Abstract
Plant responses to cold stress are mediated by a transcriptional cascade, in which the transcription factor ICE1 and possibly related proteins activate the expression of C-repeat (CRT)-binding factors (CBFs), leading to the transcription of downstream effector genes. The variant RING finger protein high expression of osmotically responsive gene (HOS)1 was identified genetically as a negative regulator of cold responses. We present evidence here that HOS1 is an E3 ligase required for the ubiquitination of ICE1. HOS1 physically interacts with ICE1 and mediates the ubiquitination of ICE1 both in vitro and in vivo. We found that cold induces the degradation of ICE1 in plants, and this degradation requires HOS1. Consistent with enhanced cold-responsive gene expression in loss-of-function hos1 mutant plants, overexpression of HOS1 represses the expression of CBFs and their downstream genes and confers increased sensitivity to freezing stress. Our results indicate that cold stress responses in Arabidopsis are attenuated by a ubiquitination/proteasome pathway in which HOS1 mediates the degradation of the ICE1 protein.
Keywords: cold stress, RING finger protein
In response to low temperatures, numerous genes are induced in plants (1). Three transcription factors known as C-repeat (CRT)-binding factors (CBFs) or dehydration-responsive element (DRE)-binding protein can bind to CRT/DRE cis elements in the promoters and activate transcription of many of the cold-responsive genes (2–4). The CBF genes are transiently induced by low temperature, and this induction precedes that of the downstream cold-responsive genes (4–6). ICE1 is an upstream constitutively expressed transcription factor that controls the cold induction of CBF3 (7).
Previously, a genetic screen using Arabidopsis plants that express the firefly luciferase reporter gene driven by the CRT/DRE element-containing RD29A promoter led to the identification of HOS1 (high expression of osmotically responsive gene 1), a negative regulator of the CBF regulon (8). CBFs and their downstream genes show enhanced cold induction in loss-of-function hos1 mutant plants (8). HOS1 was predicted to encode a 915-aa protein containing a short motif near the N terminus that is similar to the RING finger domain found in a group of animal proteins known as inhibitor of apoptosis (9). Recently, several plant proteins containing RING finger domain have been shown to function as ubiquitin E3 ligases in hormonal signaling and development (10).
Transfer and covalent ligation of the 76-aa protein ubiquitin to target proteins is a central part of the ubiquitination pathway, involving three enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). E3 is responsible for recruiting specific target proteins for ubiquitination (10). Polyubiquitinated proteins are recognized by the 26S proteasome and degraded. Arabidopsis has two E1 isoforms (11) and at least 37 E2 enzymes (12). In contrast, >1,300 genes are predicted to encode for E3 ubiquitin ligases (10). The large number and diversity of E3 ligases confer specificity upon the ubiquitination pathway. On the basis of subunit composition and action mode, four E3 types have been described in plants: homology to E6AP C terminus; RING/U box; a complex of Skp1, CDC53, and F-box protein (SCF); and anaphase-promoting complex. Arabidopsis contains at least 469 predicted RING-domain-containing proteins (13). However, the physiological functions for most of them are not known. The importance of RING proteins in plant responses to cold stress through the ubiquitin/proteasome pathway has not been investigated. In fact, the involvement of the ubiquitin/proteasome pathway in cold stress has not been explored in any system.
In this study, we have found that cold response in Arabidopsis is attenuated by the proteasome pathway. We demonstrated that the Arabidopsis HOS1 is a functional RING finger protein that has ubiquitin E3 ligase activity. We found that HOS1 physically interacts with ICE1. Both in vitro and in vivo ubiquitination assays showed that HOS1 is required for ICE1 ubiquitination. Furthermore, we discovered that cold induces degradation of the ICE1 protein, and this degradation is blocked by hos1 mutation. Overexpression of HOS1 in transgenic Arabidopsis reduces the expression of CBF genes and decreases plant-freezing tolerance. Together, our results suggest an important role of HOS1 as a ubiquitin E3 ligase in mediating the degradation of ICE1 and possibly other regulators to attenuate cold responses in Arabidopsis.
Results
The Effect of Proteasomal Inhibitors on RD29A-LUC Expression Suggests an Involvement of the Ubiquitin/Proteasome Pathway in Plant Cold Responses.
