Abstract
We investigated the role of SIRT7, one of the seven members of the mammalian sirtuin family. We show that SIRT7 is a widely expressed nucleolar protein that is associated with active rRNA genes (rDNA), where it interacts with RNA polymerase I (Pol I) as well as with histones. Overexpression of SIRT7 increases Pol I-mediated transcription, whereas knockdown of SIRT7 or inhibition of the catalytic activity results in decreased association of Pol I with rDNA and a reduction of Pol I transcription. Depletion of SIRT7 stops cell proliferation and triggers apoptosis. Our findings suggest that SIRT7 is a positive regulator of Pol I transcription and is required for cell viability in mammals.
Keywords: Sirtuin, RNA polymerase I, transcription, rDNA, cell proliferation
Transcription of rDNA by RNA polymerase I (Pol I) accounts for up to 65% of total transcription in metabolically active mammalian cells (Grummt 2003). Pol I transcription is highly coordinated with cellular metabolism and cell proliferation (Grummt 1999). Nutrient starvation, growth factor deprivation, DNA damage, and other conditions that slow down cellular division decrease pre-rRNA synthesis (Grummt 2003), whereas conditions that stimulate cell growth and proliferation increase pre-rRNA synthesis (Zhao et al. 2003).
The Sir2 family of enzymes, termed sirtuins, is conserved from bacteria to humans and regulates a wide range of biological processes such as gene silencing, aging, cellular differentiation, and metabolism (Blander and Guarente 2004). Sirtuins contain a conserved catalytic core domain conferring NAD+-dependent protein deacetylase and ADP-ribosyltransferase activity. The mammalian genome encodes seven homologs of SIR2, termed SIRT1–7 (Frye 2000). Several physiological SIRT1 substrates have been identified including p53, TAFI68, MyoD, FOXO3, PPARγ, NF-kB, and HIV-TAT (Blander and Guarente 2004), but little is known about the biological functions of the other mammalian sirtuins. It has been demonstrated that SIRT6 exhibits exclusively ADP-ribosyltransferase activity (Liszt et al. 2005), and SIRT6−/− mice show a premature aging phenotype due to a defect in base excision repair (Mostoslavsky et al. 2006). The functional properties of the remaining members of the mammalian SIRT family are not known. Here we analyze the biological function of SIRT7, the only mammalian sirtuin that preferentially localizes to nucleoli (Michishita et al. 2005).
Results and Discussion
SIRT7 is a nucleolar protein that is associated with transcriptionally active rRNA genes
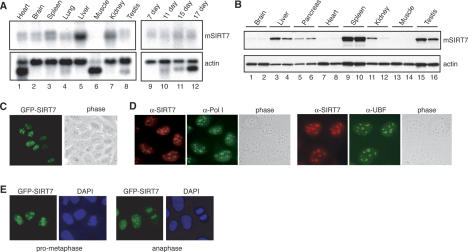
SIRT7 mRNA was expressed in all mouse tissues examined, except skeletal muscle, and was most abundant in liver (Fig. 1A). Likewise, SIRT7 mRNAs were detectable in mouse embryos between embryonic days 7 and 17 (E7 and E17). To monitor SIRT7 protein levels, antibodies were raised against the N terminus of mSIRT7, which is not conserved in SIRT1–6. These antibodies specifically recognize SIRT7, a 45-kDa polypeptide that is present in nuclear but not cytoplasmic extracts of mouse and human cells (Supplementary Fig. S1). As shown in Figure 1B, the amount of SIRT7 was very different in individual tissues. SIRT7 levels were extremely low in muscle, heart, and brain (i.e., in nonproliferating tissues), but high in metabolically active tissues, such as liver, spleen, and testis.
Figure 1.
SIRT7 is a broadly expressed nucleolar protein. (A) RNA was isolated from the indicated tissues (left panels) and embryos (right panels) in wild-type adult mice, resolved by electrophoresis, and subjected to Northern blotting. Blots were probed with 32P-labeled cDNA specific to SIRT7 (top panels) or actin (bottom panels). Muscle actin found in heart and skeletal muscle samples migrated as a distinct band. (B) Proteins extracted from the indicated wild-type mouse tissues were resolved by SDS-PAGE and analyzed by Western blotting using antibodies specific to SIRT7 (top panel) or actin (bottom panel). For each tissue type, samples from two different mice were included. (C) Nucleolar localization of GFP-SIRT7 in live human U2OS cells. (D) Endogenous SIRT7, Pol I, and UBF were visualized by immunofluorescence microscopy in U2OS cells using α-SIRT7 (red), α-Pol I (green), and α-UBF (green) antibodies. The DNA was counterstained with Hoechst 33342. (E) During mitosis GFP-SIRT7 is associated with condensed chromatin in live U2OS cells stained with Hoechst 33342.
