Bacterial cells have intricate mechanisms to ensure that genetic material is copied, proofread, and accurately partitioned into daughter cells. Partitioning now appears to also occur for some cytoplasmic proteins. Previously, using chromosomal fluorescent protein fusions, we demonstrated that a subset of Rhodobacter sphaeroides chemotaxis proteins colocalize to a discrete region within the bacterial cytoplasm. Using TlpT-yellow fluorescent protein as a marker for the position of the cytoplasmic protein clusters, we show most cells contain either one cluster localized at mid-cell or two clusters at the one-fourth and three-fourths positions of cell length. The number and positioning of these protein clusters depend on a previously unrecognized bacterial protein positioning factor, PpfA, which has homology to bacterial type I DNA partitioning factors. These data suggest that there is a mechanism involved in partitioning some cytoplasmic proteins upon cell division that is analogous to a mechanism seen for plasmid and chromosomal DNA.
Keywords: chemoreceptor, chemotaxis, partition, Rhodobacter
The chemotaxis pathway of Escherichia coli and Salmonella enterica serovar Typhimurium is one of the best understood molecular signaling systems in biology. The biochemistry of this two-component signaling system is well characterized, and atomic resolution structures are available for most of the individual signaling proteins. For a review of bacterial chemotaxis, see ref. 1. Central to the chemotaxis system are the autophosphorylating histidine kinase CheA (2) and the response regulators that receive phosphoryl moieties from CheA. Phosphorylation of response regulators acts as a molecular switch to regulate their specific activities. The chemotactic bacterial cell employs transmembrane receptors, the methyl-accepting chemotaxis proteins (MCPs), to regulate the rate of phosphotransfer reaction (3, 4) in response to changes in environmental stimuli. The MCPs use a robust adaptation mechanism to temporally sense changes in the concentrations of attractants and repellents in their environment (5). MCPs, in conjunction with CheA, integrate multiple different stimuli, and the pathway displays sensitivity over a wide dynamic range (6, 7). The sensory phenomenon of signal amplification and gain is thought to involve lateral communication between MCPs, CheW linker proteins (8), and CheA molecules within organized, dynamic lattice structures (9, 6) that are localized at the cell poles (10, 11). The polar localization of these chemotaxis protein complexes has been observed in all species studied so far (12). As cells grow, the amount of protein at the older pole increases while nascent polar clusters become apparent at the new cell pole. In this way, when a cell divides, each daughter cell inherits a complete but different sized polar cluster of chemotaxis proteins.
The purple, non-sulfur bacterium Rhodobacter sphaeroides contains homologues of the majority of chemotaxis proteins found in the enteric system. There are three main chemotaxis loci, cheOp1, cheOp2, and cheOp3, each of which encodes an ostensibly complete chemosensory pathway. cheOp2 and cheOp3 are both essential for chemotactic responses under laboratory conditions (13). R. sphaeroides has 13 putative chemoreceptors, nine of which are transmembrane MCPs. The remaining four putative receptors, the transducer-like proteins (Tlps), lack transmembrane domains. Evidence suggests that the R. sphaeroides membrane-spanning MCPs localize to the poles of the cell with most of the chemosensory homologues encoded in cheOp2 (14–16), whereas Tlps localize to a discrete region in the cytoplasm with chemosensory homologues encoded in cheOp3 (16, 17). Three-dimensional image reconstructions of the cheOp3 components indicate that these clusters are not directly associated with the membrane (data not shown). These complexes are referred to as cytoplasmic chemotaxis protein clusters.
The presence of a set of proteins that apparently colocalize within the bacterial cytoplasm introduces a novel set of problems for the bacterium, namely, how is this cluster of proteins positioned within the cytoplasm, and what happens when the cell divides? Most textbooks would suggest that the bacterial cytoplasm is a relatively homogeneous mix of soluble proteins and other compounds, with both daughter cells inheriting a roughly equal amount after septation. However, if some proteins form large, higher-order structures, a specific apparatus may be required to ensure these protein clusters are properly partitioned upon cell division.