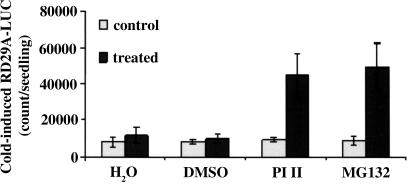
As a first step in studying the potential involvement of ubiquitin/proteasome-mediated protein degradation in plant cold responses, we investigated the effect of proteasomal inhibitors on cold-induced gene expression using transgenic Arabidopsis plants harboring the cold-responsive reporter gene RD29A-LUC. To ensure an accurate comparison, seedlings grown in an agar plate were divided into two halves; one half (control) was untreated, and the other half (treated) was sprayed with the proteasomal inhibitor PI II, MG132, water, or DMSO. The entire plates were exposed to 0°C for 24 h to induce RD29A-LUC expression in the seedlings. As shown in Fig. 1, cold-induced RD29A-LUC expression was significantly increased with either proteasomal inhibitor PI II or MG132 treatment. In contrast, treatment with water or DMSO had no effect (Fig. 1). Without cold treatment, no RD29A-LUC expression was detected in water, DMSO, or proteasomal inhibitor-treated plants (data not shown). The results suggest an involvement of ubiquitin/proteasome-mediated protein degradation in negatively regulating plant responses to cold stress.
Fig. 1.
Proteasome inhibitors enhance cold-induced expression of RD29A-LUC. Arabidopsis seeds (wild type containing RD29A-LUC) were germinated and grown on Murashige and Skoog plates for 10 days. Half of the seedlings were pretreated for 24 h in a Petri dish with H2O, 2% DMSO, or 40 μM proteasome inhibitors (PI II and MG132, Calbiochem) in 2% DMSO (treated). The untreated seedlings were used as controls. Luminescence was analyzed after cold treatment at 0°C for 24 h. Shown are the mean values + SE from 20 individual seedlings.
The RING Finger Protein HOS1 Is a Functional E3 Ligase for Ubiquitination.
HOS1 contains a variant C3HC4 RING finger domain (9), with the first Cys in the domain replaced by Leu (Fig. 2A). To test whether this variant RING finger protein may have an E3 ligase activity, HOS1 was expressed in Escherichia coli as a fusion with maltose-binding protein (MBP). In the presence of ubiquitin, E1, E2, and purified MBP-HOS1, polyubiquitinated proteins were formed (Fig. 2B). This ubiquitination activity depended on the presence of MBP-HOS1 as well as E1 and E2 (Fig. 2B). The E3 ligase was active at both warm (Fig. 2 B and C) and cold (4°C, Fig. 2D) temperatures. To test a possible requirement of the RING domain of HOS1 for its ubiquitin ligase activity, we constructed two mutant proteins, HOS1(H63Y) and HOS1(C77S), in which the conserved amino acid His-63 was mutated to Tyr-63 and Cys-77 to Ser-77, and the RING finger domain was presumably disrupted (Fig. 2A). Neither mutant protein had significant ubiquitin ligase activity (Fig. 2C), which suggests that an intact RING domain is needed for E3 activity.
Fig. 2.
E3 activity of HOS1. (A) Amino acid sequence comparison of HOS1, HOS1(H63Y), and HOS1(C77S) in the RING motif. The mutated residues in HOS1(H63Y) and HOS1(C77S) are underlined. (B) MBP-HOS1 was assayed for E3 activity in the presence of E1, E2, and 32P-labeled ubiquitin (Ub), as indicated. Radiolabeled bands corresponding to free ubiquitin (32P-Ub) and polyubiquitinated MBP-HOS1 ((Ub)n-HOS1) are indicated. (C) MBP-HOS1 E3 activity depends on its RING motif. Wild-type MBP-HOS1 and MBP-HOS1 mutants (H63Y and C77S) were assayed for self-ubiquitination. (D) Ubiquitination assay of HOS1 E3 ligase at 4°C in the presence of E1, E2, and 32P-labeled ubiquitin (Ub), as indicated.
To determine whether the RING finger domain might be sufficient for E3 ligase activity, we purified the MBP fusions of N-terminal 1–550 and 1–210 amino acids of HOS1 (truncated HOS1 proteins containing the RING finger domain; Fig. 3A). Our ubiquitination assays revealed that the mutant protein HOS1 (1–550) was as active as the full-length HOS1, but the HOS1 (1–210) did not yield any detectable ubiquitinated products (Fig. 3B). It is not known whether the truncated HOS1 (1–210) folded properly and assembled with Zn. Nevertheless, the results suggest that the RING finger domain of HOS1 is essential but not sufficient for ubiquitin ligase activity; the region between amino acid residues 210 and 550 is also required.
Fig. 3.
