Abstract
An important phenylalanine-derived volatile compound produced by plants is 2-phenylethanol. It is a major contributor to flavor in many foods, including fresh fruits, such as tomato, and an insect-attracting scent in roses and many other flowers. Despite the centrality of 2-phenylethanol to flavor and fragrance, the plant genes responsible for its synthesis have not been identified. Here, we describe a biosynthetic pathway for 2-phenylethanol and other phenylalanine-derived volatiles in tomato fruits and a small family of decarboxylases (LeAADC1A, LeAADC1B, and LeAADC2) that can mediate that pathway's first step. These enzymes each catalyze conversion of phenylalanine to phenethylamine and tyrosine to tyramine. Although tyrosine is the preferred substrate in vitro, phenylalanine levels in tomato fruits far exceed those of tyrosine, indicating that phenylalanine is a physiological substrate. Consistent with this view, overexpression of either LeAADC1A or LeAADC2 in transgenic tomato plants resulted in fruits with up to 10-fold increased emissions of the products of the pathway, including 2-phenylacetaldehyde, 2-phenylethanol, and 1-nitro-2-phenylethane. Further, antisense reduction of LeAADC2 significantly reduced emissions of these volatiles. Besides establishing a biosynthetic route, these results show that it is possible to change phenylalanine-based flavor and aroma volatiles in plants by manipulating expression of a single gene.
Keywords: metabolic engineering, phenylalanine, taste
Human perception of flavor involves integration of multiple chemical stimuli from taste and olfactory receptors. Whereas taste receptors respond to a limited set of cues, olfactory receptors respond to thousands of chemicals and provide the diversity of unique food flavors. For example, there are ≈15–20 volatile compounds that, together, constitute the unique flavor of fresh tomatoes (1–4). These volatiles are derived from various precursors, including fatty acids, carotenoids, and amino acids. Several of the most important tomato aroma volatiles, including 2-phenylacetaldehyde and 2-phenylethanol, are derived from phenylalanine (2). The latter, 2-phenylethanol is also a major flavor constituent of such diverse foods as cheese, bread, wine, and olive oil. Both 2-phenylacetaldehyde and 2-phenylethanol have pleasant fruity, floral odors and are major contributors to scent in many flowers, e.g., 2-phenylacetaldehyde in hyacinths and 2-phenylethanol in roses (5). Because of its desirable aroma and association with flowers, 2-phenylethanol is the most used fragrance chemical in cosmetic products (6). As a consequence, there is much interest in natural sources of 2-phenylethanol for the flavor and fragrance industry.
Both 2-phenylacetaldehyde and 2-phenylethanol have important biological functions in plants. 2-Phenylethanol has long been known to have antimicrobial properties (7) and its presence in plant reproductive structures suggests a protective role for flowers and fruits. Both 2-phenylacetaldehyde and 2-phenylethanol are potent insect attractants (see www.pherobase.com), and each attracts different sets of pollinating and predatory insects (8). The presence of 2-phenylacetaldehyde and 2-phenylethanol in ripening fruits is also probably related to their attractiveness to mammals and other seed dispersers (9). Such multiple roles in defense and reproduction suggest that regulation of their synthesis is likely to be critical to the plant.
Despite the importance of 2-phenylacetaldehyde and 2-phenylethanol to flavor and aroma, it is not clear how plants synthesize them. The yeast Saccharomyces cerevisiae produces 2-phenylethanol from phenylalanine via phenylpyruvate and 2-phenylacetaldehyde (10). Deuterium-labeling studies in rose (Rosa damascena Mill.) indicated that there might be as many as four pathways of synthesis (10). In addition to the yeast pathway, Watanabe et al. (11) reported synthesis via a phenethylamine/2-phenylacetaldehyde route and a trans-cinnamic acid/phenyllactate pathway. Because plants contain many aromatic l-amino acid decarboxylases (AADCs) (12), a pathway that begins with phenylalanine decarboxylation is a reasonable assumption. Here we demonstrate that tomato (Solanum lycopersicum), indeed, uses a pathway whose first step is decarboxylation of phenylalanine to phenethylamine. This reaction is catalyzed by a set of related AADCs. Overexpression of the corresponding genes in transgenic tomato plants led to accumulation of significantly higher levels of 2-phenylacetaldehyde and 2-phenylethanol as well as the related compounds 2-phenylacetonitrile and 1-nitro-2-phenylethane.
Results
The Pathway for Synthesis of 2-Phenylethanol in Tomato Fruits.