Bacterial DNA is accurately segregated upon cell division (18). Several systems are known to be involved in this partitioning; one of the best studied is the type I (ParAB) partitioning system for plasmids (19). All type I partitioning loci contain three elements: an ATPase (ParA), a DNA-binding protein (ParB), and a cis-acting centromere-like region of DNA, parS/parC (18). All ParA-like ATPases encode a deviant Walker ATPase motif (20). Analyses show that plasmids are positioned either at mid-cell or at the one-fourth and three-fourths positions of cell length, which correspond to the cell centers of nascent daughter cells (21–23). In slowly growing cells, plasmids typically remain at mid-cell until late in the cell cycle and then move bidirectionally toward the middle of each new daughter cell (23), whereas in faster growing cells, plasmid foci are separated and positioned at the one-fourth and three-fourths positions before septation (22, 23). The ParA ATPase is involved in positioning plasmids at the mid-cell and in separating these foci and ejecting them into the daughter cells in slow growing cells or positioning them at the one-fourth and three-fourths positions in faster growing cells. Interestingly, cells prevented from dividing by the antibiotic cephalexin still actively segregate plasmids to discrete regions within the filamentous cell (22). Partitioning of plasmids by type I partitioning homologues is better understood than the function of their chromosomal counterparts, such as Soj (ParA homologue) and SpoOJ (ParB homologue) found in Bacillus subtilis (18).
This study focuses on TlpT, which is encoded on the R. sphaeroides cheOp3 and colocalizes to a discrete chemotaxis protein cluster in the cytoplasm with other chemotaxis proteins from this operon (13, 16). TlpT is essential for chemotaxis and for the integrity of the cytoplasmic cluster (24). An in-frame chromosomal replacement of tlpT with a C-terminal fusion to yellow fluorescent protein (YFP), tlpT-yfp, was used to visualize the position of the cytoplasmic chemotaxis protein cluster. A detailed analysis of cluster position throughout the cell cycle using fluorescence microscopy shows a complex level of protein organization and dynamics within the bacterial cytoplasm. Further, we reveal a role for an ATPase, protein positioning factor A (PpfA) (previously called Slp) (13). PpfA has homology to DNA partitioning factors of the ParA family (22, 25) and the cell-cycle regulator MinD (26) and appears to regulate the number and position of the cytoplasmic chemotaxis protein clusters in R. sphaeroides, indicating a protein partitioning system related to that previously identified for plasmids.
Results
Cells Contain Either One or Two Cytoplasmic Clusters and This Affects Their Positioning Within the Cell.
Previous work in our laboratory has shown that most of the proteins encoded on the R. sphaeroides cheOp3 colocalize to a discrete cluster within the bacterial cytoplasm (16, 17). To visualize the cytoplasmic cluster, a strain of R. sphaeroides was used in which the chromosomal copy of tlpT, encoding an essential constituent of the cytoplasmic chemotaxis protein cluster, was replaced with a fusion to YFP (yfp), producing TlpT-YFP (16). Although the TlpT-YFP fusion shows reduced activity, immunoelectron microscopy and colocalization studies confirmed that it localizes to the cytoplasmic chemotaxis protein cluster (16, 17). Analysis of >500 cells showed that most (99%) of the TlpT-YFP cells contained either one or two cytoplasmic chemotaxis protein clusters (Table 1). To investigate the positioning of these clusters within the bacterial cell, the distance from each cluster to the farthest cell pole was measured along with the cell length. The cluster position was then defined as the distance between the cluster and the farthest cell pole as a proportion of total cell length. Interestingly, cells containing a single cytoplasmic cluster positioned this cluster around mid-cell (Fig. 1A), whereas those cells which contained two clusters positioned these at the one-fourth and three-fourths positions (Fig. 1B). Statistical analysis (one-way ANOVA, P < 0.05) confirmed that the relative position of clusters in cells containing either one or two cytoplasmic clusters was significantly different. These data suggest that the position of the cytoplasmic protein cluster within the R. sphaeroides cell is strictly regulated and that its precise location differs in cells containing either one or two clusters.
Table 1.