Next, we investigated the subcellular localization of human SIRT7. Both GFP-tagged SIRT7 (GFP-SIRT7) (Fig. 1C) and endogenous SIRT7 (Fig. 1D) were detected in nucleoli colocalizing with Pol I, and the nucleolar transcription activator UBF (Fig. 1D). During M phase, when nucleoli disintegrate, SIRT7 was not retained at the nucleolus organizer region (NOR), but remained bound to the condensed mitotic chromatin (Fig. 1E), suggesting that SIRT7 is associated with chromatin throughout the cell cycle.
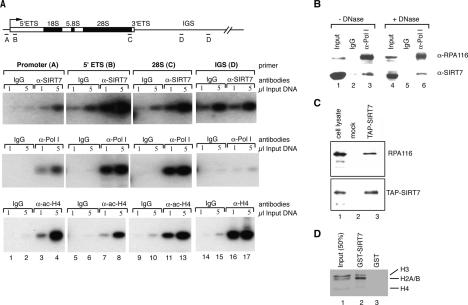
The nucleolar localization raised the possibility that SIRT7 is associated with rDNA. We therefore monitored rDNA occupancy of SIRT7 by chromatin immunoprecipitation (ChIP). Cross-linked chromatin was precipitated with SIRT7 antibodies, and precipitated DNA was analyzed by PCR using primer sets that amplify the rDNA promoter, two regions within the transcribed sequence of human rDNA (5′-ETS and 28S rRNA coding region), and a repeated sequence within the nontranscribed intergenic spacer (IGS). As shown in Figure 2A (top panel), SIRT7 was associated with both the rDNA promoter and the transcribed region. No signal was detected in the IGS above the IgG control, suggesting that SIRT7 is associated with transcriptionally active rRNA genes. In support of this, the distribution of SIRT7 on rDNA was identical to that of Pol I as revealed by ChIP using antibodies that recognize the second largest subunit of Pol I (RPA116) (Fig. 2A, middle panel), albeit with lower background binding than SIRT7. In contrast, acetylated histone H4 is found bound across the entire rDNA repeat (Fig. 2A, bottom panel). The high background seen with the SIRT7 antibody leaves open the possibility that SIRT7 is bound at low levels to the IGS. However, the equal nonspecific signal seen with control IgG and α-SIRT7 antibodies at the IGS does serve as a negative control for the specific binding of SIRT7 observed in the transcribed and promoter regions of the rDNA. We conclude that SIRT7 is associated with the rDNA coding sequence and promoter.
Figure 2.
SIRT7 is associated with the rDNA and RNA Pol I. (A) Results of ChIP analysis showing the localization of SIRT7 and Pol I within the rDNA gene repeats. Cross-linked chromatin from 293T cells was precipitated by the indicated antibodies and analyzed by PCR with the indicated primer pair. (B) Endogenous SIRT7 interacts with RNA Pol I. Partially purified human nuclear extract was immunoprecipitated with control nonspecific human antibodies (lanes 2,5) and α-Pol I antibodies (lanes 3,6). Before immunoprecipitation, the protein fraction used was treated with DNase (lanes 4–6) or left untreated (lanes 1–3). The precipitated proteins were analyzed by Western blotting with α-SIRT7 and α-RPA-116 antibodies as indicated. The input lanes contain 10% of the protein fraction used for IP. (C) Pol I is a component of the TAP-SIRT7 complex. U2OS cell lines harboring the empty vector or TAP-SIRT7 were established. TAP-tagged complexes were purified on IgG-Sepharose and Calmodulin-agarose resins, and the eluted protein complexes were tested for copurifying proteins by Western blot analysis. (Lane 1) Fifty micrograms of nuclear extract (10% of the Input). (Lane 2) Eluate from mock-transfected cells. (Lane 3) Eluate from a cell line expressing TAP-IRT7. The Western blot was probed with an antibody specific for RPA-116 (top panel) and α-SIRT7 (bottom panel). (D) SIRT7 interacts with histones. Histones were isolated from butyric acid-treated HeLa cells by ion exchange chromatography, incubated with GST-SIRT7-Sepharose and GST-Sepharose beads. Bound proteins were eluted and separated by SDS-PAGE and visualized by staining with Coomassie.