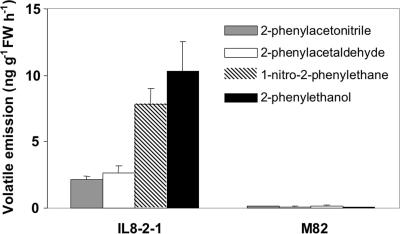
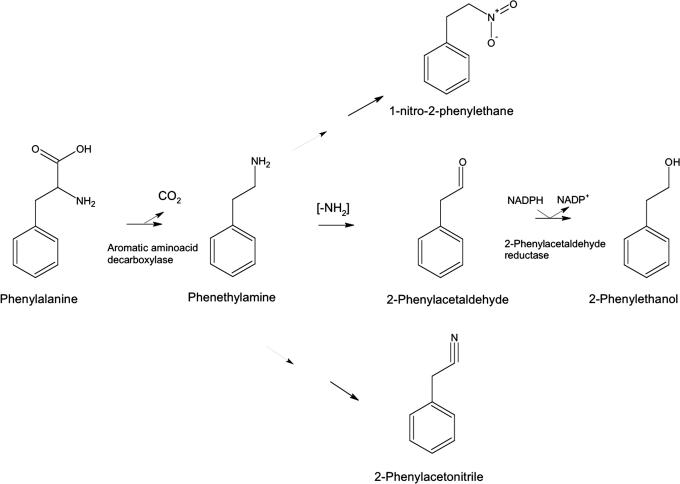
Because 2-phenylethanol comprises a benzene ring with a two-carbon side chain, it may be derived from phenylalanine by removal of the carboxyl and amino groups. If the carboxyl group is lost first, the predicted initial reaction product is phenethylamine, whereas, if the amino group is lost first, the product is phenylpyruvate. To determine the preferred tomato pathway, we examined fruits for the presence of possible intermediates. Tadmor et al. (13) described a line, IL8–2-1, that contains a single introgressed portion of chromosome 8 derived from the wild relative Solanum pennellii. This line contains a locus, malodorous, that is associated with a large increase in emissions of 2-phenylacetaldehyde and 2-phenylethanol. We have routinely observed >1,000-fold increases in these two volatiles over multiple growing seasons (14). Because of this robust behavior, we used IL8–2-1 and its near isogenic parent M82 for subsequent analyses. In addition to these two volatiles, IL8–2-1 emitted greatly increased amounts of two other phenylalanine-derived volatiles, 2-phenylacetonitrile and 1-nitro-2-phenylethane (Fig. 1). These latter two volatiles are almost surely derived from phenethylamine rather than phenylpyruvate, because they have a nitrogen atom in the side chain. Therefore, the most likely pathway for synthesis of 2-phenylethanol is via phenethylamine (Fig. 2).
Fig. 1.
Levels of phenylalanine-derived volatile emissions from M82 and S. pennellii IL8–2-1 fruit (mean of 16 replicates ± SE).
Fig. 2.
Proposed pathway for production of the volatile compounds 2-phenylacetaldehyde and 2-phenylethanol in plants. Phenylalanine is decarboxylated to phenethylamine by an AADC. Phenethylamine is converted to 2-phenylacetaldehyde by removal of the amine group, followed by conversion to 2-phenylethanol by phenylacetaldehyde reductase. The volatile compounds 1-nitro-2-phenethane and 2-phenylacetonitrile are coordinately synthesized and are likely derived from further metabolism of phenethylamine.
To validate the hypothesized pathway, fruit pericarp discs from M82 and IL8–2-1 were fed [13C]phenylalanine. After 4 h, discs were examined for the presence of and flux through [13C]phenethylamine. Labeled phenethylamine was detected in both lines with more product in IL8–2-1 (Table 1) (0.02 and 0.06 μmol·g−1 fresh weight in M82 and IL8–2-1, respectively). When the flux was estimated from the molar fractional labeling of the phenylalanine and phenethylamine pools, it was necessary to invoke a substantially larger flux from phenylalanine to phenethylamine in IL8–2-1 to account for label dilution effects (Table 1). The samples were also examined for the presence of labeled phenylpyruvate and phenyllactate. Neither product was detected. By defining the limits of detection for each compound, it was determined that levels of phenylpyruvate must be at least 10-fold lower and phenyllactate 100-fold lower than that of phenethylamine, if present at all.
Table 1.
Metabolic flux through phenethylamine in tomato
| M82 | IL8-2-1 | |
|---|---|---|
| Labeled PEA, μmol·g−1 FW | 0.02 ± 0.01 | 0.06 ± 0.01 |
| MFE PHE, % | 20.6 ± 0.02 | 20.3 ± 0.01 |
| MFE PEA, % | 1.1 ± 0.01 | 1.8 ± 0.04 |
| FluxPEA (PHE), nmol·g−1·h−1 | 0.16 ± 0.12 | 0.87 ± 0.32 |
Amount of labeled phenethylamine (PEA), molar fractional enrichments (MFE) of phenylalanine (PHE) and PEA, and estimated flux through PEA after [13C]phenylalanine feeding of M82 and IL8-2-1 pericarp discs for 4 h. Data are means of four replicates ± SE. FW, fresh weight.
M82 and IL8–2-1 were then examined for the capacity to decarboxylate phenylalanine in vivo. Pericarp discs were fed [14C]phenylalanine for 8 h and the amount of 14CO2 generated over the period was determined. The amount of [14C]phenethylamine in the tissues was also measured (Table 2). Both samples decarboxylated phenylalanine, and both contained [14C]phenethylamine with significantly higher quantities of each product generated by IL8–2-1, consistent with higher decarboxylation activity in IL8–2-1. The nonstoichiometric amounts of recovered CO2 and phenethylamine can be attributed to further metabolism of the phenethylamine to 2-phenylacetaldehyde and 2-phenylethanol. Taken together, the above data are consistent with the major, if not exclusive, route of synthesis of 2-phenylethanol in tomato fruits via a phenethylamine pathway that is more active in IL8–2-1.
Table 2.
Phenylalanine decarboxylase activity of M82 and IL8-2-1 pericarp disks
| Line | nCi CO2 | nCi phenethylamine |
|---|---|---|
| M82 | 0.96 ± 0.05 | 0.36 ± 0.02 |
| 8-2-1 | 5.16 ± 2.36 | 2.08 ± 0.39 |
Pericarp disks were fed 1 μCi (2.17 nmol) of uniformly labeled [14C]phenylalanine for 8 h, and amounts of 14CO2 and [14C]phenethylamine produced were determined. Data are means of four replicates ±SE. Note that because the phenylalanine contains nine carbon atoms and was uniformly labeled, a 1:1 molar ratio of CO2/phenethylamine corresponds to a 1:8 ratio of 14C.
