Abstract
Many fungi undergo a developmental transition from a unicellular yeast form to an invasive filamentous form in response to environmental cues. Here we describe a quorum signaling pathway that links environmental sensing to morphogenesis in Saccharomyces cerevisiae. Saccharomyces cells secrete aromatic alcohols that stimulate morphogenesis by inducing the expression of FLO11 through a Tpk2p-dependent mechanism. Mutants defective in synthesis of these alcohols show reduced filamentous growth, which is partially suppressed by the addition of these aromatic alcohols. The production of these autosignaling alcohols is regulated by nitrogen: High ammonia restricts it by repressing the expression of their biosynthetic pathway, whereas nitrogen-poor conditions activate it. Moreover, the production of these aromatic alcohols is controlled by cell density and subjected to positive feedback regulation, which requires the transcription factor Aro80p. These interactions define a quorum-sensing circuit that allows Saccharomyces to respond to both cell density and the nutritional state of the environment. These same autoregulatory molecules do not evoke the morphological switch in Candida albicans, suggesting that these molecular signals are species-specific.
Keywords: Aromatic alcohols, morphogenesis, quorum sensing, fungi
In microbes, the switch from one morphological state to another is controlled by environmental signals that evoke a novel genetic program. The environmental signals can arise from alterations in the concentration of nutrients or autoinductive molecules that are secreted by cells. In bacteria, autoinductive molecules produced in a cell density-dependent fashion are termed quorum-sensing (Bassler 2002). In eukaryotes, diverse small signaling molecules have also been shown to be critical for many processes during cellular differentiation and development. For instance, upon starvation the free-living amoeba Dictyostelium discoideum secretes cAMP, which activates a signal transduction cascade to trigger individual cells to aggregate and subsequently undergo differentiation and morphogenesis to form a multicellular fruiting body (Aubry and Firtel 1999). Mammalian cells release and respond to retinoic acid (RA), a key signaling molecule that binds to its cognate nuclear receptor (RAR/RXR) and regulates the expression of genes essential for cellular proliferation, morphogenesis, and differentiation during embryogenesis and development (Mangelsdorf et al. 1995).
Fungi also undergo a developmental switch in response to environmental changes. Many fungi can grow either as a unicellular yeast form or as a filamentous form (Lengeler et al. 2000). In both nonpathogenic (Saccharomyces cerevisiae) and pathogenic (Candida albicans, Candida glabrata, and Cryptococcus neoformans) fungi, the ability to switch between the yeast and filamentous form is dependent on environmental conditions. All of these organisms switch from the yeast to the filamentous form when starved for nitrogen (Gimeno et al. 1992; Wickes et al. 1996; Csank and Haynes 2000; Biswas and Morschhauser 2005). In S. cerevisiae, nitrogen starvation induces pseudohyphal growth in diploid cells (Gimeno et al. 1992) and enhances invasive growth in haploid cells cultured on synthetic minimal medium (H. Chen and G. Fink, unpubl.). The low-nitrogen signal that evokes these morphogenetic transitions is transmitted by elements of the highly conserved mitogen-activated protein kinase (MAPK) and protein kinase A (PKA) pathways, resulting in the enhanced transcription of genes required for filamentation (Liu et al. 1993, 1994; Lengeler et al. 2000; Gancedo 2001; Liu 2001). However, the nature of the afferent signal generated by low nitrogen is unknown.
The morphological transition in many fungi is also controlled by autoregulatory molecules through quorum-sensing mechanisms (Kugler et al. 2000; Hornby et al. 2001, 2004; Chen et al. 2004). For instance, in C. albicans, the ability to switch between the yeast and the filamentous form depends on cell density. At high densities, Candida is in the yeast form, whereas at low densities, the yeast-form cell develops a filamentous protrusion known as a germ tube. The germ tube formation at high density is inhibited by farnesol, which accumulates in the medium during growth (Hornby et al. 2001). Another quorum-sensing molecule with a role in the regulation of growth and morphogenesis in C. albicans is tyrosol, which reduces the length of lag phase of Candida cells upon dilution and promotes filamentation under permissive conditions (Chen et al. 2004). These findings indicate that the morphogenesis in C. albicans is under complex positive and negative control by environmental conditions. However, the quorum-sensing phenomenon has not been explored in S. cerevisiae.
In this report, we show that S. cerevisiae cells release aromatic alcohols as quorum-sensing molecules capable of inducing the morphogenetic switch in response to nitrogen starvation. Nitrogen starvation results in the secretion of these alcohols, which stimulate morphogenesis by up-regulating the level of Flo11p, a GPI-anchored cell surface protein (Lambrechts et al. 1996; Rupp et al. 1999; Guo et al. 2000; Reynolds and Fink 2001). The autostimulatory effects of aromatic alcohols on morphogenesis require Tpk2p, a key component of the PKA signal transduction pathway. Moreover, Saccharomyces mutants defective in producing these alcohols show reduced filamentous growth, which can be suppressed by the addition of aromatic alcohols. The secretion of aromatic alcohols is tightly controlled by cell density and subjected to autostimulation, revealing a feedback autoregulatory circuit akin to the quorum-sensing phenomena in bacteria. This autosignaling pathway allows Saccharomyces to respond both to cell density and the nutritional state of the environment. The aromatic alcohols appear to be species-specific signaling molecules because C. albicans manifests different responses to these autoregulators.
Results
S. cerevisiae secretes aromatic alcohols with morphogenetic properties
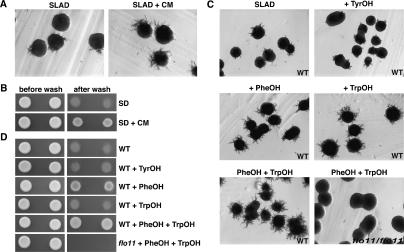
Conditioned medium (CM) from S. cerevisiae stationary-phase cultures strongly induced diploid pseudohyphal (Gimeno et al. 1992) and haploid invasive growth in S. cerevisiae (Fig. 1A,B; Roberts and Fink 1994). In agreement with its morphogenetic effects, CM stimulated the expression of FLO11, a gene essential for filamentous growth in S. cerevisiae, by approximately fivefold (data not shown). Fractionation of the CM by HPLC indicated two peaks that could stimulate filamentation and adhesion. By comparing their chemical properties—including HPLC elution profiles, thin-layer chromatography (TLC) retention profiles, and UV absorption spectra—to chemically synthesized standards, these peaks were identified as phenylethanol and tryptophol (Supplementary Fig. 1A), aromatic alcohols derived from phenylalanine and tryptophan (Sentheshanmuganathan and Elsden 1958; Dickinson et al. 2003). Using TLC and pure aromatic alcohols as standards, we found that Saccharomyces cells were capable of converting radioactive phenylalanine and tryptophan, as well as tyrosine, into the corresponding aromatic alcohols (Supplementary Fig. 1B). The identification of phenylethanol and tryptophol, together with previously identified tyrosol, a quorum-sensing molecule in C. albicans (Chen et al. 2004), reveals a unique class of autoregulatory signaling molecules in fungi.