To examine whether SIRT7 is part of the Pol I complex, we monitored the association of SIRT7 with Pol I. Coimmunoprecipitation assays revealed that antibodies against Pol I, but not control antibodies, coprecipitated endogenous SIRT7 from a fractionated nuclear extract (Fig. 2B, lanes 1–3). Treatment of the protein fraction used with DNase I did not abrogate this interaction (Fig. 2B, lanes 4–6). Likewise, Pol I was coprecipitated with α-Flag (M2) antibodies from cells overexpressing Flag-tagged SIRT7 (data not shown). Intriguingly, a significant amount of Pol I copurified with SIRT7 after tandem affinity purification from cells expressing TAP-tagged SIRT7 indicating that SIRT7 and Pol I were contained within a stable protein complex (Fig. 2C). Comparison of the amounts of purified TAP-SIRT7 (Fig. 2C, bottom panel, lane 3), copurified Pol I (Fig. 2C, top panel, lane 3), and the amount of the relevant proteins in the cell lysate (Fig. 2C, lane 1) suggested that ∼25% of the SIRT7 complexes contained Pol I. Finally, we analyzed the interactions of SIRT7 with nucleosome components. In pull-down experiments, glutathione S-transferase (GST)-SIRT7 bound all core histones isolated from HeLa cells exhibiting a preferential association with histones H2A/B (Fig. 2D; Supplementary Fig. S2).
SIRT7 activates Pol I transcription
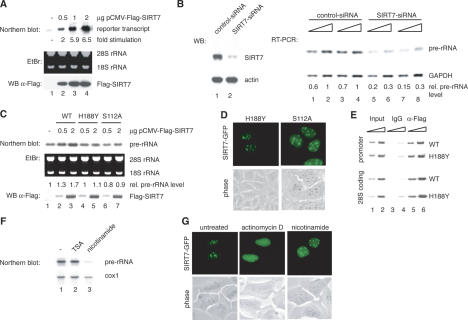
The finding that SIRT7 is enriched in metabolically active tissues, interacts with Pol I, and is mostly localized at transcribed rDNA sequences suggests that SIRT7, unlike SIRT1 (Muth et al. 2001), may have a stimulatory rather than an inhibitory effect on Pol I transcription. To test this, HEK293T cells were cotransfected with a human rDNA reporter plasmid and an expression vector encoding Flag-tagged SIRT7. Transcripts from the rDNA reporter were analyzed on Northern blots. Elevation of SIRT7 levels strongly stimulated transcription of the reporter gene in a dose-dependent manner (Fig. 3A). SIRT7 overexpression also activated rDNA transcription in U2OS and NIH3T3 cells (data not shown) but did not enhance a Pol II-dependent luciferase reporter gene (data not shown), indicating that activation is specific for RNA Pol I.
Figure 3.
SIRT7 is an activator of Pol I transcription. (A) Overexpression of SIRT7 stimulates Pol I transcription from an rDNA minigene reporter. 293T cells were cotransfected with a Pol I reporter plasmid and increasing amounts of an expression plasmid encoding Flag-SIRT7. RNA was isolated from transfected cells. The amount of reporter transcripts was determined by Northern blot analysis and quantified using a PhosphorImager. (Bottom panel) Western blot of the transfected cells probed with α-Flag antibody. (B) Knockdown of SIRT7 by RNAi impairs rRNA synthesis. (Left panel) Western blot of extracts from U2OS cells transfected with control-dsRNA (lane 1) or SIRT7-specific dsRNA (lane 2). (Right panel) Pol I transcription was analyzed by RT–PCR. RNA was isolated from cells treated with the respective dsRNAs. Increasing amounts of cDNA were used for PCR with primers that amplify a region of the 5′ external transcribed spacer of the human pre-rRNA. Results from two independent experiments are shown (one in lanes 1,2,5,6 and the other in lanes 3,4,7,8). As a control, RT–PCR was performed to analyze expression of GAPDH. The PCR-products were quantified using a PhosphorImager. (C) Overexpression of SIRT7H188Y and SIRT7S112A does not augment cellular Pol I transcription. (Top panel) Northern blot analysis of pre-rRNA transcripts from 293T cells transfected with different amounts of plasmids expressing wild-type SIRT7 or the point mutants H188Y and S112A. Pre-rRNA levels were determined by PhosphorImager. (Bottom panel) Expression levels of Flag-SIRT7, Flag-SIRT7H188Y, and