Percentage of midlogarithmic-phase, aerobically grown wild-type and ΔppfA cells containing different numbers of cytoplasmic protein clusters
| Cell type | No. of protein clusters |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 0 | |
| Wild type (n = 547) | 72.6 | 26.6 | 0.7 | 0.1 |
| ΔppfA (n = 516) | 72 | 0 | 0 | 28 |
Values are expressed as percentages.
Fig. 1.
Positioning of the cytoplasmic protein clusters. Histograms show the distance of clusters from the farthest cell pole as a function of the cell length. Data are symmetrically distributed around mid-cell because the distance to the farthest cell pole was always measured. (A) Wild-type cells with one cluster found around mid-cell. (B) Wild-type cells with two clusters localized to the one-fourth and three-fourths positions. (C) ΔppfA cells never contained more than one cluster, which was randomly positioned within the cell.
Protein Clusters Appear to Be Segregated Before Cell Division.
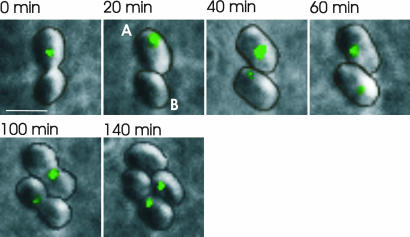
To investigate how the number and position of cytoplasmic chemotaxis protein clusters varied throughout the ≈2-h cell cycle, time-lapse microscopy was performed with fluorescence images obtained every 20 min for 3–4 h (Fig. 2 and Fig. 7, which is published as supporting information on the PNAS web site). In ≈35% of the cells, the following pattern of protein cluster position was observed. In a newly formed cell, a single cluster was visible at about mid-cell. Then, this cluster became two clusters, which migrated to positions at one-fourth and three-fourths of the cell length and remained in these positions until the cell divided (Fig. 2, cell A). This positioning resulted in each daughter cell inheriting a cytoplasmic protein cluster at their mid-cell. In the remaining cells (≈65%), one cluster was again observed at around mid-cell in a newly formed cell. However, this cluster remained around the mid-cell and moved to the nascent mid-cell of one daughter cell before division. A new cluster, however, became visible in the mid-cell of the other daughter cell shortly after septation (Fig. 2, cell B).
Fig. 2.
Time-lapse images of the cytoplasmic protein cluster protein, TlpT-YFP. One cell containing two clusters divides into cells A and B. The single A cluster becomes two clusters early in cell growth, and each cluster migrates rapidly to positions at one-fourth and three-fourths of the cell length, where they remain until the cell divides. This positioning results in each daughter cell inheriting a cluster at about mid-cell. The B cell contains a single cluster throughout most of the cell cycle. This cluster is inherited by one of the daughter cells, and a new cluster is observed in the other daughter cell soon after septation. Contour lines showing the outlines of the cells are superimposed (the raw images are shown in Fig. 7). (Scale bar: 2 μm.)
Intriguingly, the one-fourth and three-fourths positioning of foci has been observed with plasmids and chromosomes partitioned by type I DNA partitioning systems (18, 19, 22). Similarly, plasmid foci may also remain positioned at the mid-cell until close to cell division (a pattern which has been linked to slower rates of cell growth). Just before division, these plasmid foci, positioned at the mid-cell, separate, “ejecting” foci into the two daughter cells as they divide (23). A similar event may occur with the cytoplasmic protein clusters as the cells approach division, although one of the two clusters may be present at a level that is below the detection sensitivity of our fluorescence microscope until some time after cell division.
The Number and Position of Clusters Are Regulated by PpfA.
Most of the constituents of the cytoplasmic chemotaxis protein cluster are encoded in a single operon (cheOp3) (16). Surprisingly, this operon also contains a gene encoding a deviant Walker-type ATPase protein (20) with homology to type I DNA partitioning factors such as ParA (22, 25, 27, 28) and the cell division regulator MinD (26, 29). Although this gene shows no homology to any known chemotaxis protein, it is required for a normal chemotactic response on swarm plates (13). The homology to DNA partitioning factors, a putative role in chemotaxis, and the location of this gene (within cheOp3) led us to investigate whether this protein influences the observed positioning of the cytoplasmic chemotaxis protein clusters in R. sphaeroides. Cells deleted for this gene, ppfA (previously named slp) (13), never contained more than one cytoplasmic protein cluster, and a significant number (≈30%) contained no apparent clusters (Table 1).