Phenylethanol and tryptophol stimulate morphogenesisin S. cerevisiae
Phenylethanol and tryptophol, but not tyrosol, stimulated diploid pseudohyphal growth in S. cerevisiae (Fig. 1C). On solid low-ammonium dextrose (SLAD) agar plates (Gimeno et al. 1992), a relatively low concentration (≥20 μM) of either phenylethanol or tryptophol stimulated filamentation. The addition of both alcohols gave greater stimulation of pseudohyphal growth than either alcohol alone (Fig. 1C). However, aromatic alcohols failed to stimulate filamentation in diploid cells cultured on the synthetic minimal dextrose medium (SD) plates that contain abundant ammonium sulfate (37 mM), indicating that the presence of these autosignaling molecules is not sufficient to induce pseudohyphal growth in S. cerevisiae.
Phenylethanol increased haploid invasive growth of S. cerevisiae, whereas neither tryptophol nor tyrosol had any effect (Fig. 1D). However, the addition of phenylethanol and tryptophol together resulted in slightly more invasive growth, suggesting that there is some synergy between phenylethanol and tryptophol (Fig. 1D).
Autostimulation of filamentation requires FLO11
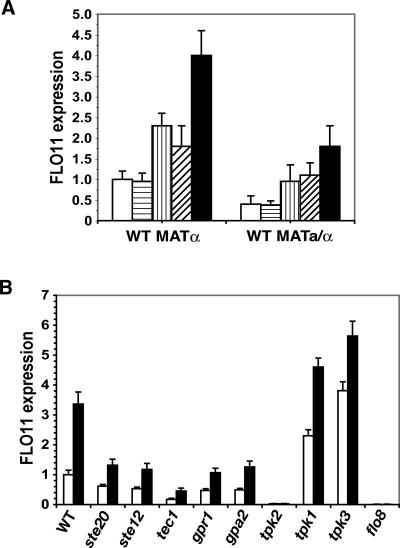
The induction of morphogenesis in S. cerevisiae by phenylethanol and tryptophol requires the function of Flo11p. Saccharomyces flo11 mutants were completely defective in diploid pseudohyphal and haploid invasive growth, and this defect was not suppressed by the addition of these autostimulatory alcohols (Fig. 1C,D). Moreover, phenylethanol and tryptophol each increased the level of FLO11 mRNA by approximately twofold (approximately fourfold when added together) as measured by either quantitative real-time RT–PCR (qRT–PCR) or β-galactosidase assays using a FLO11∷lacZ reporter (Fig. 2A; Rupp et al. 1999).
Figure 2.
Aromatic alcohols stimulate FLO11 expression through a Tpk2p-dependent mechanism. (A) Tryptophol and phenylethanol induce the expression of FLO11. Wild-type (WT) haploid and diploid cells were inoculated at OD600 0.2 in SD with or without the specified aromatic alcohol (500 μM each). The cultures were incubated for 4 h at 30°C. RNA was extracted and analyzed by qRT–PCR. The haploid strain without the externally added aromatic alcohols was used as the baseline for comparison (Flo11 level was normalized to 1). (White bars) SD alone; (bars with horizontal stripes) tyrosol; (bars with vertical stripes) phenylethanol; (bars with diagonal stripes) tryptophol; (black bars) phenylethanol and tryptophol. The β-galactosidase assays using a FLO11∷lacZ reporter show the same results. (B) The autoinduction of FLO11 expression requires TPK2 and FLO8, but not elements of the MAPK pathway. Wild-type (WT) and mutant cells were inoculated at OD600 0.2 in SD with (black bars) or without (white bars) aromatic alcohol (500 μM phenylethanol and tryptophol). The cultures were incubated for 4 h at 30°C. RNA was extracted and analyzed by qRT–PCR. The relative levels of FLO11 transcript in mutant cells with or without the externally added alcohols were compared with that of wild-type cells without alcohols, which was normalized to 1. The data shown here were obtained from haploid strains. The diploid strains show the same pattern in FLO11 expression in response to aromatic alcohols.
The autoinduction of FLO11 expression and morphogenesis by aromatic alcohols depends on Tpk2p, the catalytic subunit of PKA essential for filamentous growth in S. cerevisiae (Robertson and Fink 1998; Pan and Heitman 1999). A loss-of-function mutation in TPK2, but not TPK1 and TPK3, the filamentation-inhibitory subunits of PKA, abolished the stimulatory effects of aromatic alcohols on FLO11 expression (Fig. 2B). Moreover, the autoinduction required Flo8p, the transcription factor downstream of Tpk2p (Pan and Heitman 2002), but not Gpa2p and Gpr1p, the G-protein and G-protein-coupled receptor upstream of Tpk2p (Lorenz and Heitman 1997; Xue et al. 1998; Lorenz et al. 2000b), suggesting that Tpk2p, together with Flo8p, may play a role in sensing these signaling molecules (Fig. 2B). Consistent with the reduction in FLO11 mRNA, Saccharomyces strains missing either TPK2 or FLO8 were nonfilamentous in the presence of aromatic alcohols (data not shown). However, mutations in components of the MAPK pathway such as TEC1, STE12, and STE20 (Liu et al. 1993; Gavrias et al. 1996; Madhani and Fink 1997) had only small effects on FLO11 induction by aromatic alcohols, suggesting that these autosignaling alcohols exert their effects primarily through the PKA, but not the MAPK, pathway (Fig. 2B).
Mutants defective in aromatic alcohol biosynthesis are defective in morphogenesis
To determine the role of these aromatic alcohols in morphogenesis, we constructed Saccharomyces strains defective in the biosynthesis of aromatic alcohols. In view of the considerable redundancy of the enzymes that convert aromatic amino acids into corresponding alcohols (Fig. 3A), we made an aro8 aro9 double mutant, which lacks two transaminases required for aromatic amino acid catabolism (Iraqui et al. 1998; Dickinson et al. 2003). This double-mutant strain was defective in utilizing aromatic amino acids as indicated by reduced growth when aromatic amino acids were the sole nitrogen source (Fig. 3B). Moreover, the aro8 aro9 mutant secreted much lower levels of aromatic alcohols into the medium than wild-type cells (Fig. 3C).
Figure 3.