The RING domain of HOS1 is essential but not sufficient for its E3 ligase activity. (A) Diagram of the HOS1 proteins used for the activity assay. The RING domain is indicated in wild-type HOS1 and truncated HOS1 mutants. (B) Self-ubiquitination assay of wild-type MBP-HOS1 and MBP-HOS1 truncated mutants (1–210, 1–550). MBP is included as a negative control. (C) E3 ligase activity of HOS1 and CBL. The reaction temperatures (30°C or 4°C) are indicated.
We compared the E3 ligase activity of HOS1 with that of an Arabidopsis protein similar to the mouse E3 ligase CBL (Casitas B-lineage lymphoma protooncogene), which is involved in plant cell death and defense responses (ref. 14 and Q.X., unpublished data). The result from ubiquitination assays under warm (30°C) and cold (4°C) temperatures (Fig. 3C) indicates that HOS1 is not the only E3 ligase active under cold conditions.
HOS1 Interacts with ICE1.
Because both HOS1 and ICE1 function in controlling cold-responsive gene expression (7, 9), we used yeast two-hybrid assays to test whether there might be an interaction between HOS1 and ICE1. A combination of the bait plasmid pAS2-HOS1 with the empty prey vector pACT2 did not activate transcription of the β-gal reporter gene (Fig. 4A). Positive β-gal activity was observed when the HOS1 bait (pAS2-HOS1) was combined with ICE1 in the prey vector (pACT2-ICE1) (Fig. 4A). By using truncated HOS1 mutants for the yeast two-hybrid assays, we found that the C-terminal part of HOS1 (amino acids 450–842) conferred a very strong interaction with ICE1, compared with the HOS1 N-terminal part (amino acids 1–450) or full-length HOS1 (Fig. 4A). The HOS1 domain responsible for interaction with ICE1 likely lies in its C-terminal region, and the N-terminal region appears to hinder the interaction.
Fig. 4.
RING finger protein HOS1 interacts with ICE1. (A) HOS1 interacts with ICE1 in the yeast two-hybrid assay. Wild-type HOS1 interacts with ICE1 weakly, but the HOS1 C-terminal region (amino acids 450–842), not the N-terminal region (amino acids 1–450), interacts with ICE1 strongly. Yeast strains containing the pAS2-HOS1, pAS2-HOS1 (1–450), and pAS2-HOS1 (450–842) bait and the pACT2 or pACT2-ICE1 prey were assayed for β-gal activity (blue color). The pAS2-HOS1/pACT2, pAS2-HOS1(1–450)/pACT2, and pAS2-HOS1(450–842)/pACT2 combinations were used as negative controls. Yeast grown on synthetic complete (SC) plate (Left) and the corresponding β-gal filter assay (Right) are shown. (B) HOS1 binds to ICE1 in vitro. Recombinant MBP-HOS1 but not MBP could pull down in vitro-translated 35S-ICE1. The first lane from the left shows in vitro-translated 35S-ICE1. (C) HOS1 interacts with ICE1 in vivo. pUCSPYNE-ICE1 was transiently coexpressed with pUCSPYCE-HOS1 or the pUCSPYCE vector, and pUCSPYCE-HOS1 was transiently coexpressed with the pUCSPYNE vector in wild-type Arabidopsis protoplasts. YFP fluorescence was detected only when pUCSPYNE-ICE1 was coexpressed with pUCSPYCE-HOS1.
To further test the interaction between ICE1 and HOS1, we carried out an in vitro pull-down assay using in vitro-translated35S-labeled ICE1 protein. ICE1 was found to bind to the MBP-HOS1 but not the MBP control Sepharose beads (Fig. 4B). An in vivo interaction between HOS1 and ICE1 was verified by a bimolecular fluorescence complementation assay (15, 16). ICE1 was fused translationally with the N-terminal 155-aa portion of yellow fluorescent protein (YFP) (pUCSPYNE-ICE1), and HOS1 was fused with C-terminal 86-aa portion of YFP (pUCSPYCE-HOS1). pUCSPYNE-ICE1 and pUCSPYCE-HOS1, pUCSPYNE-ICE1 and the pUCSPYCE vector, or pUCSPYCE-HOS1 and the pUCSPYNE vector were cotransformed into Arabidopsis protoplasts. YFP fluorescence was imaged after 6-h cold treatment using confocal laser-scanning microscopy. Fig. 4C shows that YFP fluorescence was detected in the nucleus of the cell cotransformed with both pUCSPYNE-ICE1 and pUCSPYCE-HOS1 but not from protoplasts cotransformed with one of the constructs together with either vector. These results indicate that HOS1 and ICE1 can interact in vivo in plant cells.
HOS1 Mediates the Ubiquitination of ICE1 in Vitro and in Vivo.