We analyzed the position of these single clusters within ppfA mutant cells as we did for the wild-type tlpT-yfp strain and discovered that the observed single clusters in these mutants did not localize to the mid-cell position or to the one-fourth or three-fourths position but appeared randomly positioned throughout the cell (Fig. 1C). Statistical analysis (one-way ANOVA, P < 0.05) confirmed that the position of single clusters was significantly different in cells deleted for this gene compared with the wild-type background. These data suggest that PpfA is involved in positioning the cytoplasmic protein cluster to specific regions within the bacterial cell.
Fluorescence images of the ppfA mutant obtained every 20 min (Fig. 3 and Fig. 8, which is published as supporting information on the PNAS web site) reveal that these single clusters were inherited by one daughter cell upon division and that a new cluster did not appear in the “empty” daughter cell until 30–40 min after septation. This observation suggests that in the mutant strain there is only one cluster to inherit upon cell division, and one daughter cell has to synthesize a new cytoplasmic cluster de novo. Cells lacking a chemotaxis cluster would be nonchemotactic immediately after division, and this deficiency probably explains the reduced chemotaxis shown by populations of PpfA mutants (13). PpfA may therefore assist in somehow segregating cytoplasmic protein clusters within wild-type cells or in initiating the nucleation of a new cluster. If this segregation is lacking, newly synthesized components of the cluster will probably agglomerate into the existing single cluster, and only when this cluster is not present, after cell division, will the proteins start to form a new cytoplasmic protein cluster.
Fig. 3.
Time-lapse images of the cytoplasmic protein cluster protein, TlpT-YFP, in cells deleted for the putative ATPase, PpfA. ΔppfA cells contain a maximum of one cytoplasmic cluster. After cell division, one daughter cell inherits the single cluster, and a new cluster becomes apparent in the other cell sometime later. Contour lines showing the outlines of the cells are superimposed (the raw images are shown in Fig. 8). (Scale bar: 2 μm.)
PpfA Anchors the Cytoplasmic Clusters Within the Cell.
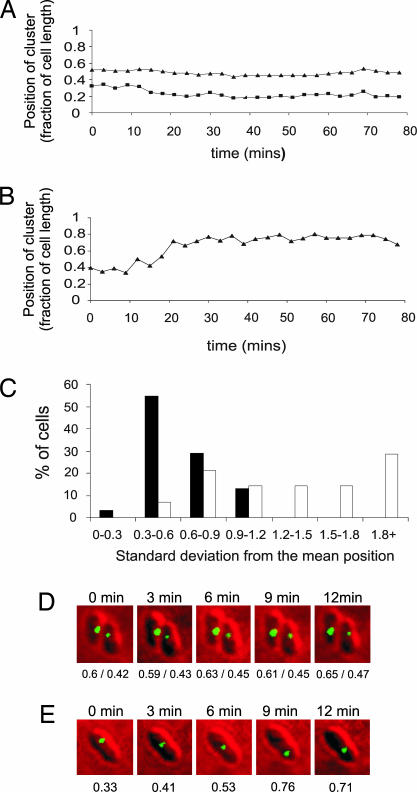
To characterize the mobility of the TlpT-YFP cluster and the role of PpfA in the process, short-interval time-course experiments were undertaken. Wild-type and mutant PpfA cells containing bright TlpT-YFP foci were chosen, and their positions were recorded every 3 min over an 80-min period for cells containing or lacking PpfA. Using these data, we calculated the position of clusters as a fraction of cell length for each time point. The mean position and standard deviation for clusters from each time course were calculated for individual cells (Fig. 4). Statistical analysis of cluster mobility (as measured by the standard deviation of cluster position) in strains containing or lacking PpfA demonstrate that cytoplasmic clusters are more mobile in the ppfA mutant (Wilcoxon rank-sum test, P < 0.05). These results support our earlier evidence suggesting a loss of specific cluster positioning in ppfA mutants.
Fig. 4.