Saccharomyces mutants missing the ARO8 and ARO9 genes are defective in the production of aromatic alcohols and morphogenesis. (A) The biosynthetic pathways of aromatic alcohols. (B) The Saccharomyces aro8 aro9 double mutant is unable to utilize the nitrogen of aromatic amino acids and grows poorly on plates containing aromatic amino acids (400 μM each) as the sole nitrogen source. The growth defect of the aro8 aro9 strain is specific to aromatic amino acids. The mutant grows well on plates containing other amino acids (such as 400 μM Leu and Pro) as the sole nitrogen source. The aro8/aro8 aro9/aro9 diploid mutant shows the same growth defect as the aro8 aro9 haploid mutant. (C) The aro8 aro9 double mutant is defective in producing aromatic alcohols. Wild-type (WT) and aro8 aro9 mutant cells were incubated in YNB + 2% glucose in the presence of each 14C-aromatic amino acid. CM was prepared and subjected to TLC analysis. The aro8/aro8 aro9/aro9 diploid mutant shows the same defect in the production of aromatic alcohols as the haploid mutant. (D) The Saccharomyces aro8 aro9 mutant is defective in haploid invasive growth, and the defect can be suppressed by phenylethanol (PheOH). The strong adhesion by wild-type (WT) haploid cells in the absence of any alcohol presumably results from the presence of tyrosine and phenylalanine (100 μM each) in the plates, which may induce the production of endogenous aromatic alcohols. (E) The Saccharomyces aro8/aro8 aro9/aro9 diploid strain is defective in pseudohyphal growth, and the defect can be suppressed by tryptophol (TrpOH) and phenylethanol (PheOH). Wild-type (WT) and mutant diploid strains were grown on SLAD plates with or without the specified aromatic alcohols (100 μM each). (F) The aro8 aro9 strain has a lower level of FLO11 transcript than wild type, and the defect in FLO11 expression can be partially restored by the addition of tryptophol (TrpOH) and phenylethanol (PheOH). Wild-type (WT) and mutant cells were incubated at OD600 0.2 in SD with or without the specified aromatic alcohols (500 μM each) for 4 h. RNA was purified and analyzed by qRT–PCR. The wild-type strain without the addition of any aromatic alcohol was used as the baseline for comparison. Only the results from haploid strains are shown. The corresponding diploid strains have the same pattern of FLO11 expression. (White bars) SD alone; (bars with horizontal stripes) tyrosol; (bars with vertical stripes) phenylethanol; (bars with diagonal stripes) tryptophol; (black bars) phenylethanol and tryptophol.
The aro8 aro9 double mutant was defective in haploid invasive growth and diploid pseudohyphal formation as compared with isogenic wild-type strains (Fig. 3D,E). The adhesion defect of the haploid aro8 aro9 strain could be suppressed by the addition of phenylethanol, but not by tryptophol and tyrosol (Fig. 3D). The addition of either tryptophol or phenylethanol to the diploid aro8/aro8 aro9/aro9 double mutant suppressed its defect in diploid pseudohyphal filamentation, and the combination gave greater stimulation than either alcohol alone (Fig. 3E). Moreover, the addition of these two alcohols to the medium enhanced the filamentous growth of wild-type cells (Fig. 1C,D). These observations strongly suggest that the presence of aromatic alcohols is necessary for morphogenesis in S. cerevisiae.
Consistent with these morphogenetic effects, the aro8 aro9 mutant (both haploids and diploids) had a lower level of FLO11 transcript than wild type (Fig. 3F). The addition of phenylethanol and tryptophol induced FLO11 expression in aro8 aro9 cells (Fig. 3F), suggesting that phenylethanol and tryptophol are autostimulatory molecules that alter morphogenesis of Saccharomyces by elevating FLO11 expression. However, the addition of aromatic alcohols to aro8 aro9 mutant cells did not completely restore the level of FLO11 transcript to the level of wild-type cells with alcohols (Fig. 3F). The fact that the addition of the alcohols only partially suppresses the transcription and filamentation defect of the aro8 aro9 mutant suggests that these transaminases may be required for the synthesis of other inductive molecules.
Ammonium represses both filamentation and aromatic alcohol production
The fact that filamentation is suppressed by both high ammonia and defects in the biosynthesis of aromatic alcohols suggests that the biosynthetic pathway might be repressed by ammonia. Direct measurement by TLC analysis showed that growth in the presence of abundant ammonium sulfate (>500 μM) dramatically reduced the synthesis of aromatic alcohols (Fig. 4A). The alcohols were secreted at their highest levels when the ammonium sulfate concentration was ≤50 μM (Fig. 4A), which is also optimal for pseudohyphal growth (Gimeno et al. 1992). The production of tryptophol was under more strict nitrogen control than that of tyrosol and phenylethanol (Fig. 4A).
Figure 4.
The production of aromatic alcohols is controlled by nitrogen source. (A) The concentration of aromatic alcohols accumulated in the media increases with decreasing ammonium concentration. Wild-type (WT) haploid cells were incubated in YNB + 2% glucose containing specified concentrations of ammonium sulfate for 4 h at 30°C before TLC analysis. The concentration of ammonium sulfate used in each lane (from left to right) is 37 mM; 5 mM; 500 μM; 50 μM; and none, but with 5 mM L-pro. Wild-type diploid cells show the same pattern as haploid cells. (B) High ammonium condition represses the expression of ARO9, ARO10, and PDC6 genes. Wild-type haploid or diploid cells were incubated in YNB + 2% glucose with either 37 mM (SD, white bars) or 50 μM NH4+ (SLAD, black bars) at OD600 0.5 for 2 h before being collected for RNA preparation and qRT–PCR assay. The fold change in the specified gene expression was calculated as the ratio between SLAD and SD. The representative results from haploid cells are shown. The expression patterns of these genes in diploid cells are essentially the same.
The regulation of ARO9 and ARO10 genes by ammonium provides a missing connection between morphogenesis, aromatic alcohol production, and nitrogen availability. In the presence of abundant nitrogen, such as in SD, the expression of ARO9 and ARO10 was repressed by ∼20- to 30-fold, and PDC6 by ∼200-fold (Fig. 4B; Supplementary Fig. 2A,B). High concentrations of ammonium also repressed the expression of ARO8, a gene constitutively expressed at high levels in most conditions (Iraqui et al. 1999), by approximately twofold (data not shown). Independent expression analysis using whole-genome microarrays confirmed this observation and further showed that a large number of additional dehydrogenase genes were also induced by ammonium limitation (Supplementary Fig. 2C; the whole microarray data are available at http://jura.wi.mit.edu/fink_public), suggesting that nitrogen control of aromatic alcohol production is achieved by regulating the expression of these genes.
The regulation of aromatic alcohol production by ammonium is not directly mediated by known regulators of nitrogen metabolism or by carbon source. Neither single nor double mutations in GAT1, DAL80, and GZF3—genes encoding transcription factors involved in nitrogen regulation (Magasanik and Kaiser 2002)—affected the expression of ARO9, ARO10, and PDC6 in response to nitrogen starvation, whereas both a single mutation in GLN3 or double mutations in GLN3 and GAT1 increased the level of these transcripts (Supplementary Fig. 3), indicating that these factors are not required for the transcriptional stimulation of aromatic alcohol biosynthetic genes when nitrogen is limited. Moreover, low concentrations of glucose (such as 0.02% and 0.002%, cf. standard 2%) or alternative carbon sources such as glycerol and galactose had no effect on the expression of ARO9, ARO10, and PDC6 or the production of aromatic alcohols, suggesting that the biosynthetic pathway is primarily controlled by the availability of nitrogen, but not carbon source.