Flag-SIRT7S112A were monitored on Western blots using α-Flag antibodies. (D) The SIRT7 mutants H188Y and S112A were expressed as GFP-tagged proteins in U2OS cells and their cellular localization was examined in live cells. (E) SIRT7H188Y is associated with rDNA. Flag-SIRT7 wild type (WT) and Flag-SIRT7H188Y mutant (H188Y) were overexpressed in 293T cells and assayed for binding to rDNA by ChIP. Immunoprecipitations were performed with mouse IgGs (lanes 3,4) and α-Flag (M2) antibodies (lanes 5,6); 0.5 and 1.5 μL of whole-cell extract DNA (lanes 1,2) and precipitated DNA (lanes 3–6) were amplified with rDNA primers A (promoter, top panel) and C (28S coding region, bottom panel). (F) Nicotinamide represses rRNA synthesis. NIH3T3 cells were cultured for 6 h in medium containing 40 nM TSA or 5 mM nicotinamide, and pre-rRNA levels were measured by Northern blot analysis using a 32P-labeled riboprobe specific for the 5′-transcribed external spacer. The blot was subsequently reprobed for cytochrome c oxidase (cox 1) mRNA. (G) Treatment with actinomycin D, but not with nicotinamide, releases SIRT7-GFP from nucleoli. U2OS cells expressing SIRT7-GFP were cultured in the presence of either 50 ng/mL of actinomycin D for 2 h to inhibit Pol I activity or 5 mM nicotinamide for 6 h to inhibit NAD+-dependent deacetylase activity. Localization of SIRT7-GFP was examined in live cells.
If SIRT7 is involved in activation of Pol I transcription, then knockdown of SIRT7 should inhibit pre-rRNA synthesis. Thus, 45S pre-rRNA levels were measured in cells that were depleted of SIRT7 by small interfering RNA (siRNA)-mediated gene silencing. Cellular SIRT7, but not actin, was drastically reduced in cells transfected with SIRT7-specific siRNA (Fig. 3B, left). Strikingly, knockdown of SIRT7 markedly reduced pre-rRNA levels, whereas expression of GAPDH was not affected (Fig. 3B, right). Thus, consistent with SIRT7 being a positive regulator of Pol I transcription, depletion of SIRT7 decreases cellular rRNA synthesis.
Individual sirtuins including SIRT1, SIRT2, SIRT3, and SIRT5 exhibit deacetylase and ADP-ribosyltransferase activity on acetylated histone H4 as substrate (North et al. 2003). In contrast, SIRT6 shows no deacetylase, but a robust ADP-ribosyltransferase activity (Liszt et al. 2005). To examine whether SIRT7-mediated activation of Pol I transcription requires any enzymatic activity, we generated two mutants, SIRT7S112A and SIRT7H188Y, which are mutations in conserved residues that inactivate other mammalian sirtuins (Frye 1999; Vaziri et al. 2001; Liszt et al. 2005). HEK293T cells were transfected with expression vectors encoding wild-type or mutant SIRT7, and 45S pre-rRNA levels were analyzed on Northern blots. Notably, pre-rRNA synthesis was only stimulated by expressing wild-type SIRT7, but not SIRT7S112A or SIRT7H188Y (Fig. 3C). Both wild-type and mutant SIRT7 localized to the nucleolus (Fig. 3D) and associated with the rDNA promoter and coding regions (Fig. 3E).
We next monitored pre-rRNA levels in cells treated with nicotinamide, a potent inhibitor of sirtuins (Denu 2005). Treatment of NIH3T3 cells with nicotinamide strongly inhibited pre-rRNA synthesis, whereas trichostatin A (TSA), an inhibitor of class I and II histone deacetylases (HDACs), had no effect (Fig. 3F). The localization of GFP-SIRT7 was nearly identical in the presence or absence of nicotinamide demonstrating that inhibition of Pol I transcription is not due to cellular redistribution of SIRT7 (Fig. 3G). In contrast, low doses of actinomycin D that selectively inhibit Pol I transcription caused the release of both GFP-SIRT7 and endogenous SIRT7 from the nucleoli (Fig. 3G; Supplementary Fig. S3), indicating that nucleolar localization of SIRT7 is tightly linked to ongoing Pol I transcription.