Short-interval time-course data on cytoplasmic cluster movement. Representative traces of the positions of cytoplasmic clusters of TlpT-YFP as a fraction of cell length in a wild-type background (A) and ΔppfA (B) are shown. The histogram (C) bins the results depending on the standard deviation from the mean position. Filled bars represent wild-type cells (n = 31), and open bars represent ΔppfA cells (n = 15). A low standard deviation of cluster position indicates a relatively fixed position within the cell. Clusters in the wild type are clearly less mobile than those in ΔppfA cells (Wilcoxon rank-sum test, P = 0.000133). Two example time courses are also shown in cells containing (D) and deleted for (E) ppfA. Underneath each cell is the position of each cluster as a fraction of cell length.
PpfA Does Not Regulate Expression of Cytoplasmic Cluster Components.
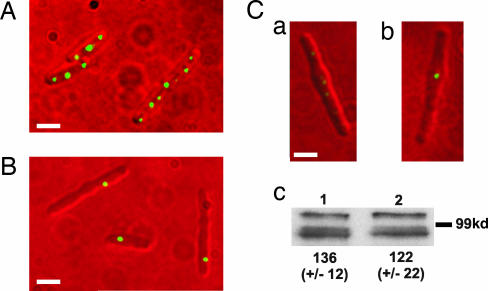
To further characterize the cytoplasmic chemotaxis protein clusters, we made filamentous R. sphaeroides cells. Early-logarithmic-phase cultures were treated with the antibiotic cephalexin at 2.5 μg/ml for 4 h to produce long, nonseptate, filamentous cells. Strikingly, wild-type cells contained multiple relatively evenly spaced cytoplasmic chemotaxis protein clusters, separated by an average distance of 1.13 ± 0.48 μm (Fig. 5A). Untreated R. sphaeroides cells grow from just below 2 μm to almost 3.5 μm during the cell cycle (30), thus the spacing of ≈1.13 μm would enable the partitioning of a single cytoplasmic cluster into each daughter cell upon division. In the ΔppfA strain, however, irrespective of the length of the filamentous cells, only one cytoplasmic chemotaxis protein cluster was observed in any cell filament (Fig. 5B). Analyses of multiple images of filamentous cells acquired in an identical fashion (ensuring subsaturating emission intensities) for both the wild-type and ppfA mutant demonstrated that the TlpT-YFP clusters from the ppfA mutant cells were brighter and larger in size. This is visually apparent in the images (Fig. 5 Ca and Cb). The average filamentous cell length measured for TlpT-YFP cells was 4.25 ± 1.3 μm, and on average, the cells contained 3.3 clusters. The ΔppfA mutants showed a similar average cell length of 4.26 ± 1.8 μm and never contained more than one cytoplasmic cluster per cell.
Fig. 5.
Fluorescence images of the cytoplasmic cluster position in filamentous cells. (A) Wild-type background. (B) ΔppfA. (C) Images have been treated in an identical manner and clearly show that the single cluster present in the ΔppfA strain (Cb) is brighter than any of the six clusters present in the wild-type background (only five clusters are visible in Ca, but six are observed with longer exposure times; data not shown). (Scale bars: 2 μm.) (Cc) Western blot analysis using anti-GFP antibody to crossreact with TlpT-YFP in filamentous cells containing (Cc1) and deleted for (Cc2) ppfA. Values represent the mean band intensity from six independent experiments (arbitrary units). The same samples were also probed with an antibody to CheA2, a component of the polar chemotaxis protein cluster whose level remained constant in both strains (data not shown).
Quantitative Western blot analysis of six independent filamentous cell samples of both the wild-type and ΔppfA strains was carried out by using anti-GFP antibody (BD Living Colors; Clontech) to crossreact with TlpT-YFP. Analysis of band intensities (imagequant; Molecular Dynamics) demonstrated no significant difference in the level of TlpT-YFP expression in either strain (Fig. 5Cc). These samples were also probed in a quantitative Western blot by using antibody to the cheOp2-encoded polar chemotaxis protein CheA2 (16), whose expression should be unaffected by deleting ppfA. The band intensities were not significantly different in either strain, confirming equal loading of all samples (data not shown). Therefore, in the absence of PpfA, there is one large cytoplasmic chemotaxis protein cluster per filamentous cell, containing all the protein that would normally be segregated between multiple clusters in a wild-type filamentous cell. This finding confirms that PpfA does not regulate the expression of TlpT-YFP but does regulate the number of clusters formed and their positioning within the R. sphaeroides cell.