Identification of AADC Candidate Genes by Screening in Escherichia coli.
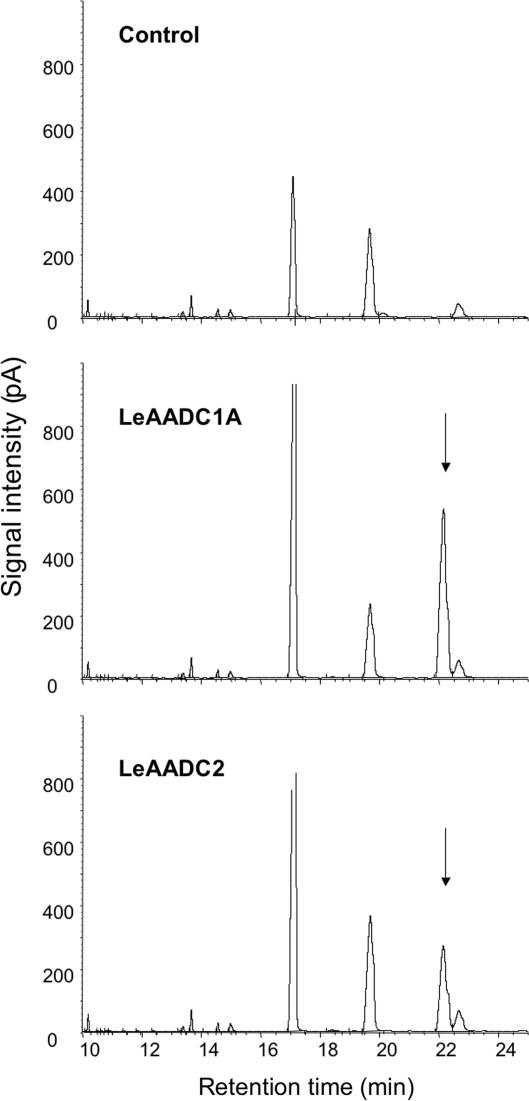
The tomato EST database (www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=tomato) contains many clones annotated as amino acid decarboxylases, based on homology to known enzymes. Several full-length cDNA clones representing different subgroups of enzymes were isolated and expressed as recombinant proteins in E. coli. Bacterial cultures were grown in media supplemented with phenylalanine. After growth, cells and culture media were extracted with hexanes and assayed for the presence of phenethylamine. Cultures expressing proteins annotated as histidine decarboxylases (clones cLEC73K23 and cLEC75E21) readily converted phenylalanine to phenethylamine; those expressing proteins annotated as tyrosine/dopa decarboxylases and a control expressing β-glucuronidase did not (Fig. 3).
Fig. 3.
Gas chromatographic profiles of volatile compounds extracted from E. coli cultures expressing the tomato LeAADC1A and LeAADC2 cDNAs. Only cultures expressing these genes produce phenethylamine (arrows) when grown in media supplemented with phenylalanine. The control contained the identical vector expressing E. coli β-glucuronidase.
Further screening of the available EST clones indicated that there are two highly related genes, designated LeAADC1A (DQ458998) and LeAADC1B (DQ458999). The proteins encoded by these two genes are 95% identical. Both genes map to the same position on chromosome 8 (www.sgn.cornell.edu), indicating that they may have arisen by a duplication event. A third cDNA encoding an enzyme with phenylalanine decarboxylase activity (DQ459000) with 81% identity to LeAADC1A was named LeAADC2 (Fig. 6, which is published as supporting information on the PNAS web site).
Activities of AADC Enzymes.
Histidine-tagged versions of LeAADC1A and LeAADC1B were produced in E. coli, purified, and kinetically characterized with tyrosine and phenylalanine as substrates. Tyrosine was preferred, giving far greater activity than phenylalanine at low substrate concentrations (Table 3). Because exploratory results with LeAADC1A and LeAADC1B were similar, only LeAADC1A was investigated in detail. The Km, Vmax, and Kcat values for tyrosine are 0.92 mM, 815 nmol/min/mg, and 0.7 sec−1, respectively. Substrate inhibition was apparent at higher concentrations (Fig. 7, which is published as supporting information on the PNAS website). Because of substrate inhibition, kinetic parameters were determined by the method of Cleland (15). The Km for phenylalanine was clearly much higher, because saturation was not reached at 40 mM, the highest concentration tested (Fig. 7). Partial characterization of LeAADC2 likewise indicated a strong preference for tyrosine (Table 3). Neither enzyme acted on histidine (data not shown). In connection with the substrate preference, it should be noted that tomato fruits contain much more phenylalanine than tyrosine (10.5 vs. 0.76 μmol·g−1 fresh weight, respectively) (16).
Table 3.
Comparison of the activities of LeAADC1A and LeAADC2 with tyrosine or phenylalanine as substrates
| Enzyme | Tyrosine activity, nmol CO2·min−1·mg−1 protein |
Phenylalanine activity, nmol CO2·min−1·mg−1 protein |
||
|---|---|---|---|---|
| 1 mM | 10 mM | 1 mM | 10 mM | |
| LeAADC1A | 120 | 100 | 0.32 | 3.5 |
| LeAADC1B | 200 | 280 | 1.5 | 16 |
| LeAADC2 | 0.90 | 0.54 | 0.0054 | 0.035 |
Activities were measured at pH 7.9 by means of the 14CO2 release assay. The LeAADC1A and LeAADC1B proteins were purified by Ni2+ affinity chromatography. The LeAADC2 protein was extracted from E. coli cells.