The interaction between HOS1 and ICE1 suggests that HOS1 may ubiquitinate ICE1 and target it for proteasomal degradation. To test whether ICE1 can be ubiquitinated by HOS1, we carried out an in vitro ubiquitination assay using in vitro-translated35S-labeled ICE1 protein. In the presence of ubiquitin and E1, E2, and HOS1, an apparently polyubiquitinated form of ICE1 was detected (Fig. 5A). As negative controls, when either of the enzymes was omitted from the reaction, no ubiquitinated form of ICE1 could be found (Fig. 5A).
Fig. 5.
Ubiquitination of ICE1 in vitro and in vivo. (A) Ubiquitination of ICE1 by HOS1 in vitro. 35S-ICE1 was assayed for in vitro ubiquitination in the presence of E1, E2, and E3 (MBP-HOS1). Polyubiquitinated ICE1 is indicated as (Ub)n-ICE1. (B) Detection of in vivo polyubiquitinated GFP-ICE1 in wild-type and hos1 plants after cold treatment. Seedlings of GFP-ICE1 in wild-type (WT/GFP-ICE1) or hos1 mutant (hos1/GFP-ICE1) background were pretreated with the proteasome inhibitor MG132 (50 μM) at 4°C for 15 h. Total protein extract from 30 seedlings was immunoprecipitated with antibody against GFP and analyzed by immunoblotting with antibody against ubiquitin. A Coomassie blue-stained nonspecific band from a duplicate gel is shown as loading control.
To determine whether ICE1 is ubiquitinated in vivo, we immunoprecipitated GFP-ICE1 using anti-GFP antibodies from transgenic plants expressing GFP-ICE1 driven by the cauliflower mosaic virus 35S promoter (7). Wild-type plants expressing GFP-ICE1 were crossed with the hos1 mutant, and the resulting homozygous line of GFP-ICE1 in a hos1 background (hos1/GFP-ICE1) was obtained from the F2 population. These plants were treated with the proteasome inhibitor MG132 (50 μM) and subjected to cold treatment (4°C) for 15 h before the immunoprecipitation experiment to prevent degradation of ubiquitinated protein. The immunoprecipitate was separated by SDS/PAGE, blotted, and probed with antiubiquitin antibodies. In wild-type plants expressing GFP-ICE1, high-molecular-weight polypeptide bands corresponding to apparently polyubiquitinated forms of GFP-ICE1 were detected (Fig. 5B, left lane). These bands were absent in hos1 mutant plants expressing GFP-ICE1 (Fig. 5B). The results suggest that GFP-ICE1 is ubiquitinated in vivo, and this ubiquitination requires HOS1.
HOS1 Is Required for Cold-Stress-Induced Degradation of ICE1.
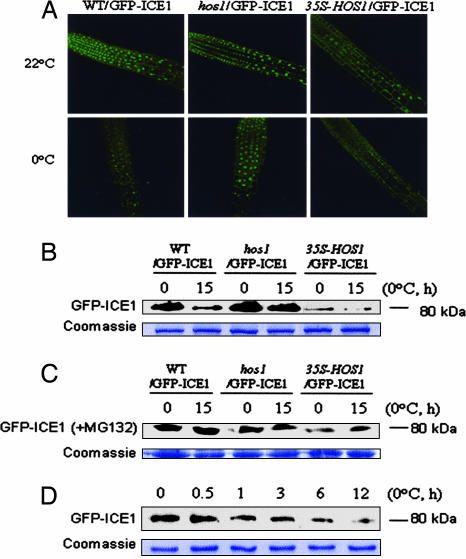
HOS1 mediating in vivo ubiquitination of ICE1 prompted us to investigate the in vivo regulation of ICE1 by HOS1 using GFP-ICE1 transgenic plants. In wild-type GFP-ICE1 transgenic plants (WT/GFP-ICE1), the fusion protein was present in nuclei at high levels (Fig. 6A). Disruption of HOS1 in the hos1 mutant affected neither the nuclear localization nor the abundance of GFP-ICE1 under normal growth conditions (22°C) (Fig. 6A). Interestingly, cold treatment (0°C) substantially reduced the level of nuclear GFP-ICE1 protein in the wild-type (WT/GFP-ICE1) but not in the hos1 mutant (hos1/GFP-ICE1) (Fig. 6A). Overexpression of HOS1 (35S-HOS1, line 6; see next section) substantially reduced the GFP-ICE1 (from the GFP-ICE1 crossed into 35S-HOS1 plants) protein level even under normal growth conditions (22°C) (Fig. 6A). These observations indicate that cold stress triggers protein degradation of nuclear GFP-ICE1 protein, and the RING finger protein HOS1 is required and may also be a limiting factor for degradation.