The biosynthesis of aromatic alcohols is controlled by cell density
The accumulation of aromatic alcohols in the medium was growth-phase dependent and reached its highest level (5–8 μM phenylethanol and tyrosol, and 1–2 μM tryptophol) after cultures entered stationary phase (Fig. 5A). This growth-phase-dependent production of aromatic alcohols was independent of ammonium regulation since the measurements were performed in an ammonium-free medium (L-proline as the sole nitrogen source). To test whether cell density controls the production of these signaling molecules, cells from a high-density culture (108 cells/mL) were diluted to either low (5 × 105 cells/mL) or high density (5 × 107 cells/mL) in fresh SLAD medium. The amount of aromatic alcohols secreted by cells was assessed by TLC. After normalizing for the difference in cell number, we found that CM taken from a high-density culture contained a much greater concentration of aromatic alcohols than CM from a low-density culture (Fig. 5B), indicating that cells at high density produce more aromatic alcohols per cell than do cells at low density. These results strongly suggest that aromatic alcohols are quorum-sensing molecules in S. cerevisiae.
Figure 5.
The production of aromatic alcohols is controlled by cell density. (A) The accumulation of aromatic alcohols in the medium increases with increasing cell density. Saccharomyces haploid cells were inoculated at 105 cells/mL in minimum medium (YNB) containing 5 mM L-Pro as nitrogen source. At indicated time points the cell density (◆) was measured by OD600 and the concentration of aromatic alcohols (●, phenylethanol; ▴, tyrosol; ◼, tryptophol) was determined by HPLC. The diploid cells have very similar growth and alcohol production curves to haploid cells. (B) The rate of aromatic alcohol production is controlled by cell density. Wild-type (WT) haploid cells were incubated in SLAD containing 50 μM specific aromatic amino acids either at low density (5 × 105 cells/mL) or high density (5 × 107 cells/mL) for 2 h. One milliliter of CM from the low-density culture and 10 μL of CM from the high-density culture were lyophilized and loaded on the TLC silica gel. Wild-type diploid cells show similar results. (C) High cell density up-regulates the level of ARO9 and ARO10 transcripts, and the induction requires ARO80. Wild-type (WT) and aro80 cells were incubated in liquid SLAD medium either at low (105 cells/mL, white bars) or high (5 × 107 cells/mL, black bars) cell densities for 60 min. Cells were collected by filtration and centrifugation. Total RNA was purified and subjected to qRT–PCR analysis. The transcript level in wild-type cells at low density was used as the baseline for comparison. The results shown here were obtained from haploid strains. The diploid strains show the same pattern. (D) Tryptophol, but not tyrosol and phenylethanol, induces the expression of ARO9 and ARO10 genes, and the autostimulation depends on ARO80. Wild-type (WT) and aro80 cells were cultured at OD600 0.2 in SD with or without the specified aromatic alcohol (500 μM) for 4 h. RNA was isolated and analyzed by qRT–PCR. The transcript level of the wild-type strain in SD was the baseline for calculating fold changes. Tryptophol has the same effects on the expression of ARO9 and ARO10 in both haploid and diploid strains. The results shown here were obtained from haploid strains. (White bars) SD alone; (light-gray bars) tyrosol; (dark-gray bars) phenylethanol; (black bars) tryptophol. (E) Tryptophol (TrpOH) stimulates the growth of a reporter strain PARO9-URA3 on uracil-minus (SD) plates, and the stimulation requires ARO80. Five microliters of 10-fold titrated Saccharomyces cells (L8199 for wild-type and L8220 for aro80 mutant) was spotted on specified plates for 2 d at 30°C. (F) Tryptophol up-regulates the transcript levels of ARO9 and ARO10 in cells at low density (only ARO9 is shown). Wild-type (WT) or aro80 cells were incubated in SLAD with (black bars) or without (white bars) 500 μM tryptophol at low (105 cells/mL) or high (5 × 107 cells/mL) cell densities for 60 min. RNA was isolated and analyzed by qRT–PCR. The transcript level of ARO9 in wild-type cells at low density without tryptophol was the baseline for comparison. Both haploid and diploid strains show the same pattern of gene expression. Only the results from haploid strains are shown. (G) The Saccharomyces aro80 mutant is defective in the biosynthesis of aromatic alcohols. Saccharomyces wild-type (WT) and aro80 cells were incubated in YNB + 2% glucose with either 37 mM (lanes 1,3) or 50 μM ammonium sulfate (lanes 2,4). CM was analyzed by TLC. The aro80/aro80 mutant shows the same defect in the production of aromatic alcohols as the aro80 haploid mutant. The results from haploid strains are shown.
High cell density stimulates the production of aromatic alcohols by up-regulating the expression of ARO9 and ARO10, two key genes required for the biosynthesis of aromatic alcohols. The level of ARO9 and ARO10 transcripts in cells at low density (105 cells/mL) was very low, even when these cells were cultured in low-ammonium SLAD medium (Fig. 5C). However, incubation at high density (5 × 107 cells/mL) in the same SLAD medium significantly stimulated their expression (∼20- to 50-fold, Fig. 5C). Moreover, the cell density-dependent regulation of ARO9 and ARO10 expression depends on Aro80p, a transcription factor required for the transcriptional induction of ARO9 and ARO10 by aromatic amino acids (Iraqui et al. 1999). The aro80 cells expressed comparable low levels of ARO9 and ARO10 transcripts whether cultured at high or low cell density (Fig. 5C), suggesting a key role for Aro80p in sensing cell density.
The biosynthesis of aromatic alcohols is autoregulated
Tryptophol stimulates the production of all three aromatic alcohols, whereas tyrosol and phenylethanol do not. This autoregulation is achieved by regulating the transcript levels of ARO9 and ARO10 genes. The addition of tryptophol to cells, but not tyrosol and phenylethanol, dramatically induced the expression of ARO9 and ARO10 (∼25- to 30-fold) (Fig. 5D). The autoinduction by tryptophol required the function of the transcription activator Aro80p, because tryptophol failed to induce the expression of ARO9 and ARO10 in an aro80 mutant (Fig. 5D). The presence of this positive feedback loop could be independently confirmed in a reporter strain in which the only copy of the URA3 gene is under the control of the native ARO9 promoter at the ARO9 locus (PARO9-URA3). This construct rendered the strain Ura− in the absence of tryptophol, and Ura+ in the presence of tryptophol (Fig. 5E). Moreover, this strain required a functional ARO80 gene for the expression of PARO9-URA3 because an aro80 mutant was Ura− even in the presence of tryptophol (Fig. 5E).
The positive feedback regulation by tryptophol could explain the cell density-dependent production of aromatic alcohols. The addition of tryptophol, but not tyrosol and phenylethanol, to Saccharomyces cells cultured in SLAD at low density (105 cells/mL) up-regulated the expression of ARO9 and ARO10 to a level comparable to that of cells at high density (5 × 107 cells/mL) (Fig. 5F). Tryptophol did not further stimulate the expression of ARO9 and ARO10 in cells cultured in SLAD at high density (Fig. 5F). These results suggest that the low levels of ARO9 and ARO10 transcripts in cells at low density is due to the absence of autoregulation by tryptophol. Moreover, both autoregulation and cell density-dependent regulation require the same transcription factor, Aro80p. Consistent with these observations, the Saccharomyces aro80 mutant made lower levels of aromatic alcohols, particularly tryptophol, than the wild-type strain (Fig. 5G). These findings reveal an Aro80p-dependent molecular circuitry that links cell density sensing and aromatic alcohol production.