To assess the stoichiometry of the reaction, [U-14C]phenylalanine was used as substrate for LeAADC1A and LeAADC1B. The molar ratio of the two products, CO2 and phenethylamine, was determined, assuming uniform distribution of the 14C within the phenylalanine molecule. This ratio was found to be 1.04 ± 0.04 (mean of five replicates ± SE) for LeAADC1A and 1.06 ± 0.02 (mean of six replicates ± SE) for LeAADC1B.
In Vivo Functions of AADC Gene Products.
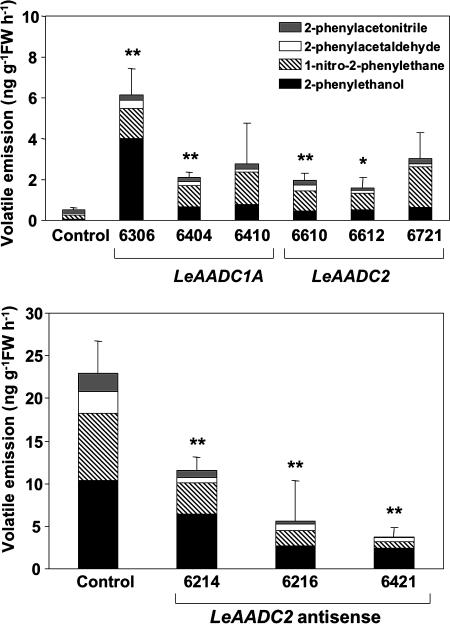
The above in vitro data indicated that the LeAADC enzymes convert phenylalanine to phenethylamine. To validate these observations in vivo, expression vectors containing full-length LeAADC1A and LeAADC2 cDNA clones under control of the constitutive 35S promoter were constructed, and transgenic tomato plants were produced. Multiple independent lines overexpressing each cDNA were identified, and the volatile profiles of ripe fruits from each line were determined. The results showed that overexpression of either enzyme significantly enhanced production of multiple phenylalanine-derived volatiles (Fig. 4). Each of the lines overexpressing either gene product had substantially higher emissions of 2-phenylethanol and 1-nitro-2-phenylethane. The 2-phenylacetaldehyde levels were usually not significantly elevated. Tomato fruits typically accumulate only small quantities of 2-phenylacetaldehyde, suggesting rapid conversion in vivo to 2-phenylethanol. We have produced transgenic plants with greatly elevated phenylacetaldehyde reductase expression. These plants do not have elevated 2-phenylethanol synthesis, indicating that this enzyme is not normally limiting for 2-phenylethanol synthesis (D.T., Holly M. Loucas, David G. Clark, and H.J.K. unpublished work). The levels of phenethylamine in selected transgenic lines were also determined. Ratios of phenethylamine in red ripe fruits from five lines were 1.68 ± 0.25-, 1.1 ± 0.18-, 1.67 ± 0.27-, 1.3 ± 0.15-, and 1.35 ± 0.11-fold higher than control M82 fruit.
Fig. 4.
Volatile emissions from fruits overexpressing LeAADC1A or LeAADC2 (Upper), or reduced in LeAADC2 expression (Lower). The overexpression constructs for LeAADC1A and LeAADC2 were introduced into M82, whereas the antisense LeAADC2 construct was introduced in to IL8–2-1. For lines with ∗ or ∗∗, total volatile emission differed significantly (P < 0.05 or P < 0.01, respectively) from those of nontransformed controls.
To unequivocally demonstrate a role for an AADC enzyme in the 2-phenylethanol pathway in vivo, an antisense LeAADC2 construct was introduced into IL8–2-1. Three independent lines with significantly reduced phenylalanine-derived volatile emissions were identified (Fig. 4). The coordinated reduction in emissions of these volatiles indicates that LeAADC2 catalyzes, at least in part, their synthesis in ripening fruits. Reductions in volatile emissions were also observed in antisense LeAADC1A lines, although the values were not significantly different from controls (data not shown).
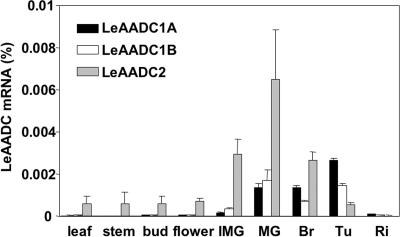
Real-time RT-PCR analysis of expression patterns for each of the genes indicated that all three are expressed in ripening fruit tissues (Fig. 5). LeAADC2 is the predominantly expressed gene in vegetative tissues. Because multiple AADC genes are expressed in ripening fruit tissues, the pathway for synthesis has redundancy. Consistent with this redundancy, single gene loss-of-function transgenic lines did not completely lose the ability to synthesize the phenylalanine-derived volatiles.
Fig. 5.
LeAADC expression in different tissues of M82 tomato plants. Fruit samples from immature green (IMG), mature green (MG), breaker (Br), turning (Tu), and fully ripe (Ri) stages were sampled. Levels of mRNA were determined with TaqMan real-time quantitative RT-PCR. Values are averages for two measurements from each of three independent biological replicates ± SE.