Fig. 6.
Cold-induced degradation of GFP-ICE1. (A) Visualization of GFP-ICE1 fusion protein. Ten-day-old seedlings of wild-type (WT/GFP-ICE1), hos1 (hos1/GFP-ICE1), and 35S-HOS1 (35S-HOS1/GFP-ICE1) plants grown on agar plates were treated (0°C, 15 h) or not treated (22°C) with cold stress. Seedling roots were observed immediately after cold stress under a confocal microscope. (B) Detection of GFP-ICE1 protein by Western blot analysis. Wild-type (WT/GFP-ICE1), hos1 (hos1/GFP-ICE1), or 35S-HOS1 (35S-HOS1/GFP-ICE1) seedlings were treated with cold, as described above. GFP-ICE1 protein (80-kDa) levels were analyzed with anti-GFP. (C) Detection of GFP-ICE1 protein levels in the presence of MG132. Wild-type (WT/GFP-ICE1), hos1 (hos1/GFP-ICE1), or 35S-HOS1 (35S-HOS1/GFP-ICE1) seedlings were treated with cold in the presence of MG132 (50 μM). GFP-ICE1 protein (80 kDa) was analyzed with anti-GFP. (D) Kinetics of GFP-ICE1 degradation upon cold treatment. Ten-day-old wild-type (WT/GFP-ICE1) seedlings were treated at 0°C for different times (0, 0.5, 1, 3, 6, or 12 h), as indicated. Levels of the GFP-ICE1 protein (80 kDa) were analyzed with anti-GFP. In B–D, a nonspecific Coomassie blue-stained band from a duplicate gel shows that similar amounts of proteins were loaded.
The reduction in GFP-ICE1 fluorescence appeared to happen gradually, and at least 12-h exposure to cold was necessary before this reduction became very obvious under the confocal microscope. To further verify the GFP-ICE1 protein degradation triggered by low temperature, proteins from wild-type, hos1 mutant, and 35S-HOS1 plants expressing GFP-ICE1 were probed with anti-GFP antisera. Cold treatment in wild-type plants caused a substantial reduction in the GFP-ICE1 protein level (Fig. 6B). However, this reduction was not observed in hos1 mutant plants (Fig. 6B). HOS1 overexpression also caused a drastic reduction in the GFP-ICE1 protein level (Fig. 6B). In the presence of MG132 (50 μM), cold treatment did not cause a reduction in the GFP-ICE1 protein level in wild-type, hos1 mutant, or 35S-HOS1 plants (Fig. 6C). This result is consistent with the notion that the reduction in the GFP-ICE1 protein level in wild-type plants was caused by proteasome-mediated degradation.
To study the kinetics of GFP-ICE1 degradation upon cold treatment, 10-day-old wild-type (WT/GFP-ICE1) seedlings were treated at 0°C for different time periods, and the levels of the GFP-ICE1 protein were analyzed (Fig. 6D). The results indicate that, although the reduction in GFP-ICE1 fluorescence did not become obvious later (e.g., 15 h; Fig. 6A) during cold treatment, cold-induced degradation of GFP-ICE1 protein level could be detected after 1 h of cold stress by the immunoblot method (Fig. 6B).
HOS1 Overexpression Reduces Cold-Induced Gene Expression and Confers Increased Sensitivity to Freezing Stress.
ICE1 is an upstream transcriptional activator controlling cold induction of CBF3 and other genes (7). Overexpression of ICE1 in transgenic Arabidopsis plants enhances the expression of CBF genes in response to cold stress and improves freezing tolerance (7). Our results above suggest that HOS1 ubiquitinates ICE1 in vivo, thereby promoting its degradation via the ubiquitin/proteasome pathway. Therefore, we would expect overexpression of HOS1 to reduce the transcript levels of downstream CBFs and to decrease plant cold tolerance. Northern blot analysis identified several transgenic Arabidopsis plant lines showing high levels of HOS1 transcripts (Fig. 7A). One of these lines (no. 6) was used for further analysis of downstream cold-induced gene expression. In plants overexpressing HOS1, the expression of CBF1, -2, and -3 was substantially lower than in wild type (Fig. 7B). Expression of the CBF downstream target genes (COR15, COR47, and RD29A) was also lower in HOS1 overexpression plants (Fig. 7B).
Fig. 7.