Discussion
Two of three of the R. sphaeroides chemosensory pathways are essential for chemotaxis to a range of attractants under laboratory conditions (13). Each of these operons encodes its own subset of chemosensory homologues that form large, distinct sensory complexes, one at the pole of the cell and the other in the cytoplasm (16).
Clustering of chemosensory proteins may ensure the correct stoichiometry for sensitivity and gain within the system (6). The physical separation of the complexes at the poles and in the cytoplasm (16) may prevent unwanted crosstalk between the two pathways. Importantly, this information raises the notion that bacteria may spatially organize large cytoplasmic protein complexes.
Here we have demonstrated that the cytoplasmic cluster of chemotaxis proteins is specifically localized to discrete regions of the cell and that its precise positioning is determined by the number of clusters present within the cell. In cells with a single cluster, it tends to be positioned at mid-cell, whereas when two clusters are present, they tend to be positioned at the one-fourth and three-fourths positions. Interestingly, the positioning is reminiscent of that reported for some low-copy-number plasmids, which are segregated by an active partitioning system upon cell division (21, 23, 31).
It seems likely that large protein complexes in the cytoplasm of bacterial cells require special mechanisms to ensure that they are localized to specific regions of the cytoplasm. If this conjecture is true, then there may also exist a mechanism to ensure that upon cell division, these large protein factories are properly segregated to the two daughter cells. It is possible that these mechanisms for positioning and potentially segregating protein clusters may be analogous to those previously observed for partitioning of the genetic components of bacterial cells.
A ParA homologue, PpfA, which is encoded within the same chemotaxis operon as most of the constituents of the cytoplasmic protein cluster, is required for the correct positioning of the chemotaxis protein clusters to specific regions of the bacterial cytoplasm. As with ParA, the ATPase activity of PpfA is essential to its function (unpublished data). PpfA is contained within a locus of chemotaxis proteins with no ParB homologue or centromere-like region. It is likely that PpfA, like ParA, will have partner proteins involved in positioning the cluster within the cell. These proteins are currently unidentified but may include a component of the chemosensory cluster and/or a component of the bacterial cytoskeleton. Neither PpfA nor any other known components of the cytoplasmic chemotaxis protein cluster contain recognizable DNA-binding motifs. Unfortunately, R. sphaeroides chromosomes do not condense, making colocalization studies with DAPI staining uninformative, and techniques for visualizing chromosomal regions (32, 33) are undeveloped for R. sphaeroides. Therefore, at the present we are unable to conclusively demonstrate whether the cytoplasmic clusters or PpfA associate with the DNA.
Cells lacking PpfA failed to produce more than a single cytoplasmic protein cluster, even when the cells were made filamentous, and newly synthesized protein appeared to be continually incorporated into this single cluster. After cell division, therefore, only one daughter cell inherits the chemotaxis cluster, and a new cluster must form in the empty cell. Fig. 6 summarizes the observed positioning of cytoplasmic chemotaxis clusters and the effects of deleting ppfA. Deletion of ppfA results in a small but significant reduction in chemotaxis on swarm plates (13). This reduction in the rate of chemotaxis is probably the result of the ≈30% of cells that have recently divided and therefore lack a visible cytoplasmic chemotaxis cluster. These cells will be nonchemotactic until they have synthesized a new cluster. In contrast, <1% of wild-type cells lack visible clusters.
Fig. 6.
The positioning of cytoplasmic chemotaxis protein clusters in the presence and the absence of PpfA. (A) Wild-type cells have a single cluster at around mid-cell. Before cell division, this cluster becomes two clusters that move to the one-fourth and three-fourths positions, resulting in each daughter cell inheriting a cluster at about mid-cell. When this happens late in the cell cycle, the second cluster may not be visible until after cell division. (B) In the absence of PpfA, cells never contain more than one cluster. This cluster appears mobile and increases in size as protein synthesis continues. Upon division, one daughter cell inherits the single cluster, and the other daughter cell synthesizes a new cluster de novo.