Discussion
The results presented here establish the major pathway for synthesis of 2-phenylethanol from phenylalanine in tomato fruits. This pathway involves enzymatic decarboxylation of phenylalanine by aromatic amino acid decarboxylases to produce phenethylamine. Although we have not identified genes encoding the second step, phenethylamine is presumably converted by an amine oxidase, dehydrogenase, or transaminase to 2-phenylacetaldehyde. There are at least a dozen candidate ESTs in the tomato database. The final step in the pathway to 2-phenylethanol is catalyzed by 2-phenylacetaldehyde reductase (D.T., Holly M. Loucas, David G. Clark, and H.J.K., unpublished work). Analysis of introgression and transgenic lines overproducing AADC enzymes further indicates common intermediates for synthesis of the related volatiles 1-nitro-2-phenylethane and 2-phenylacetonitrile. These latter two volatiles are most likely derived from phenethylamine (Fig. 2). Both have been previously identified in tomato fruits, and 1-nitro-2-phenylethane is considered to be a major contributor to tomato flavor, as are 2-phenylacetaldehyde and 2-phenylethanol (1, 2). Although we cannot rule out alternative pathways to the synthesis of 2-phenylethanol, we were unable to detect either phenylpyruvate or phenyllactate in fruit tissue. Based on the limits of detection of these compounds, we can say that these latter pathways for 2-phenylethanol synthesis are relatively minor, if they exist at all in tomato fruits.
The first step in 2-phenylethanol synthesis is catalyzed by a family of aromatic amino acid decarboxylases. Although currently annotated in databases as histidine decarboxylases, they are active against phenylalanine and tyrosine and do not decarboxylate histidine. This initial misannotation is not surprising because both histidine and aromatic amino acid decarboxylases belong to the same class of pyridoxal phosphate-dependent amino acid decarboxylases (Fig. 8, which is published as supporting information on the PNAS website) (17). In tomato, there are two highly homologous AADC enzymes (LeAADC1A and LeAADC1B) as well as a third distinct enzyme (LeAADC2). Each enzyme catalyzes conversion of phenylalanine to phenethylamine in vitro. All three genes are expressed at comparable levels in fruits, whereas LeAADC1B and LeAADC2 are also expressed in leaves. We cannot rule out the existence of additional AADC genes in the tomato genome.
Kinetic analysis of the AADC enzymes indicates that they have substantial activity against both phenylalanine and tyrosine, the affinity for tyrosine being higher (Table 3). It is not unusual that decarboxylases are not specific for one substrate and have Km values in the millimolar range (12). Moreover, a strawberry O-methyltransferase catalyzing synthesis of the flavor volatile 2,5-dimethyl-4-methoxy-3(2H)-furanone is a multifunctional enzyme that utilizes several substrates (18). It has been proposed that this methyltransferase functions in both lignin and flavor volatile synthesis. Whereas the Km for the lignin precursor caffeic acid is 0.145 mM, the Km for the furanone precursor is 5 mM. More generally, enzymes involved in flavor volatile synthesis would be expected to have Km values above the physiological concentrations of their substrates, because the volatiles are emitted at concentrations far below the substrate concentrations. For 2-phenylethanol, the measured concentration of phenylalanine in M82 fruits is 10.5 μmol·g−1 fresh weight (FW) (16), whereas emissions of 2-phenylethanol are ≈1 pmol·g−1 FW·h−1.
Most aromatic amino acid decarboxylases convert their substrates to the corresponding amine. However, a petunia AADC that converts phenylalanine directly to phenylacetaldehyde, having amine oxidase as well as decarboxylase activity, has been identified (N. Dudareva, personal communication). Although petunia and tomato are quite closely related (both belong to the family Solanaceae), all evidence indicates that the tomato enzymes described here simply convert phenylalanine to phenethylamine. Specifically: (i) the tomato enzymes LeAADC1A and LeAADC1B produce CO2 and phenethylamine in a 1:1 molar ratio; (ii) representative plants overproducing LeAADC enzymes have expanded pools of phenethylamine; (iii) two volatile compounds, 1-nitro-2-phenethane and 2-phenylacetonitrile, which are produced at very high levels in IL8–2-1 and to a lesser extent in the transgenic lines, are almost certainly derived from phenethylamine. In our analyses of petunia volatiles, we have not detected emissions of 1-nitro-2-phenylethane or 2-phenylacetonitrile. It, therefore, appears that petunia and tomato have evolved distinct pathways to 2-phenylethanol that start with different enzymes, the petunia enzyme being unusually specialized.
AADC activity appears to exert major control over the flux from phenylalanine to multiple volatile compounds. IL8–2-1 has significantly higher AADC activity as well as emissions of 2-phenylacetaldehyde, 2-phenylethanol, 1-nitro-2-phenylethane, and 2-phenylacetonitrile. This increased flux occurs in the absence of any change in the pool of free phenylalanine in IL8–2-1 relative to M82 (19). Transgenic plants overexpressing either LeAADC1A or LeAADC2 also exhibited increases in these volatiles. However, none of the transgenic plants synthesized these volatiles to levels even remotely approaching those of IL8–2-1. Although LeAADC1A and LeAADC1B map to the segment of chromosome 8 corresponding to the S. pennellii introgression, we have no evidence that the AADC orthologues are responsible for the malodorous phenotype.
The high activity against tyrosine exhibited by the tomato enzymes indicates that they have the potential to synthesize tyramine as well as phenethylamine in vivo. Tyramine is a precursor for many plant alkaloids (12) and may have a role in synthesis of metabolites associated with defenses against pathogenic organisms (20). The IL8–2-1 fruits would be expected to have elevated levels of tyrosine-derived alkaloids. Expression of LeAADC2 in vegetative tissues may be tied to other defense-related secondary metabolites.