Gene expression analysis and freezing sensitivity of HOS1-overexpression plants. (A) Overexpression of HOS1 in transgenic Arabidopsis plants. Total RNA was prepared from wild-type (WT) and 35S-HOS1 transgenic seedlings. The blot was probed with labeled HOS1 cDNA or 25S rRNA as a control. (B) Transcript levels of CBF and the CBF downstream target genes in WT and HOS1-overexpression plants (35S-HOS1). Gene expression was analyzed by Northern blot hybridization with total RNA prepared from 2-week-old WT and HOS1-overexpression transgenic Arabidopsis plants after cold treatment. The blot was probed with labeled CBF3, CBF2, CBF1, RD29A, COR47, COR15A, and 25S rRNA, respectively. (C) Decreased survival of HOS1-overexpression transgenic plants (35S-HOS1) after a freezing treatment at −8°C. (D) Quantification of the survival rates for WT and HOS1-ovexpression transgenic plants (35S-HOS1) upon freezing treatment, after 48-h cold acclimation at 4°C. Data shown are mean values with standard errors (n = 7). The two genotypes are significantly different (χ2 test, P < 0.01) at all of the tested temperatures.
We then investigated the effect of HOS1 overexpression on plant-freezing tolerance. Twelve-day-old seedlings of HOS1 overexpression and wild-type genotypes grown on separate halves of the same agar plates were cold-treated at 4°C for 48 h and then subjected to a freezing tolerance assay. The HOS1-overexpression seedlings were less tolerant to freezing than the wild type, particularly at −8°C and −10°C freezing temperatures (Fig. 7 C and D). For example, freezing at −8°C killed ≈67% of the HOS1-overexpression plants (line no. 6) but only ≈24% of wild-type plants. These results show that HOS1 overexpression has a negative impact on the expression of CBF genes, CBF downstream target genes, and freezing tolerance.
Discussion
The Arabidopsis HOS1 contains a variant RING finger domain and was identified in a genetic screen for mutants with deregulated cold-responsive gene expression (9). In this study, we show that HOS1 is a functional E3 ligase in targeting ICE1 for ubiquitination-mediated protein degradation. The RING finger of Arabidopsis HOS1 can be classified as a C3HC4-type RING, but the first Cys in the RING domain of HOS1 is replaced with Leu (9). Our results indicate that the RING finger in HOS1 is functional, and that the conserved amino acids His-63 and Cys-77 in the RING finger domain are essential for HOS1 as a functional E3 ligase.
Cold stress triggers transcription of the CBF family of transcription factors, which in turn activate the transcription of downstream genes containing the CRT/DRE promoter element (1). The CBF target genes include a number of transcription factors (17). Therefore, cold signaling for acquired freezing tolerance requires a cascade of transcriptional regulations. As an upstream transcription factor of this cascade, ICE1 positively regulates plant cold responses. The ICE1 transcript is constitutively expressed and slightly up-regulated by cold stress (7). ICE1 protein binds to the CBF3 promoter and activates its transcription (7). Overexpression of ICE1 enhances the expression of CBF3 in the cold and confers increased tolerance to freezing stress in transgenic plants (7). We found here that HOS1 interacts with ICE1 and mediates its ubiquitination, resulting in degradation of ICE1. Ubiquitin/proteasome-mediated proteolysis of cold signaling components is also supported by our results showing that proteasomal inhibitor treatments increased the expression level of the RD29A-LUC reporter gene in Arabidopsis plants (Fig. 1). Direct evidence demonstrating that HOS1 mediates the ubiquitination of ICE1 comes from experiments detecting polyubiquitinated GFP-ICE1 in vivo in wild-type but not hos1 mutant plants (Fig. 5B). Furthermore, we found that the level of GFP-ICE1 protein declines after cold treatment, perhaps reflecting ICE1 degradation for the attenuation of cold response. Importantly, our results suggest that this decline or degradation of ICE1 protein in the cold depends on the presence of HOS1.
HOS1 negatively regulates the expression of cold-responsive genes (8, 9). The CBF transcription factor genes and downstream cold-regulated genes such as RD29A, COR47, COR15A, and KIN1 are induced by cold at higher levels in the hos1 mutant (8, 9). Our analysis of cold-responsive gene expression in transgenic Arabidopsis plants overexpressing HOS1 showed that the expression levels of CBFs and their target genes decreased. The freezing tolerance of these plants was also reduced (Fig. 7).