Aromatic alcohols up-regulate the expression of stationary-phase genes
To identify Saccharomyces genes other than FLO11 that are regulated by aromatic alcohols, we performed whole-genome microarray expression analysis, comparing the transcription profiles of log-phase cells cultured either in the absence or presence of each individual aromatic alcohol or all three molecules together. Aromatic alcohols regulated the expression of a small set of genes: Approximately 150 genes were induced (twofold or greater) by these alcohols (Supplementary Fig. 4; the complete microarray data are available at http://jura.wi.mit.edu/fink_public). The genes whose transcript levels fluctuated in response to the treatment of aromatic alcohols encode proteins involved in a diverse range of functions, such as metabolic enzymes and stress regulators. Strikingly, ∼70% of the genes up-regulated by aromatic alcohols, particularly by tryptophol and phenylethanol, are also up-regulated upon the entry of cells into stationary phase (Gasch et al. 2000; Andalis et al. 2004). These genes are predominantly expressed in cells in stationary phase but not during exponential growth. The list contains a number of key signature genes of stationary phase, including HSP12, YGP1, CTT1, and SPI1, etc. The induction of stationary-phase-expressed genes by aromatic alcohols suggests that these quorum-sensing molecules may also play a role in signaling the proper entry of Saccharomyces cells into stationary phase by modulating the physiology of cells.
C. albicans filamentation is not stimulated by tryptophol or phenylethanol
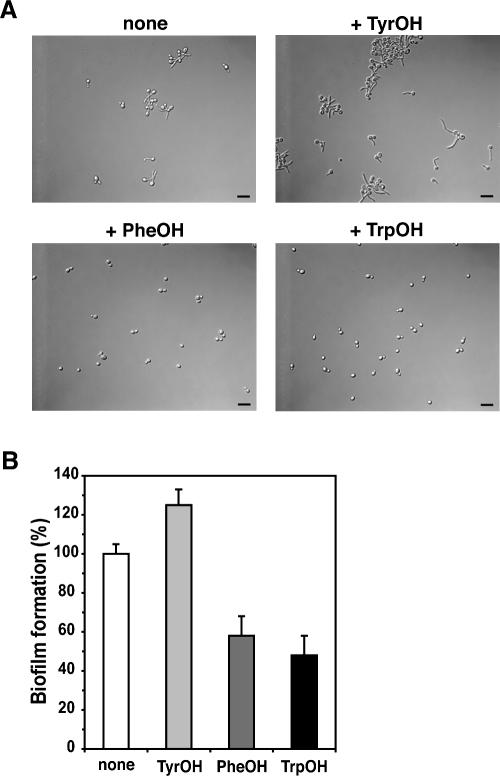
The fungal pathogen C. albicans secreted micromolar levels of aromatic alcohols into the environment during growth (Supplementary Fig. 1B). The only aromatic alcohol with an effect at relatively low concentrations (≥20 μM) is tyrosol, which accelerates the morphological conversion of Candida yeast-form cells to germ tubes (Fig. 6A; Chen et al. 2004). However, the production of all three alcohols is tightly controlled by ammonium and cell density (H. Chen and G. Fink, unpubl.), suggesting some as yet unknown roles for these alcohols in C. albicans.
Figure 6.
Aromatic alcohols evoke different morphological effects in C. albicans. (A) phenylethanol (PheOH) and tryptophol (TrpOH) at high concentrations (≥500 μM) inhibit the filamentous growth of Candida, whereas tyrosol (TyrOH) stimulates it. The final concentration of aromatic alcohols used in the assay was 1 mM. Bar, 25 μm. (B) Phenylethanol (PheOH) and tryptophol (TrpOH) at high concentrations (≥500 μM) inhibit biofilm formation in C. albicans, whereas tyrosol (TyrOH) promotes it. The final concentration of aromatic alcohols used in the assay was 1 mM. OD420 reading of Candida cells without aromatic alcohols was normalized to 100%. The data represent the averages of three independent measurements. The error bars represent standard deviations.
High concentrations of tryptophol and phenylethanol (≥500 μM) reduced germ tube formation in C. albicans (Fig. 6A). This inhibition of germ tube formation was not due to the inhibition of growth, because at concentrations of 500 μM to 5 mM, phenylethanol and tryptophol didn’t dramatically affect the growth of Candida cells. The aromatic alcohols also affected the ability of Candida to form a biofilm in a manner that parallels their effect on filamentation. Tyrosol slightly stimulated the formation of a biofilm, whereas tryptophol and phenylethanol inhibited it (Fig. 6B). As in the case of filamentation, so in biofilm formation the effects required relatively high concentrations of these compounds (≥500 μM).
Discussion
The results described here show that aromatic alcohols are key quorum-sensing molecules capable of stimulating the developmental transition from the unicellular yeast to filamentous form in S. cerevisiae. Although the morphological switch from yeast to filamentous form (pseudohyphal growth in diploids and invasive growth in haploids) was known to be induced by nitrogen starvation, the molecular events signaled by starvation were not understood. Our data provide evidence for a feedback control mechanism that connects environmental sensing with the secretion of aromatic alcohols to morphogenesis in S. cerevisiae (Fig. 7).
Figure 7.
A quorum signaling pathway linking environmental sensing to morphogenesis and entry into stationary phase in S. cerevisiae. Aromatic alcohols are autosignaling molecules capable of stimulating morphogenesis in Saccharomyces by elevating the level of Flo11p through a Tpk2p-dependent mechanism. The production of aromatic alcohols is tightly controlled by environmental conditions. High ammonium represses their production by inhibiting the transcription of key genes in the biosynthesis pathway (such as ARO9 and ARO10), whereas low ammonium activates it. Cell density also controls the production of aromatic alcohols. High density stimulates the production by activating the expression of ARO9 and ARO10, whereas low cell density inhibits it. Moreover, the production of aromatic alcohols is under positive feedback control by tryptophol. Both the cell density-dependent and feedback regulation require transcription factor Aro80p. Aromatic alcohols also regulate the expression of many stationary-phase genes, suggesting a potential role in modulating the entry of Saccharomyces cells into stationary phase.
Strong support for a connection between morphogenesis and the synthesis of aromatic alcohols is that a Saccharomyces strain defective in the first step of aromatic alcohol formation (aro8 aro9) is also defective in switching into filamentous growth. An important corollary is that the filamentation defect of the aro8 aro9 double mutant can be suppressed by addition of phenylethanol and tryptophol to the medium. Moreover, the aro8 aro9 mutant expresses low levels of FLO11, a cell surface protein that is required for and tightly associated with filamentous growth. The addition of aromatic alcohols to the medium induces the expression of FLO11, and this autoinduction requires a key subunit of PKA, Tpk2p, and its downstream effector Flo8p (Pan and Heitman 2002), but not elements of MAPK pathway. It is possible that Tpk2p plays an important role in sensing these autosignaling molecules by modulating the activity of Flo8p, a master regulator of FLO11 transcription.