The availability of transgenic plants synthesizing a range of phenylalanine-derived volatiles should facilitate evaluation of their roles in many plant-related processes. Both 2-phenylethanol and 2-phenylacetaldehyde attract pollinating insects and repel feeding insects (8). It will also be possible to critically evaluate the roles of these volatiles in human taste preferences. Increased production of these important volatiles can be accomplished by expression of a single gene, opening the possibility for engineering enhanced scent production in flowers such as rose, where scent has been lost in many varieties in the course of breeding. Finally, these genes will likely be useful as markers for flavor and scent in breeding programs aimed at quality improvement of food and ornamental crops.
Materials and Methods
Volatile Collection.
Tomato (S. lycopersicum cv. M82), and the S.pennellii-derived introgression line IL8–2-1 (21) were grown in the greenhouse or field under standard conditions. Tomato fruit volatiles were collected from ≈100 g of chopped ripe tomato fruit with nonyl acetate as an internal standard, as described in ref. 22. Fruits were enclosed in glass tubes. Air filtered through a hydrocarbon trap (Agilent Technologies, Palo Alto, CA) flowed through the tubes for 1 h with the aid of a vacuum pump. Volatiles were collected on a Super Q column and subsequently eluted with methylene chloride. Volatiles were separated on an Agilent Technologies DB-5 column and analyzed on an Agilent Technologies 6890N gas chromatograph (GC); retention times were compared to known standards. Identities of volatile peaks were confirmed by GCMS, as described in ref. 23. Standards were purchased from Sigma-Aldrich.
In Vivo Phenylalanine Decarboxylase Assays.
Tomato (M82 or IL8–2-1) pericarp discs were incubated with 1 μCi (1 Ci = 37 GBq; 460 mCi/mmol) [U-14C]phenylalanine (Amersham Pharmacia Biosciences, Piscataway, NJ) for 8 h in sealed flasks, each with a 1-cm diameter filter paper disk impregnated with 20 μl of 2 M KOH suspended in the head-space. 14CO2 trapped on the filter paper was quantified by scintillation counting. [14C]phenylalanine and [14C]phenethylamine were extracted from the tissue and separated on an AG-1 (OH−) column in series with a BioRex-70 (H+) column, as described in ref. 24. The identity of [14C]phenethylamine was confirmed by comigration with an authentic standard in thin layer chromatography on silica gel 60 F254 plates in methylene chloride/methanol/triethylamine (80:10:1 vol/vol/vol).
LeAADC Expression in E. coli.
Full-length LeAADC cDNAs were identified by sequencing putative clones from the TIGR database. After sequence analysis, the full-length coding sequences were PCR-amplified and cloned into vector pENTR/D-TOPO. The coding regions were then cloned into vector pDEST15 containing a GST tag (Invitrogen) by recombination and introduced into E. coli BL21-AI (Invitrogen) for inducible protein expression. Control E. coli BL21-AI strains contained pDEST15 with a β-glucuronidase gene inserted. Production of recombinant protein was confirmed by protein blotting with anti-GST antibodies. Activity of each enzyme was determined by growing E. coli expressing each LeAADC in Luria–Bertani medium containing 20 mM phenylalanine. Volatile compounds were extracted from the cultures with an equal volume of hexanes. Extracts were concentrated and analyzed on an Agilent 6890N gas chromatograph. Identification of phenethylamine was confirmed by cochromatography with an authentic standard and by GC-MS (21). The samples produced the characteristic mass-spectral fragments of m/z 103, 91, 65, 58, 51, 39, and 30 as well as the parental ion of m/z = 121.
Protein Purification and Enzyme Assays.
The coding region of LeAADC1A, LeAADC1B, or LeAADC2 was cloned into vector pENTR/D-TOPO (Invitrogen). The coding region was then recombined into vector pDEST17 containing a His tag (Invitrogen) and transformed into E. coli strain BL21-AI (Invitrogen) for inducible protein expression. To purify His-tagged protein, bacterial cultures were centrifuged at 5,000 × g for 5 min, followed by sonication for 1 min in 1× PBS buffer and centrifugation at 10,000 × g for 15 min. The His-tagged protein was bound to Ni-NTA resin (Invitrogen) according to the manufacturer's instructions with the inclusion of 200 μM pyridoxal 5′-phosphate in all solutions. After elution from the Ni-NTA column, the protein was desalted with PD-10 columns (Amersham Pharmacia Biosciences) equilibrated with 50 mM Tris·HCl, pH 8.0, containing 200 μM pyridoxal 5′-phosphate. Protein purity was analyzed by SDS/PAGE, followed by staining with Coomassie brilliant blue and protein blotting with anti-His tag antibodies. LeAADC2 was assayed as a crude extract from E. coli. E. coli containing the control β-glucuronidase gene had no detectable activity. Decarboxylase activity was determined by the method of Facchini et al. (25) with [U-14C]phenylalanine or [U-14C]tyrosine as a substrate and measuring the release of 14CO2. It was first determined that the reaction was linear for 4 h at 30°C; subsequent assays were run for 3 h. To determine the stoichiometry of the reaction, [14C]phenethylamine was separated from the reaction mix with the two-column system described above. [14C]phenethylamine data were corrected for recovery from the columns; the recovery was determined to be 60.3% (mean of triplicate observations) with authentic [14C]phenethylamine.
RNA Expression Analysis.