The attenuation of cold response is suggested by the transient nature of the expression of CBF genes in the cold (4–6). The ICE1 and also perhaps related proteins that control the expression of CBFs are present without cold stress. These proteins probably undergo certain posttranslational modification(s) (e.g., phosphorylation) in response to cold stress, to become active in switching on the expression of CBFs (7). This active modified form of ICE1 may be more efficiently recognized by HOS1 and then degraded through the ubiquitination/proteasome pathway. In addition, although cold stress does not change the HOS1 expression level substantially, it promotes the nuclear localization of HOS1 (9). This may also contribute to cold-induced degradation of ICE1. In the hos1 mutant, ICE1 is not degraded, and the expression of CBFs is more sustained. Intriguingly, in HOS1 overexpression plants, ICE1 protein levels appear reduced even at warm temperatures. The reasons for this reduction in ICE1 protein level at warm temperatures are unclear at the present time. At least in vitro, HOS1 can be an active E3 ligase at both warm and cold temperatures and, compared with other E3 ligases, it does not appear to be preferentially functional at cold temperatures.
In summary, our results suggest a mechanism by which HOS1 attenuates cold signaling by mediating the degradation of ICE1 and possibly other regulators of low-temperature responses through the ubiquitin/proteasome pathway. Our work establishes HOS1 as a ubiquitin E3 ligase and identifies ICE1 as a target of this ligase activity. HOS1 may also mediate the ubiquitination and degradation of other regulators of CBF genes, because ICE1 has a major role in the expression of CBF3 only (7), but CBF1 and CBF2 expression is also altered in the hos1 mutant (9). Additionally, HOS1 may also target regulator(s) of the flowering time gene FLC (18, 19), because the hos1 mutant is early-flowering and shows decreased expression of FLC (9).
Materials and Methods
Plant Materials.
Transgenic Arabidopsis plants harboring constructs of CBF3-LUC, RD29A-LUC, HOS1, GFP-HOS1, and GFP-ICE1 were grown as described (7, 9). The GFP-ICE1 transgenic line was crossed with the hos1 mutant or the HOS1 overexpression line (35S-HOS1). Homozygous lines of 35S-HOS1/GFP-ICE1 and hos1/GFP-ICE1 were obtained from the F2 generation. Subcellular localization of GFP-ICE1 was analyzed by a confocal laser-scanning microscopy [Leica (Deerfield, IL) TCS SP2].
Plant-Freezing Test.
Tests for plant-freezing sensitivity were as described (7). Briefly, 12-day-old seedlings of HOS1-overexpression and wild-type genotypes grown on separate halves of the same agar plates were cold-acclimated at 4°C for 48 h under continuous light. These plates were placed on ice in darkness in a freezing chamber set to −1°C for 16 h. Ice chips were sprinkled on the plants before the chamber was programmed to cool at −1°C per hour. Petri dishes of plants were removed after being frozen at desired temperatures for 2 h, thawed at 4°C overnight, and then transferred to a growth chamber at 22°C. Survival of the seedlings was scored visually (plants with green leaves) after 2 days.
Yeast Two-Hybrid Interaction and Bimolecular Fluorescence Complementation Assays.
Yeast two-hybrid experiments were performed as described (20). A bimolecular fluorescence complementation assay (15, 16) was conducted by using constructs of pUCSPYNE-ICE1 and pUCSPYCE-HOS1 in the vector pUCSPYNE or pUCSPYCE. Plasmids were cotransformed into Arabidopsis protoplasts, and biofluorescence was imaged after 6-h cold treatment (4°C) under a confocal laser-scanning microscope (Leica TCS SP2).
In Vitro Binding Assays.
[35S]methionine-labeled ICE1 was generated by in vitro transcription and translation with the use of a T7/T3-coupled TnT kit (Promega). For in vitro binding, 5 μl of the translation mix was added to 200 μl of binding buffer containing 50 mM Hepes (pH 7.4), 1 mM EDTA, 150 mM NaCl, 10% glycerol, 0.1% Tween 20, and 0.5 mM DTT, and the mixture was incubated at room temperature for 1 h. The reaction mixture was incubated with amylose resin beads, which were then washed five times with washing buffer (50 mM Tris, pH 7.5/150 mM NaCl/0.2% Nonidet P-40). In vitro-translated products and HOS1-interacting proteins were analyzed by SDS/PAGE.
Ubiquitination Assays.
32P-labeled ubiquitin was prepared as described (21). Unlabeled ubiquitin was purchased from Sigma. For in vitro ubiquitination, MBP fusions of wild-type HOS1 and HOS1 mutants were immobilized on amylose resin beads. Ubiquitination assays were conducted in the presence of E1, E2, and 32P-labeled ubiquitin in a buffer containing 50 mM Tris (pH 7.4), 2 mM ATP, 5 mM MgCl2, and 2 mM DTT (21). Wheat (Tritium aestivum) E1 and Arabidopsis thaliana E2 (AtUBC8, Q.X., unpublished results) were used for the assay. One microliter of crude extract of E1 or E2 (≈40 ng) was used in a mixture of 30 μl containing ≈1 μg of E3. The reaction mixture was incubated for 2 h at 30°C or 6 h at 4°C with agitation. Samples were then heated to 95°C in a buffer containing 2-mercaptoethanol before separation by SDS/PAGE.