The effects of these aromatic alcohols on haploid adhesion and diploid filamentation are specific; i.e., butanol and isoamyl alcohol have no effect on either of these morphogenetic changes in our strain background, although in some genetic backgrounds they stimulate filamentation in haploid strains (Dickinson 1996; Lorenz et al. 2000a). Moreover, only aromatic alcohols, but not butanol and isoamyl alcohol, suppress the filamention defect of aro8 aro9 mutant strains and induce the expression of FLO11 (Lorenz et al. 2000a).
A further clue to the circuitry is that the production of aromatic alcohols is dependent on nitrogen limitation. High ammonium concentrations repress the expression of transaminases as well as the decarboxylases and dehydrogenases required for the production of aromatic alcohols (Supplementary Fig. 2). The transcript levels of genes required for the catabolism of nonaromatic amino acids (other transaminases and decarboxylases) are either not affected or are repressed by ammonium starvation (Supplementary Fig. 2), suggesting that under ammonium-limiting conditions, aromatic alcohols are preferentially produced.
We propose that these aromatic alcohols are fungal autosignaling molecules akin to the quorum-sensing molecules of bacteria that also monitor environmental conditions and regulate diverse cellular behaviors. The hallmark of quorum sensing in bacteria is that the concentration of autoinducers released by cells increases with increasing cell density. We show that the production of these aromatic alcohols is also cell density-dependent: In high-density cultures, the amount of aromatic alcohols in the medium is much higher than that in low-density cultures. Cell density regulates the biosynthesis of aromatic alcohols by controlling the levels of ARO9 and ARO10 transcripts, which are significantly greater in cells at high density than in those at low density. Moreover, the production of aromatic alcohols is autostimulated by tryptophol: The addition of tryptophol activates the biosynthetic pathway by up-regulating the expression of ARO9 and ARO10. Both cell density-dependent regulation and autostimulation of ARO9 and ARO10 transcription and subsequent aromatic alcohol production require the key transcription factor Aro80p, which directly controls the expression of ARO9 and ARO10 genes (Iraqui et al. 1999; Lee et al. 2002). These observations reveal a cell density-dependent autoinduction loop for the production of quorum-sensing molecules, which allows an exponential increase in the biosynthesis of signaling molecules when it is activated (Fig. 7). Similar autoinduction pathways have also been documented in bacteria. For instance, in the Gram-negative bacterium Vibrio fischeri, quorum-sensing N-acylhomerserine lactone (AHL) induces its own production by binding to transcription factor LuxR, which activates the transcription of the luxICDABE operon (luxI encodes the enzyme for the biosynthesis of AHL) (Engebrecht et al. 1983; Miller and Bassler 2001).
The tight regulation of aromatic alcohol production by nitrogen source indicates an intimate relationship between quorum sensing and nutrient sensing. Quorum-sensing and nutrition-sensing pathways are also interconnected in bacteria (Lazazzera 2000). For example, in the Gram-positive bacterium Bacillus subtilis, the production of competence and sporulation factor (CSF), a quorum-sensing peptide, is regulated by starvation through a specific promoter recognition subunit of RNA polymerase, σH, which is activated by starvation and the entry into stationary phase (Healy et al. 1991; Lazazzera et al. 1999). In many Gram-negative bacteria, such as Pseudomonas aeruginosa and Ralstonia solanacearum, the production of AHL depends on σS, an RNA polymerase σ factor activated by starvation conditions (Flavier et al. 1998; You et al. 1998).
Aromatic alcohols induce the transcription of many stationary-phase genes when added to exponentially growing Saccharomyces cells. Similar patterns of regulation have also been reported in other microorganisms (Lazazzera 2000). In B. subtilis, CSF activates Spo0A, the key transcription factor involved in the expression of stationary-phase and early sporulation genes (Grossman 1995). In P. aeruginosa, AHL controls the expression of stationary-phase genes by regulating the expression of σS (You et al. 1998). Quorum sensing has also been shown to regulate the expression of stationary-phase genes in Escherichia coli (Baca-DeLancey et al. 1999). These findings suggest that microorganisms have evolved an effective way to integrate quorum sensing and nutrient sensing by using the same set of signaling molecules that regulate not only “specialized” processes such as morphogenesis, genetic competence, bioluminescence, and virulence, but also the general physiology of cells.
The finding that these aromatic alcohols exert different effects on morphogenesis in S. cerevisiae and C. albicans shows that these quorum-sensing molecules have distinct species-specific effects. In C. albicans, tyrosol promotes the formation of germ tubes and biofilms, whereas phenylethanol and tryptophol inhibit these developmental states (Fig. 6; Chen et al. 2004). In S. cerevisiae, tyrosol has no detectable effects, whereas phenylethanol and tryptophol are powerful inducers of morphogenesis. Although the production of these molecules is regulated similarly in both organisms by environmental conditions, the differential responses to these signals in nonpathogenic S. cerevisiae and pathogenic C. albicans must reflect the fundamental difference between two organisms in sensing and processing these signals. The differential effects of alcohols between these organisms suggest that fungi, like bacteria (Miller and Bassler 2001), have evolved molecular signals that evoke species-specific behaviors.
Materials and methods
Strains, media, reagents, and genetic manipulations
Yeast strains are listed in Table 1. All strains are derived from isogenic Σ1278b strain background. Standard yeast media, yeast transformations, and genetic manipulations were performed as previously described (Rose et al. 1990). Yeast cells were regularly grown on YPD. Chemically defined media were made from yeast nitrogen base without ammonium sulfate and amino acids (YNB) supplemented with 2% glucose and specified nitrogen source. Pure aromatic alcohols were from Sigma-Aldrich. L-[U-14C]-Tyr and Phe were from Amersham Biosciences. L-[3-14C]-Trp was from New England Nuclear. For gene deletion in the Σ1278b strain background, the KanMX4 cassette plus ∼500 base pairs (bp) of gene-specific DNA sequences upstream and downstream of the marker were amplified from the corresponding EUROSCARF deletion strain. The HphMX4 cassette was amplified from plasmid pAG32 using primers containing 60 nucleotides (nt) of gene-specific sequences and 20 nt of common sequences: —CGGATCCCCGGGTTAATTAA (forward) and —GAATTCGAGCTCGTTTAAAC (reverse) (Goldstein and McCusker 1999). PCR products were used to transform haploid strains to obtain G418- or hygromycin-resistant colonies, respectively. The diploid strains were made by mating L4996 (MATα ura3-52) or its derivatives with L5243 (MATa trp1∷hisG) or its derivatives. The PARO9-URA3-SPHIS5 strains were constructed in strain L7554 (MATα ura3-52 his3∷hisG leu2∷hisG) by directly replacing the ARO9 coding region with the URA3-SPHIS5 cassette of pUG27-URA3 (S. Chan and G. Fink, unpubl.). SPHIS5 encodes the Schizosaccharomyces pombe HIS5 gene, a functional homolog of the Saccharomyces HIS3 gene. All strains were confirmed by PCR.