Total RNA was extracted with a Qiagen total RNA extraction kit, followed by DNase treatment to remove any contaminating DNA. LeAADC mRNA levels were measured by real-time quantitative RT-PCR with TaqMan One-Step RT-PCR reagents and a GeneAmp 5700 Sequence Detection System (Applied Biosystems). Primers and probe for real-time PCR were as follows: LeAADC1A TaqMan probe, 5′-FAM-CCGAACGTGGACAACAAGAAACAGAAAATG-3′-BHQ; LeAADC1A forward primer, 5′-AGCGCGACGACGATTGTT-3′; LeAADC1A reverse primer, 5′-GGTCCTGCACCTGGTTGTG-3′. LeAADC1B TaqMan probe, 5′-FAM-TTTAGCACGACGAAGATTGTTTCCAAATGTG-3BHQ, LeAADC1B forward primer, 5′-GATTTTGAGCCATCACCTATGACA-3′, LeAADC1B reverse primer, 5′-TGTTCCACCTTCTGTTTTTTGTTG-3′, LeAADC2 TaqMan probe, 5′-FAM-TTGGATTGTACATTGATGAATTATATTGATACACTCACCC-3BHQ, LeAADC2 forward primer, 5′-CAGTGACGGAGCCAGGAAA-3′, LeAADC2 reverse primer, 5′-TGGATAACCGATATGATAGTTGATACG-3′. For absolute quantification of RNA, a standard curve was constructed from LeAADC RNAs. LeAADC RNAs were synthesized by in vitro transcription of the coding sequence as described in ref. 26.
GC-MS Analyses of Nonvolatile Plant Metabolites.
For [13C]-labeling studies, pericarp disks weighing ≈600 mg were supplied with 10 μmol of ring-labeled [13C6]phenylalanine (Cambridge Isotope Laboratories, Andover, MA) in 50 μl of H2O for 4 or 8 h. Metabolites were extracted from pericarp tissue and quantified as described in ref. 27, with the exception that, for low abundance metabolites, a substantially higher extract concentration (up to 1,000-fold) was injected onto the GC-MS. The absolute concentration of metabolites was determined by comparison to standard concentration curves as defined in ref. 16. For metabolite analysis, mass-spectral peaks were compared to mass-spectral tag (MST) libraries housed in the Golm Metabolome Database (28, 29). In addition, the metabolites (phenethylamine, phenylpyruvate, phenyllactate, and phenylacetaldehyde) for which no MST information was available were identified by analysis of identically derivatized authentic standards (Sigma-Aldrich). These metabolites were subsequently quantified with the 178, 117, 193, and 193 m/z ions of their derivatives, respectively. The derivatives did not coelute, hence phenyllactate and phenylacetaldehyde could both be quantified. For metabolites that could not be detected in fruit extracts, the limit of detection of the method was determined (Table 4, which is published as supporting information on the PNAS website).
Analysis of [U-13C6]Phenylalanine-Labeled Samples.
Tomato pericarp discs were extracted as described above. After centrifugation, the supernatant was dried under vacuum, and the resulting residue was derivatized for 120 min at 37°C (in 50 μl of 20 mg·ml−1 methoxyamine hydrochloride in pyridine), followed by a 30-min treatment at 37°C with 50 μl of N-trimethylsilyl-N-methyl trifluoroacetamide. GC-MS analysis of the derivatized samples was carried out as described in ref. 22. Uncorrected molar percentage enrichments of metabolites were evaluated as described in ref. 30 by comparison of the 12C spectral fragments and the isotopic spectral fractions of nonlabeled control incubations with the fragmentation patterns of the [13C]-fed tomato pericarp discs as detailed in ref. 31. The reaction rates from metabolic precursors through intermediates to end-products was estimated by dividing the amount of label accumulating in the product by the calculated average proportional labeling of the precursor pool.
Transgenic Plants.
The full-length LeAADC1A or LeAADC2 coding region was cloned in a vector in the sense or antisense orientation under the control of the Figwort Mosaic Virus 35S promoter (32) and followed by the Agrobacterium nopaline synthase (nos) 3′ terminator. The transgene was introduced into tomato cultivar M82 by the method of McCormick et al. (33), with kanamycin resistance as a selectable marker. Transgenic plants were grown to maturity in a greenhouse under standard horticultural practices and fruit collected for further analysis.
Supplementary Material
Acknowledgments
We thank Eric Schmelz for mass spectrometry of the AADC products and Steve Tanksley and his laboratory for mapping the LeAADC1A and LeAADC1B genes. This work was supported in part by the Florida Agricultural Experiment Station and by National Science Foundation Grant DBI-0501778 (to H.J.K.).
Abbreviations
- AADC
aromatic amino acid decarboxylase
- GC
gas chromatograph.