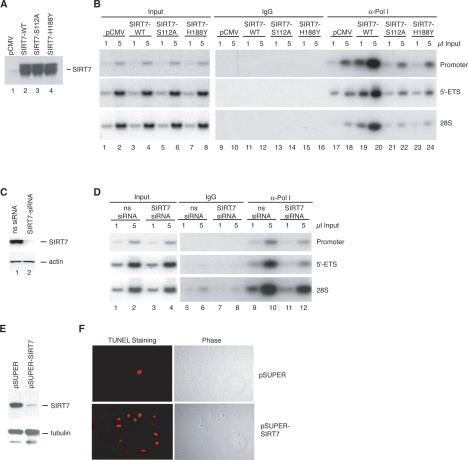
To get mechanistic insight into the role of SIRT7 in rDNA transcription, we monitored Pol I occupancy at rDNA in cell lines expressing Flag-tagged wild-type or mutant SIRT7. The level of tagged SIRT7wt, SIRT7S112A, and SIRT7H188Y was similar in the three cell lines (Fig. 4A). ChIP assays revealed that the amount of Pol I associated with promoter and coding rDNA was significantly higher in cells overexpressing SIRT7 (Fig. 4B, lanes 19,20) compared with cells expressing SIRT7S112A or SIRT7H188Y (Fig. 4B, lanes 21–24). We also analyzed the association of Pol I with rDNA in cells depleted of SIRT7 by siRNA (Fig. 4C). There was much less Pol I associated with rDNA in cells treated with SIRT7 siRNA compared with control siRNA (Fig. 4D), demonstrating that reduction of cellular SIRT7 decreased Pol I association with rDNA. These findings suggest that SIRT7 may be required for efficient elongation of rDNA transcription and depends on conserved residues within the catalytic domain of the protein for this activity.
Figure 4.
SIRT7 stimulates the association of RNA Pol I with the rDNA. (A) Western blot showing overexpression levels of SIRT7 proteins. (B) Enrichment of Pol I at the transcribed region of the rDNA genes in the presence of ectopic SIRT7. Chromatin was prepared from cells harboring empty vector (pCMV) or plasmids expressing wild-type SIRT7 (SIRT7-WT) and mutant SIRT7 (SIRT7-S112A and SIRT7-H188Y). The chromatin was precipitated with α-RPA116 antibodies and analyzed by PCR with the primer pairs indicated on the right side of the figure. (C) Following siRNA transfections, whole-cell lysates were analyzed by immunoblot for SIRT7 and β-actin. (D) ChIP analysis of Pol I levels in cells transfected with siRNAs. Chromatin was prepared from cells transfected with nonspecific (ns) or α-SIRT7 siRNAs. The chromatin was precipitated with α-RPA116 antibodies and analyzed by PCR with the primer pairs indicated on the right side of the figure. (E) Western blot of SIRT7 and α-tubulin in 20 μg of whole-cell lysates from U2OS cells transfected with the pSUPER or pSUPER–shSIRT7 vectors. (F, right) U2OS cells transfected with the indicated vectors were fixed 5 d post-transfection and visualized by phase contrast. (Left) Apoptotic cells were detected by TUNEL staining.
To analyze whether SIRT7 function is essential for cell survival, cell proliferaton was monitored in U2OS cells that expressed control or SIRT7-shRNAs (short interfering RNAs). Five days post-transfection the SIRT7 protein levels were substantially reduced (Fig. 4E). Significantly, the cell number was substantially decreased and the remaining cells stained positive in the TUNEL assay, indicating that cellular depletion of SIRT7 leads to apoptosis (Fig. 4F). Depletion of the Pol I factor TIF-IA also resulted in cell cycle arrest and apoptosis (Yuan et al. 2005). All transcription and ChIP assays in Figures 3 and 4 above in which SIRT7 was knocked down were performed prior to the onset of the apoptotic phenotype.