Table 1.
S. cerevisiae strains used in this study

aAll strains are derived from isogenic ∑1278b background.
Saccharomyces filamentation assays
To induce diploid pseudohyphal growth, Saccharomyces strains were restreaked on an ammonium-limiting SLAD agar plate, which contains 4× YNB, 2% glucose, 50 μM ammonium sulfate, and 2% washed agar (Gimeno et al. 1992). Plates were incubated for 3 d at 30°C. Colony morphology was observed and photographed under a microscope. Haploid invasive growth assay was performed on SD plates (Rose et al. 1990). Five microliters of each 10-fold titrated culture (starting from 106 cells/mL) was spotted on plates and incubated for 3–4 d at 30°C. Pictures were taken before and after the wash. For assays involving aro8 aro9 mutants, 100 μM L-Tyr and Phe were supplemented to the media since aro8 aro9 mutants are auxotrophic to Tyr and Phe (Iraqui et al. 1998).
HPLC purification
Saccharomyces cells were inoculated at 105 cells/mL in SD, or otherwise specified medium, and incubated for 3 d at 30°C. CM was collected by removing cells by centrifugation and filtration, and was stored at −20°C. For HPLC analysis, CM was concentrated 100-fold by lyophilization and fractionated by a reverse-phase column (Supelco Discovery C18, 25 cm × 4.6 mm, particle size 5 μm). At a flow rate of 1 mL/min and gradient of 0.25% acetonitrile/min, tyrosol was eluted at ∼12% acetonitrile, phenylethanol at ∼32%, and tryptophol at ∼32.5% acetonitrile.
TLC
Wild-type or mutant yeast cells were grown in SD overnight at 30°C. Cells were washed extensively with dH2O and then subcultured at 5 × 107 cells/mL (unless otherwise noted) in the specified media with ∼2 μM 14C-Tyr, 14C-Phe, or 14C-Trp, respectively. For aro8 aro9 mutants, 100 μM L-Tyr and Phe were supplemented to the media. After incubating for 4 h at 30°C, cells were removed by centrifugation and, typically, 10 μL CM was loaded on a Silica Gel 60F (EMD Chemicals). The mobile solvent for TLC contains 85% chloroform, 14% methanol, and 1% dH2O. The 14C-labeled molecules were detected by autoradiography. 14C-aromatic alcohols were identified by comparing with pure aromatic alcohol standards run side by side on TLC.
qRT–PCR
Saccharomyces cells were cultured in SD overnight at 30°C, washed extensively with dH2O, and then reinoculated in the specified medium at 30°C before cells were collected. Total RNA was purified by a MasterPure Yeast RNA Purification Kit (Epicentre). RT was performed using TaqMan Reverse Transcription Reagents (Applied Biosystems) according to the manufacturer’s protocol. The RT mixture (total 50 μL) contained 1 μg total RNA (DNase I-treated) and random hexamer as the RT primer. The RT reaction was incubated sequentially for 10 min at room temperature, for 1 h at 37°C, and for 5 min at 95°C. Quantitative PCR reactions (total 25 μL PCR mixture per reaction) contained 2.5 μL RT product, 1× SYBR Green PCR Master Mix (Applied Biosystems), and 1 μM forward and reverse primers. Primers were designed by PrimerExpress software (Applied Biosystems). The DNA sequences of primers are Flo11, CACT TTTGAAGTTTATGCCACACAAG (F) and CTTGCATATT GAGCGGCACTAC (R); ARO9, CCCAGCCTTTTCCGATGA (F) and ACCCGCCAGAAAGGATGAC (R); ARO10, AAGA TGGGTCGGCACGTG (F) and TATCCGTCGGCCGCAT (R); PDC6, TTCATGGGCCTCACGCA (F) and GGCGAGGTGAT CCCAGGT (R); and internal control ACT1, CTCCACCACT GCTGAAAGAGAA (F) and CCAAGGCGACGTAACATAG TTTT (R). The quantitative PCR reactions were performed on a 7500 Real-Time PCR System (Applied Biosystems). The thermal profile was one initial cycle of 2 min at 50°C and 10 sec at 95°C, followed by 41 cycles of 15 sec at 95°C and 1 min at 60°C, and a last cycle of 15 sec at 95°C. The relative transcript level of each gene was formatted as a percentage of ACT1 from the same sample. The fold change for a specific gene was calculated as the ratio of relative levels (percentage of ACT1) between experimental and control samples. The averages of three measurements are presented. The error bars in the graphs represent standard deviations.
Whole-genome transcription profiling analysis
Cell cultures and total RNA were prepared the same way as in the qRT–PCR assays. The whole-genome microarray analysis was performed on an Affymetrix platform as previously described (Galitski et al. 1999; Andalis et al. 2004). For each sample, 15 μg total RNA was used to produce biotinylated cRNA. Fifteen micrograms of the biotinylated cRNA was then hybridized to a Yeast genome S98 array (Affymetrix). The detailed protocols for amplification, hybridization, washing, scanning, and data analysis are available at http://www.wi.mit.edu/CMT/protocols.html. Scan-to-scan variations in intensity were corrected by applying a scaling factor to each experiment so that the overall intensity of all experiments was 150. All scaling factors were between 2 and 5. To evaluate the relative change in transcript level under different experimental conditions, a ratio was calculated by dividing the signal intensity in each experiment by that of a control experiment. Cluster analysis was performed using Cluster and visualized by TreeView (Eisen et al. 1998). Raw data of microarray analysis are available at http://jura.wi.mit.edu/fink_public.
Candida filamentation assay
Candida strain SC5314 was grown in SD (pH 4.3) overnight to early stationary phase. Cells were washed with dH2O and reinoculated at 106 cells/mL in prewarmed SD (pH 7.0) with or without the specified compound (≥500 μM). The cultures were incubated for 2 h at 37°C with shaking. Cellular morphology was observed directly under a microscope.
Candida biofilm assay
Candida biofilm formation was measured as previously described (Kruppa et al. 2004). Briefly, Candida strain SC5314 was grown in YNB (pH 7.0) plus 1% glucose overnight at 30°C. Cells were washed twice with YNB + 1% glucose. Two-hundred microliters of cells (final OD600 0.1) in YNB + 1% glucose with or without the specified compound (≥500 μM) were inoculated into a tissue culture treated 96-well polystyrene plate (flat bottom). The plate was gently spun for a few seconds to allow cells to settle on the bottom of wells and was incubated for 24 h at 37°C with gentle shaking (100 rpm). The plate was then washed four times with PBS. One-hundred-fifty microliters XTT (2.5 mg/mL in PBS) plus 2 μM menadione was added to each well and incubated at room temperature until color developed. After quickly spinning the plate, 100 μL of supernatant from each well was transferred to a new 96-well plate and OD492 was measured by a plate reader.