For in vivo detection of ubiquitinated GFP-ICE1 in the wild-type and hos1 mutant, 8-day-old seedlings of GFP-ICE1 plants in wild-type or hos1 background were pretreated with 50 μM proteasome inhibitor MG132 for 15 h at 22°C or 4°C in the light. A total of 30 seedlings for each sample were ground in a buffer containing 20 mM Tris (pH 7.4), 100 mM NaCl, 0.5% Nonidet P-40, 0.5 mM EDTA, 0.5 mM PMSF, and 0.5% protease inhibitor mixture (Sigma). The total protein extract was immunoprecipitated with antibody to GFP (Sigma) and analyzed by immunoblotting with antibody to ubiquitin (Sigma).
Western and Northern Blot Analysis.
For examination of protein levels of GFP-ICE1 in plants of genotype WT/GFP-ICE1, hos1/GFP-ICE1, or WT/GFP, total protein was extracted from 8-day-old seedlings. Proteins were separated by SDS/PAGE, blotted, probed by antibody to GFP, and visualized by using chemiluminescence as instructed by manufacturer (ECL; Amersham Pharmacia). RNA extraction and hybridization for Northern blot analysis were performed as described (7).
Acknowledgments
We thank Ms. Weiping Tang for excellent technical assistance, Dr. Richard D. Vierstra (Harvard University, Cambridge, MA) for providing the E1 construct, and Dr. Peter Howley (Harvard University, Cambridge, MA) for the pGST-Ub clone. This work was supported by National Institutes of Health Grant R01GM59138, National Science Foundation Grants IBN-0212346 and MCB-0241450, and U.S. Department of Agriculture National Research Institute Grant 2003-00751 (to J.-K.Z.), and by China Ministry of Science and Technology 863 Project 2002AA224111 and CNFS30325030/30530400 (to Q.X.).
Abbreviations
- CRT
C-repeat
- CBF
CRT-binding factor
- DRE
dehydration-responsive element
- YFP
yellow fluorescent protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Thomashow M. F. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi-Shinozaki K., Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockinger E. J., Gilmour S. J., Thomashow M. F. Proc. Natl. Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilmour S. J., Zarka D. G., Stockinger E. J., Salazar M. P., Houghton J. M., Thomashow M. F. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 6.Medina J., Bargues M., Terol J., Perez-Alonso M., Salinas J. Plant Physiol. 1999;119:463–470. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., Zhu J. K. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishitani M., Xiong L., Lee H. J., Stevenson B., Zhu J. K. Plant Cell. 1998;10:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J. K. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalle J., Vierstra R. D. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 11.Hatfield P. M., Gosink M. M., Carpenter T. B., Vierstra R. D. Plant J. 1997;11:213–226. doi: 10.1046/j.1365-313x.1997.11020213.x. [DOI] [PubMed] [Google Scholar]
- 12.Bachmair A., Novatchkova M., Potuschak T., Eisenhaber F. Trends Plant Sci. 2001;6:463–470. doi: 10.1016/s1360-1385(01)02080-5. [DOI] [PubMed] [Google Scholar]
- 13.Stone S. L., Hauksdottir H., Troy A., Herschleb J., Kraft E., Callis J. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng L. R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B. H., Leung H., Wang G. L. Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapoor A., Agarwal M., Andreucci A., Zheng X., Gong Z., Hasegawa P. M., Bressan R. A., Zhu J. K. Curr. Biol. 2005;15:1912–1918. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Walter M., Chaban C., Schutze K., Batistic O., Weckermann K., Nake C., Blazevic D., Grefen C., Schumacher K., Oecking C., et al. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 17.Fowler S., Thomashow M. F. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung S., Amasino R. M. Curr. Opin. Plant Biol. 2004;7:4–10. doi: 10.1016/j.pbi.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Henderson I. R., Dean C. Development (Cambridge, U.K.) 2004;131:3829–3838. doi: 10.1242/dev.01294. [DOI] [PubMed] [Google Scholar]
- 20.Ohta M., Guo Y., Halfter U., Zhu J. K. Proc. Natl. Acad. Sci. USA. 2003;100:11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Q., Guo H. S., Dallman G., Fang S., Weissman A. M., Chua N. H. Nature. 2002;419:167–170. doi: 10.1038/nature00998. [DOI] [PubMed] [Google Scholar]