Here, we establish that SIRT7 is a nucleolar protein, which is essential for cell survival. SIRT7 is a component of the RNA Pol I transcriptional machinery and stimulates Pol I transcription in vivo by an enzymatic mechanism. Further, the presence and enrichment of SIRT7 on the coding rDNA points to an important function during transcriptional elongation. We still have not identified the substrate targeted by SIRT7. Recombinant SIRT7 isolated from different expression systems exhibited no deacetylase or ADP-ribosyltransferase activity, when acetylated histones and various acetylated components of the Pol I transcription complex were used as substrates (data not shown). Therefore, we speculate that SIRT7 modifies (or perhaps simply binds to) a not yet identified protein of the Pol I elongation complex in a NAD+-dependent manner. Coincident with this working hypothesis, we observed that SIRT7 purified from HeLa nuclear extracts was found in several high molecular weight complexes (R. Voit and I. Grummt, unpubl.). Our data suggest a novel mechanism by which mammalian cells may regulate rDNA transcription by sensing the cellular NAD+ levels. In yeast, a change in the NAD+/NADH ratio translates the metabolic shift of calorie restriction into lifespan extension via Sir2p (Lin et al. 2004). In liver and muscle cells, changes in the NAD+/NADH ratio can arise from diet or exercise and influence SIRT1 activity (Fulco et al. 2003; Rodgers et al. 2005). This paradigm suggests that dietinduced changes in the NAD+/NADH ratio may regulate SIRT7 to couple changing energy status with levels of rRNA synthesis and ribosome production.
Sir2 homologs positively regulate longevity in yeast, worms, and flies despite extraordinary phylogenetic distance between these organisms (Hekimi and Guarente 2003). In yeast, Sir2p promotes longevity predominantly through its silencing role at the rDNA (Sinclair and Guarente 1997; Kaeberlein et al. 1999). It is interesting that the effect of sirtuins at the rDNA is complex in mammals. The nearest mammalian homolog of Sir2p, SIRT1, deacetylates TAFI68 and represses Pol I transcription in vitro (Muth et al. 2001). This indicates that mammalian SIRTs regulate Pol I transcription both positively and negatively. Since SIRT1 is enriched in nucleoli only at the onset of mitosis (R. Dischinger and R. Voit, unpubl.), it is likely that SIRT1 activity is functionally linked to mitotic repression of Pol I transcription. On the other hand, the presence of SIRT7 in nucleoli of cycling cells throughout interphase underlines the positive function of SIRT7 activity for Pol I transcription and cell growth. We suspect that a link between SIRT7 and growth may be part of a larger set of physiological changes that is induced by diet.
Materials and methods
Plasmids and antibodies
To generate pCMV-FLAG-SIRT7, human SIRT7 cDNA (ATCC-IMAGE 222518) was cloned into pCMV-TAG-4a (Stratagene). pCMV-Flag-SIRT7 served as template for site-directed mutagenesis using the Stratagene QuickChange mutagenesis kit to produce pCMV-Flag-SIRT7S112A and pCMV-Flag-SIRT7H188Y. pEGFP-SIRT7, pET-GST-SIRT7, and pTAP-SIRT7 were constructed by inserting the SIRT7 coding sequence into pEGFP-N1 (Clontech), pET-GST (kind gift from R. Marciniak, University of Texas, San Antonio, TX), and pZome-1C (Cellzome), respectively.
The N-terminal 81 amino acids of mouse SIRT7 were expressed in Escherichia coli as a GST fusion, purified on glutathione-Sepharose and injected into rabbits to produce SIRT7 antiserum. Full-length SIRT7 and SIRT71–81 proteins were bound to NHS-Sepharose according to the manufacturer instructions (Amersham). The crude serum was first passed over the SIRT71–81-NHS-Sepharose resin. After extensive washing, the antibodies were eluted with 0.1 M glycine (pH 2.5), and the eluate neutralized by the addition of one-tenth volume of 1 M Hepes (pH 7.9). The purification was repeated using full-length SIRT7-NHS-Sepharose as resin.
Other antibodies used were α-actin (C4, ICN), α-Flag (M2, Sigma), α-acetyl-histone H4 (Upstate Biotechnologies), anti-α-tubulin (Sigma), α-RPA116 (Seither et al. 1997), and α-human Pol I antiserum 52,799 (Percipalle et al. 2006).
Expression analysis
A hybridization probe containing the SIRT7 open reading frame was used to probe Multiple Tissue Northern Blots (Stratagene). The blots were stripped and reprobed with the actin probe included in the kit. SDS-solubilized proteins from the mouse FVB background were obtained as a gift from L. Bordone (Massachesetts Institute of Technology, Cambridge, MA), separated by SDS-PAGE, and analyzed by Western blotting.