Acknowledgments
We thank I. Rubin-Bejerano, C. Tseng, and R. Wheeler for their comments on the manuscript; S. Chan for providing plasmid pUG27-URA3; J. Love and G. Bell for their assistance on microarray expression analysis; and members of Fink Laboratory for useful discussions. This work was supported by National Institutes of Health Grant GM40266 (to G.R.F) and Ellison Medical Foundation Senior Scholars Award ID-SS-0038-01 (to G.R.F). G.R.F is an American Caner Society Professor of Genetics.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1411806
References
- Andalis A.A., Storchova Z., Styles C., Galitski T., Pellman D., Fink G.R. Defects arising from whole-genome duplications in Saccharomyces cerevisiae. Genetics. 2004;167:1109–1121. doi: 10.1534/genetics.104.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L., Firtel R. Integration of signaling networks that regulate Dictyostelium differentiation. Annu. Rev. Cell Dev. Biol. 1999;15:469–517. doi: 10.1146/annurev.cellbio.15.1.469. [DOI] [PubMed] [Google Scholar]
- Baca-DeLancey R.R., South M.M., Ding X., Rather P.N. Escherichia coli genes regulated by cell-to-cell signaling. Proc. Natl. Acad. Sci. 1999;96:4610–4614. doi: 10.1073/pnas.96.8.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B.L. Small talk. Cell-to-cell communication in bacteria. Cell. 2002;109:421–424. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- Biswas K., Morschhauser J. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 2005;56:649–669. doi: 10.1111/j.1365-2958.2005.04576.x. [DOI] [PubMed] [Google Scholar]
- Chen H., Fujita M., Feng Q., Clardy J., Fink G.R. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. 2004;101:5048–5052. doi: 10.1073/pnas.0401416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C., Haynes K. Candida glabrata displays pseudohyphal growth. FEMS Microbiol. Lett. 2000;189:115–120. doi: 10.1111/j.1574-6968.2000.tb09216.x. [DOI] [PubMed] [Google Scholar]
- Dickinson J.R. ‘Fusel’ alcohols induce hyphal-like extensions and pseudohyphal formation in yeasts. Microbiol. 1996;142:1391–1397. doi: 10.1099/13500872-142-6-1391. [DOI] [PubMed] [Google Scholar]
- Dickinson J.R., Salgado L.E., Hewlins M.J. The catabolism of amino acids to long chain and complex alcohols in. Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:8028–8034. doi: 10.1074/jbc.M211914200. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: Isolation and genetic analysis of functions from. Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Flavier A.B., Schell M.A., Denny T.P. An RpoS (σS) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol. Microbiol. 1998;28:475–486. doi: 10.1046/j.1365-2958.1998.00804.x. [DOI] [PubMed] [Google Scholar]
- Galitski T., Saldanha A.J., Styles C.A., Lander E.S., Fink G.R. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- Gancedo J.M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrias V., Andrianopoulos A., Gimeno C.J., Timberlake W.E. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- Gimeno C.J., Ljungdahl P.O., Styles C.A., Fink G.R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grossman A.D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in. Bacillus subtilis. Annu. Rev. Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- Guo B., Styles C.A., Feng Q., Fink G.R. A Saccharomyces gene family involved in invasive growth, cell–cell adhesion, and mating. Proc. Natl. Acad. Sci. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy J., Weir J., Smith I., Losick R. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor σH in. Bacillus subtilis. Mol. Microbiol. 1991;5:477–487. doi: 10.1111/j.1365-2958.1991.tb02131.x. [DOI] [PubMed] [Google Scholar]
- Hornby J.M., Jensen E.C., Lisec A.D., Tasto J.J., Jahnke B., Shoemaker R., Dussault P., Nickerson K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001;67:2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby J.M., Jacobitz-Kizzier S.M., McNeel D.J., Jensen E.C., Treves D.S., Nickerson K.W. Inoculum size effect in dimorphic fungi: Extracellular control of yeast-mycelium dimorphism in Ceratocystis ulmi. Appl. Environ. Microbiol. 2004;70:1356–1359. doi: 10.1128/AEM.70.3.1356-1359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui I., Vissers S., Cartiaux M., Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- Iraqui I., Vissers S., Andre B., Urrestarazu A. Transcriptional induction by aromatic amino acids in. Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:3360–3371. doi: 10.1128/mcb.19.5.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa M., Krom B.P., Chauhan N., Bambach A.V., Cihlar R.L., Calderone R.A. The two-component signal transduction protein Chk1p regulates quorum sensing in. Candida albicans. Eukaryot. Cell. 2004;3:1062–1065. doi: 10.1128/EC.3.4.1062-1065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler S., Schurtz Sebghati T., Groppe Eissenberg L., Goldman W.E. Phenotypic variation and intracellular parasitism by histoplasma Capsulatum. Proc. Natl. Acad. Sci. 2000;97:8794–8798. doi: 10.1073/pnas.97.16.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts M.G., Bauer F.F., Marmur J., Pretorius I.S. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera B.A. Quorum sensing and starvation: Signals for entry into stationary phase. Curr. Opin. Microbiol. 2000;3:177–182. doi: 10.1016/s1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- Lazazzera B.A., Kurtser I.G., McQuade R.S., Grossman A.D. An autoregulatory circuit affecting peptide signaling in. Bacillus subtilis. J. Bacteriol. 1999;181:5193–5200. doi: 10.1128/jb.181.17.5193-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in. Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lengeler K.B., Davidson R.C., D’Souza C., Harashima T., Shen W.C., Wang P., Pan X., Waugh M., Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 2001;4:728–735. doi: 10.1016/s1369-5274(01)00275-2. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C.A., Fink G.R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Liu H., Kohler J., Fink G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M.C., Cutler N.S., Heitman J. Characterization of alcohol-induced filamentous growth in. Saccharomyces cerevisiae. Mol. Biol. Cell. 2000a;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M.C., Pan X., Harashima T., Cardenas M.E., Xue Y., Hirsch J.P., Heitman J. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in. Saccharomyces cerevisiae. Genetics. 2000b;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H.D., Fink G.R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C.A. Nitrogen regulation in. Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Pan X., Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in. Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Heitman J. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 2002;22:3981–3993. doi: 10.1128/MCB.22.12.3981-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T.B., Fink G.R. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Fink G.R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes & Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Robertson L.S., Fink G.R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston F., Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor Laboratory Press; New York: 1990. [Google Scholar]
- Rupp S., Summers E., Lo H.J., Madhani H., Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentheshanmuganathan S., Elsden S.R. The mechanism of the formation of tyrosol by Saccharomyces cerevisiae. Biochem. J. 1958;69:210–218. doi: 10.1042/bj0690210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B.L., Mayorga M.E., Edman U., Edman J.C. Dimorphism and haploid fruiting in Cryptococcus neoformans: Association with the α-mating type. Proc. Natl. Acad. Sci. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Batlle M., Hirsch J.P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Fukushima J., Tanaka K., Kawamoto S., Okuda K. Induction of entry into the stationary growth phase in Pseudomonas aeruginosa by N-acylhomoserine lactone. FEMS Microbiol. Lett. 1998;164:99–106. doi: 10.1111/j.1574-6968.1998.tb13073.x. [DOI] [PubMed] [Google Scholar]