The existence of large bacterial cytoplasmic protein clusters and a putative partitioning system is unlikely to be limited to the R. sphaeroides chemotaxis system. parA homologues that are not contained within typical DNA partitioning loci, as is the case for ppfA, are common in prokaryotes, and until now, an explanation for any of these “orphan” parA genes was unknown. The presence of organized clusters of proteins within the cytoplasm of prokaryotic cells (17) and a dedicated machinery to ensure daughter cells inherit these protein clusters has profound implications for our understanding of biochemical processes within bacterial cells. Although it is well established that prokaryotes have intricate mechanisms to ensure that genetic material is accurately partitioned between daughter cells (32, 34), this work reports a system for regulating the number and positioning of large cytoplasmic clusters of proteins in bacterial cells.
Materials and Methods
Bacterial Strains and Growth Conditions.
R. sphaeroides strains were grown aerobically in succinate medium (35) at 30°C with shaking. WS8N is the wild-type strain, JPA1558 contains a chromosomal replacement of tlpT with tlpT-yfp (16), and JPA1553 (tlpT-YFP, ΔppfA) was constructed by genomic replacement of tlpT with tlpT-yfp in a strain already containing an in-frame deletion of ppfA (JPA1301) (13) as described in ref. 16.
Fluorescence Microscopy and Analysis.
Cells were immobilized on a thin layer of 0.8% agarose in succinate medium on microscope slides as described in ref. 15. Differential interference contrast microscopy and fluorescence images were acquired with a Nikon TE200 microscope and YFP filter set (Chroma, Rockingham, VT) and recorded with a cooled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan). Graticule-calibrated software (simple pci imaging software; Compix, Sewickley, PA) was used to analyze images and measure distances. The cluster position was defined as the distance between the cluster and the farthest pole, expressed as a factor of cell length, and these data were binned into 10% regions of the cell length. Statistical significance was calculated by analysis of variances with Tukey's modification. For time-lapse microscopy, fluorescence images of growing cells were obtained and analyzed either every 20 min for 4 h or every 3 min for 80 min. For time-lapse experiments, the position of the clusters was measured from one designated cell pole.
Analysis of Filamentous Cells.
Cells were grown aerobically to an OD700 of 0.2 and treated with 2.5 μg/ml cephalexin for exactly 4 h before being viewed by fluorescence microscopy (as described above). Cell lengths and distances between clusters were measured by using graticule-calibrated software (simple pci). Equal volumes of each culture were also harvested, run on 8% polyacrylamide gels, and transferred to poly(vinylidene difluoride) membrane (Bio-Rad). The membranes were probed by using anti-GFP antibody (BD Living Colors Full-Length A.v. polyclonal antibody), and band intensities were analyzed by using imagequant (Molecular Dynamics), correcting for background. The same samples were also probed with an antibody against CheA2 (a component of the polar chemotaxis protein cluster whose level should be constant across experiments) to ensure equal loading of all lanes.
Supplementary Material
Acknowledgments
We thank D. J. Sherratt and J. Errington for comments on a previous version of the manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council.