Transfections
HEK293T, U2OS, and NIH3T3 cells were cultured in DMEM supplemented with 10% FCS. Where indicated, cells were treated with 0.05 μg/mL actinomycin D, 5 mM nicotinamide, or 40 nM TSA. For transient expression, HEK293T or U2OS cells were transfected using different amounts of pCMV-SIRT7 expression plasmids and 2 μg of the rDNA reporter pHr-P2-BH, which contains the human rDNA promoter fragment (from −401 to +378) fused to a BamHI–HinfI fragment harboring two transcription terminators (T1 and T2). Reporter transcripts and pre-rRNA were analyzed on Northern blots as described (Voit et al. 1999). To monitor 45S pre-rRNA, Northern blots were hybridized with a 32P-labeled antisense RNA encompassing 5′-terminal rDNA sequences from −57 to +183. For normalization, the blots were reprobed with a radiolabeled riboprobe against cytochrome c oxidase 1 (cox 1).
For stable expression of TAP-tagged SIRT7, U2OS cells were transfected with pZome–SIRT7 or pZome alone followed by selection with 2 μg/mL puromycin. For stable expression of Flag-tagged SIRT7, HEK293T cells were transfected with pCMV-TAG-4a, pCMV-Flag-SIRT7, pCMVFlag-SIRT7S112A, or pCMV-Flag-SIRT7H188Y in combination with pBABE– PURO at a 10:1 ratio and selected with 2 μg/mL puromycin.
Immunohistochemistry
Fixation and permeabilization of U2OS cells were performed as described (Zatsepina et al. 1993). For immunostaining cells were incubated with affinity-purified α-SIRT7 (1:200–1:400) and anti-Pol I (1:1200) for 1 h at RT, followed by incubation with α-rabbit-Cy3 antibodies and α-human-FITC antibodies (Dianova). The DNA was stained with Hoechst 33342. Localization of GFP-tagged proteins was visualized with a Nikon miscroscope in live cells that had been treated with Hoechst 33342. Detection of apoptotic cells at the single-cell level was based on the TUNEL assay using the In Situ Death Detection Kit (Roche). TUNEL-positive cells were visualized with avidin-Texas-red by fluorescent microscopy.
Protein purification and interaction assays
Purification of TAP-tagged complexes was performed essentially as described (Knuesel et al. 2003). Briefly, nuclear extracts were prepared from U2OS cells stably expressing TAP-SIRT7 and incubated with IgG-Sepharose (Amersham). TAP-tagged proteins were released in the presence of AcTEV (Invitrogen) and collected on calmodulin beads (Stratagene). After washing, TAP-SIRT7 and associated proteins were eluted with 25 mM Tris-Hcl (pH 8.0), 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 20 mM EGTA, 10 mM β-mercaptoethanol, and 0.02% NP-40, and analyzed by Western blotting.
For coimmunoprecipitation, HeLa cell nuclear extracts were chromatographed on DEAE-Sepharose columns. Two-hundred micrograms of the protein fraction eluting at 280 mM was incubated with 10 U DNase I (Roche) for 10 min at RT or left untreated. Following the addition of human IgGs (Dianove) or human α-Pol I for 4 h at 4°C and protein G-Agarose for another 2 h at 4°C, beads were extensively washed in buffer AM-200 (200 mM KCl, 20 mM Tris-HCl at pH 7.9, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM EDTA), and bound proteins were analyzed by Western blotting using α-SIRT7 and α-RPA116 antibodies.
GST and recombinant GST-tagged SIRT7 were expressed in E. coli Codon Plus-RP cells (Stratagene) and affinity-purified over glutathione-Sepharose (Lin et al. 2004). Acetylated core histones were purified from butyric acid treated HeLa cells by hydroxylapatite (Ausubel et al. 1997). To assay for interaction of SIRT7 with histones, GST and GST-SIRT7 were immobilized on glutathione-Sepharose and incubated with 2 μg of purified core histones for 2 h at 4°C in buffer AM-400 (400 mM KCl, 20 mM Tris-HCl at pH 7.9, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM EDTA) supplemented with 0.2% NP-40, protease inhibitors, and ethidium bromide (10 μg/mL). After five washes in incubation buffer, bound proteins were eluted with SDS loading buffer, separated by SDS-PAGE, and stained with Coomassie.
ChIP
ChIP experiments were performed essentially as described (Weinmann et al. 2001), except that for each ChIP one 15-cm plate of HEK293T cells was grown to 75% confluency and each wash was performed twice. Increasing amounts of precipitated chromatin was used as template in quantitative PCR reactions (O’Sullivan et al. 2002).