Footnotes
References
- 1.Baldwin E. A., Scott J. W., Shewmaker C. K., Schuch W. Hortic. Sci. 2000;35:1013–1022. [Google Scholar]
- 2.Buttery R. G. In: Flavor Science: Sensible Principles and Techniques. Acree T. E., Teranishi R., editors. Washington, DC: Am. Chem. Soc; 1993. pp. 259–286. [Google Scholar]
- 3.Buttery R. G., Seifert R. M., Guadagni D. G., Ling L. C. J. Agric. Food Chem. 1971;19:524–529. doi: 10.1021/jf60166a061. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin E. A., Goodner K., Plotto A., Pritchett K., Einstein M. J. Food Sci. 2004;69:S310–S318. [Google Scholar]
- 5.Knudsen J. T., Tollsten L., Bergstron L. G. Phytochemistry. 1993;33:253–280. [Google Scholar]
- 6.Clark G. S. Perfume Flavor. 1990;15:37–44. [Google Scholar]
- 7.Berrah G., Konetzka W. A. J. Bacteriol. 1962;83:738. doi: 10.1128/jb.83.4.738-744.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J., Obrycki J., Ochieng S., Baker T., Pickett J., Smiley D. Naturwissenschaften. 2005;92:277–281. doi: 10.1007/s00114-005-0624-2. [DOI] [PubMed] [Google Scholar]
- 9.Goff S., Klee H. Science. 311:815–819. doi: 10.1126/science.1112614. [DOI] [PubMed] [Google Scholar]
- 10.Vuralhan Z., Morais M. A., Tai S.-L., Piper M. D. W., Pronk J. T. Appl. Environ. Microbiol. 2003;69:4534–4541. doi: 10.1128/AEM.69.8.4534-4541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe S., Hayashi K., Yagi K., Asai T., MacTavish H., Picone J., Turnbull C., Watanabe N. Biosci. Biotechnol. Biochem. 2002;66:943–947. doi: 10.1271/bbb.66.943. [DOI] [PubMed] [Google Scholar]
- 12.Facchini P. J., Huber-Allanach K. L., Tari L. W. Phytochemistry. 2000;54:121–138. doi: 10.1016/s0031-9422(00)00050-9. [DOI] [PubMed] [Google Scholar]
- 13.Tadmor Y., Fridman E., Gur A., Larkov O., Lastochkin E., Ravid U., Zamir D., Lewohnson E. J. Agric. Food Chem. 2002;50:2005–2009. doi: 10.1021/jf011237x. [DOI] [PubMed] [Google Scholar]
- 14.Tieman D., Zeigler M., Schmelz E., Taylor M., Bliss P., Kirst M., Klee H. J. J. Exp. Bot. 2006;57:887–896. doi: 10.1093/jxb/erj074. [DOI] [PubMed] [Google Scholar]
- 15.Cleland W. W. In: The Enzymes. 3rd Ed. Boyer P. D., editor. Vol. 2. New York: Academic; 1970. pp. 1–65. [Google Scholar]
- 16.Schauer N., Zamir D., Fernie A. R. J. Exp. Bot. 2005a;56:297–307. doi: 10.1093/jxb/eri057. [DOI] [PubMed] [Google Scholar]
- 17.Sandmeier E., Hale T., Christen P. Eur. J. Biochem. 1994;221:997–1002. doi: 10.1111/j.1432-1033.1994.tb18816.x. [DOI] [PubMed] [Google Scholar]
- 18.Wein M., Lavid N., Lunkenbein S., Lewinsohn E., Schwab W., Kaldenhoff R. Plant J. 2002;31:755–765. doi: 10.1046/j.1365-313x.2002.01396.x. [DOI] [PubMed] [Google Scholar]
- 19.Schauer N., Semel Y., Roessner U., Gur A., Balbo I., Carrari F., Pleban T., Perez-Melis A., Bruedigam C., Kopka J., et al. Nature Biotech. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- 20.Von Roepenack-Lahaye E., Newman M. A., Schornack S., Hammond-Kosack K. E., Lahaye T., Jones J. D., Daniels M. J., Dow J. M. J. Biol. Chem. 2003;278:43373–43383. doi: 10.1074/jbc.M305084200. [DOI] [PubMed] [Google Scholar]
- 21.Eshed Y., Zamir D. Euphytica. 1994;79:175–179. [Google Scholar]
- 22.Schmelz E. A., Alborn H. T., Tumlinson J. H. Planta. 2001;214:171–179. doi: 10.1007/s004250100603. [DOI] [PubMed] [Google Scholar]
- 23.Schmelz E. A., Alborn H. T., Banchio E., Tumlinson J. H. Planta. 2003;216:665–673. doi: 10.1007/s00425-002-0898-y. [DOI] [PubMed] [Google Scholar]
- 24.Rontein D., Nishida I., Tashiro G., Yoshioka K., Wu W.-I., Voelker D. R., Basset G., Hanson A. D. J. Biol. Chem. 2001;276:35523–35529. doi: 10.1074/jbc.M106038200. [DOI] [PubMed] [Google Scholar]
- 25.Facchini P. J., Yu M., Penzes-Yost C. Plant Physiol. 1999;120:653–663. doi: 10.1104/pp.120.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tieman D. M., Ciardi J. A., Taylor M. G., Klee H. Plant J. 2001;26:47–58. doi: 10.1046/j.1365-313x.2001.01006.x. [DOI] [PubMed] [Google Scholar]
- 27.Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A. R. Plant Physiol. 2003;133:84–99. doi: 10.1104/pp.103.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmuller E., Dormann P., Weckwerth W., Gibon Y., Stitt M., et al. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 29.Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner-Tunali U., Forbes M. G., Willmitzer L., et al. FEBS Lett. 2005b;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 30.Giege P., Heazlewood J. L., Roessner-Tunali U., Millar A. H., Fernie A. R., Leaver C. J., Sweetlove L. J. Plant Cell. 2003;15:2140–2151. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roessner-Tunali U., Liu J. L., Leisse A., Balbo I., Perez-Melis A., Willmitzer L., Fernie A. R. Plant J. 2004;39:668–679. doi: 10.1111/j.1365-313X.2004.02157.x. [DOI] [PubMed] [Google Scholar]
- 32.Richins R. D., Scholthof H. B., Shepard R. J. Nucleic Acids Res. 1987;15:8451–8466. doi: 10.1093/nar/15.20.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCormick S., Neidermeyer J., Fry J., Barnason A., Horsch R., Fraley R. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.