Abbreviations
- MCP
methyl-accepting chemotaxis protein
- PpfA
protein positioning factor A
- Tlp
transducer-like protein
- YFP
yellow fluorescent protein.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wadhams G. H., Armitage J. P. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 2.Bilwes A. M., Alex L. A., Crane B. R., Simon M. I. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 3.Hess J. F., Oosawa K., Kaplan N., Simon M. I. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 4.Levit M., Liu Y., Surette M., Stock J. J. Biol. Chem. 1996;271:32057–32063. doi: 10.1074/jbc.271.50.32057. [DOI] [PubMed] [Google Scholar]
- 5.Alon U., Surette M. G., Barkai N., Leibler S. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 6.Sourjik V., Berg H. C. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 7.Sourjik V., Berg H. C. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boukhvalova M., VanBruggen R., Stewart R. C. J. Biol. Chem. 2002;277:23596–23603. doi: 10.1074/jbc.M202288200. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu T. S., Le Novere N., Levin M. D., Beavil A. J., Sutton B. J., Bray D. Nat. Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 10.Gestwicki J. E., Lamanna A. C., Harshey R. M., McCarter L. L., Kiessling L. L., Adler J. J. Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skidmore J. M., Ellefson D. D., McNamara B. P., Couto M. M. P., Wolfe A. J., Maddock J. R. J. Bacteriol. 2000;182:967–973. doi: 10.1128/jb.182.4.967-973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro L., McAdams H. H., Losick R. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- 13.Porter S. L., Warren A. V., Martin A. C., Armitage J. P. Mol. Microbiol. 2002;46:1081–1094. doi: 10.1046/j.1365-2958.2002.03218.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin A. C., Wadhams G. H., Armitage J. P. Mol. Microbiol. 2001;40:1261–1272. doi: 10.1046/j.1365-2958.2001.02468.x. [DOI] [PubMed] [Google Scholar]
- 15.Wadhams G. H., Martin A. C., Armitage J. P. Mol. Microbiol. 2000;36:1222–1233. doi: 10.1046/j.1365-2958.2000.01936.x. [DOI] [PubMed] [Google Scholar]
- 16.Wadhams G. H., Warren A. V., Martin A. C., Armitage J. P. Mol. Microbiol. 2003;50:763–770. doi: 10.1046/j.1365-2958.2003.03716.x. [DOI] [PubMed] [Google Scholar]
- 17.Wadhams G. H., Martin A. C., Porter S. L., Maddock J. R., Mantotta J. C., King H. M., Armitage J. P. Mol. Microbiol. 2002;46:1211–1221. doi: 10.1046/j.1365-2958.2002.03252.x. [DOI] [PubMed] [Google Scholar]
- 18.Leonard T. A., Moller-Jensen J., Lowe J. Philos. Trans. R. Soc. London B. 2005;360:523–525. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes K., Moller-Jensen J., Jensen R. B. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 20.Koonin E. V. J. Mol. Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 21.Gordon G. S., Sitnikov D., Webb C. D., Teleman A., Straight A., Losick R., Murray A. W., Wright A. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 22.Ebersbach G., Gerdes K. Mol. Microbiol. 2004;52:385–398. doi: 10.1111/j.1365-2958.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Austin S. Mol. Microbiol. 2002;46:63–74. doi: 10.1046/j.1365-2958.2002.03156.x. [DOI] [PubMed] [Google Scholar]
- 24.Wadhams G. H., Martin A. C., Warren A. V., Armitage J. P. Mol. Microbiol. 2005;58:895–902. doi: 10.1111/j.1365-2958.2005.04880.x. [DOI] [PubMed] [Google Scholar]
- 25.Gerdes K., Moller-Jensen J., Ebersbach G., Kruse T., Nordstrom K. Cell. 2004;116:359–366. doi: 10.1016/s0092-8674(04)00116-3. [DOI] [PubMed] [Google Scholar]
- 26.de Boer P. A., Crossley R. E., Hand A. R., Rothfield L. I. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fung E., Bouet J. Y., Funnell B. E. EMBO J. 2001;20:4901–4911. doi: 10.1093/emboj/20.17.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figge R. M., Easter J., Gober J. W. Mol. Microbiol. 2003;47:1225–1237. doi: 10.1046/j.1365-2958.2003.03367.x. [DOI] [PubMed] [Google Scholar]
- 29.Lutkenhaus J., Sundaramoorthy M. Mol. Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- 30.Slovak P. M., Wadhams G. H., Armitage J. P. J. Bacteriol. 2005;187:54–64. doi: 10.1128/JB.187.1.54-64.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Austin S. Plasmid. 2002;48:174–178. doi: 10.1016/s0147-619x(02)00104-x. [DOI] [PubMed] [Google Scholar]
- 32.Sherratt D. J. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- 33.Bates D., Kleckner N. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiraga S. Annu. Rev. Genet. 2000;34:21–59. doi: 10.1146/annurev.genet.34.1.21. [DOI] [PubMed] [Google Scholar]
- 35.Sistrom W. R. J. Gen. Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.