RNA interference (RNAi)
To knock-down cellular SIRT7 by RNAi, double-stranded RNAs (dsRNAs) were synthesized as a custom “Smart Pool” against SIRT7 and “nonspecific control duplexes-XIII” (Dharmacon). The duplexes were transfected into U2OS cells using RNAifect (Qiagen). After incubation for 48 h, cells were harvested and lysed with SDS loading buffer for Western blot analysis or with GITC for isolation of cellular RNA. Cellular RNA was reverse-transcribed with random primers. The amount of copied pre-rRNA was determined by semiquantitative PCR in the presence of α-32P-dCTP with primer 1 (forward) 5′-GCTGTCCTCTGGC-3′ and primer 2 (reverse) 5′-CGGCAGGCGGCTCAAG-3′ that amplify a fragment from +9 to +120 of the external transcribed spacer of the human rDNA repeat. Radiolabeled PCR fragments were analyzed by 8% PAGE and quantified using a PhosphorImager. GAPDH-cDNA was amplified by PCR using the primer pair 5′-CCATCACCATCTTCCAGGAG-3′ and 5′-CCTGCTTCACCACCTTCTTG-3′.
HEK293T cells for ChIP assays were transfected with the above siRNAs using Oligofectamine (Invitrogen). Forty-eight hours post-transfection the cells were transfected a second time and 48 h after the second transfection the cells were harvested.
For RNAi by expression of plasmid-encoded shRNAs, 2 × 105 U2OS cells were transfected with 2.5 μg of pSUPER expressing SIRT7-specific shRNAs under the control of the Pol III H1 promoter or a control pSUPER plasmid (Brummelkamp et al. 2002). Twenty-four hours after transfection cells were selected in the presence of 1.5 μg/mL puromycin for 4 d, and the cells were analyzed for SIRT7 levels and the presence of apoptotic cells.
Acknowledgments
We thank Nadine Wagner for excellent technical assistance, and Marcia Haigis and Gil Blander for thoughtful discussion and careful reading of the manuscript. L.G. and E.F. are supported by NIH grants. R.V. and I.G. were supported by Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1399706
References
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current protocols in molecular biology. Wiley; New York: 1997. [Google Scholar]
- Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Denu J.M. The Sir 2 family of protein deacetylases. Curr. Opin. Chem. Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Frye R.A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Frye R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Fulco M., Schiltz R.L., Iezzi S., King M.T., Zhao P., Kashiwaya Y., Hoffman E., Veech R.L., Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- Grummt I. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes & Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- Hekimi S., Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel M., Wan Y., Xiao Z., Holinger E., Lowe N., Wang W., Liu X. Identification of novel protein–protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell. Proteomics. 2003;2:1225–1233. doi: 10.1074/mcp.T300007-MCP200. [DOI] [PubMed] [Google Scholar]
- Lin S.J., Ford E., Haigis M., Liszt G., Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes & Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G., Ford E., Kurtev M., Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Michishita E., Park J.Y., Burneskis J.M., Barrett J.C., Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M., et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Muth V., Nadaud S., Grummt I., Voit R. Acetylation of TAFI68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B.J., Marshall B.L., Borra M.T., Denu J.M., Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- O’Sullivan A.C., Sullivan G.J., McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P., Fomproix N., Cavellán E., Voit R., Reimer G., Krüger T., Thyberg J., Scheer U., Grummt I., Östlund-Farrants A.-K. The chromain remodeling complex WSTF–SNH2g interacts with nuclear myosin 1 and serves a role in RNA polymerase I transcription. EMBO Rep. 2006 doi: 10.1038/sj.embor.7400657. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Seither P., Zatsepina O., Hoffmann M., Grummt I. Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma. 1997;106:216–225. doi: 10.1007/s004120050242. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A., Guarente L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Voit R., Hoffmann M., Grummt I. Phosphorylation by G1-specific cdk–cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Bartley S.M., Zhang T., Zhang M.Q., Farnham P.J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Grone H.J., Schutz G., Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Zatsepina O.V., Voit R., Grummt I., Spring H., Semenov M.V., Trendelenburg M.F. The RNA polymerase I-specific transcription initiation factor UBF is associated with transcriptionally active and inactive ribosomal genes. Chromosoma. 1993;102:599–611. doi: 10.1007/BF00352307. [DOI] [PubMed] [Google Scholar]
- Zhao J., Yuan X., Frodin M., Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]