The plant hormone ethylene controls many aspects of growth and development, including organ abscission, leaf and flower senescence, and, in certain species, fruit ripening. It also has a critical role in mediating many stress responses. The importance of ethylene to many fundamental processes in plants has resulted in a great deal of research on how plants perceive and respond to this hormone. Simple genetic screens have facilitated isolation of many of the genes encoding mediators of ethylene responses, most notably the receptors (1, 2). Plants have small gene families of ethylene receptors; Arabidopsis thaliana, for example, has five. These receptors are homologous to bacterial two-component regulators and have protein kinase activities (3, 4). Genetic analyses of the receptor family are consistent with a model in which receptors actively suppress ethylene responses in the absence of the hormone. There is redundancy built into the system: single loss-of-function mutants have no visible phenotype, whereas higher-order loss-of-function mutants exhibit varying degrees of enhanced ethylene responses (5). In contrast, single amino acid changes in the ethylene-binding domain of receptors frequently result in dominant ethylene insensitivity. This insensitivity is believed to be the consequence of an inability to recognize ethylene (6). Mechanistically, little is known about how the receptors bind to ethylene and transduce that information to the downstream signaling components. The only protein that is known to physically interact with the receptors is the RAF-like protein kinase CTR1 (7).
In this issue of PNAS, two articles describe the identification of a new class of protein that genetically interacts with ethylene receptors to negatively regulate ethylene responses. Resnick et al. (8) started with a dominant ethylene insensitive Arabidopsis etr1 receptor mutant and screened for second-site suppressors that restore ethylene responsiveness. They identified a loss-of-function mutation in the REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1) gene that restores ethylene responses to close to wild-type levels, presumably because of inactivation of dominant etr1 signaling. In contrast, Barry and Giovannoni (9) characterized a tomato mutant in which fruits do not ripen. Tomato fruit ripening is absolutely dependent on ethylene responses, and this dominant mutant, Green ripe (Gr), had been previously identified as being specifically defective in fruit ethylene responses (10). When they cloned the gene associated with Gr, they discovered that the mutation was the consequence of a deletion in the 5′ flanking region of the gene. Molecular analysis revealed that the Gr deletion causes ectopic expression of GR, a phenomenon consistent with a dominant gain-of-function mutation. GR is a member of the same family as RTE1. Thus, the two groups, using two different genetic screens, identified a critical function for the same family of proteins in ethylene responses of two different plants.
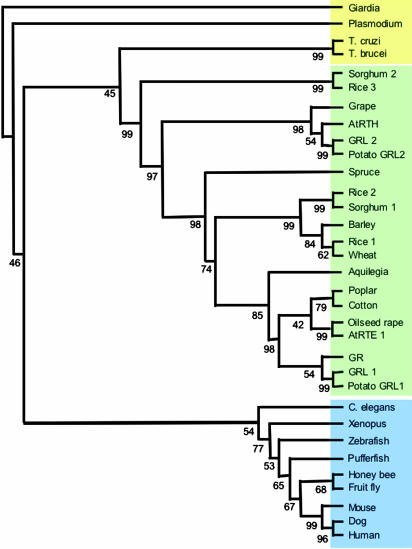
So what are RTE1 and GR? Both of these genes encode novel proteins that are highly conserved. RTE1 encodes a novel protein that is highly conserved throughout eukaryotes, with the exception of fungi, and absent in prokaryotes (Fig. 1). There are two homologous genes in Arabidopsis and three in tomato and rice. Computer modeling predicts that they should be integral membrane proteins, as are the ethylene receptors. The RTE1 sequence contains no known motifs, and no biological function has been assigned to this gene/protein in any species. However, the existence of RTE1 in a wide range of organisms suggests that it performs an essential conserved function in higher eukaryotes. It is curious that functions should be defined for this novel class of proteins in plants, where there is gene redundancy, as opposed to animals, where there is only a single gene in each species. It is also curious that the only described function of the proteins is to modify the action of a two-component regulator (the ethylene receptor), but the family is nonexistent in bacteria and fungi, where most two-component regulators exist.
Fig. 1.
Phylogenetic analysis of RTE1/GR-related proteins. A nonrooted phylogenetic tree was generated by using the phylip 3.5c suite of programs (http://evolution.genetics.washington.edu/phylip.html) using the Giardia protein as the out group. Bootstrap percentage supports are indicated. TIGR and SGN identifiers and GenBank accession numbers are as follows: tomato GR (DQ372895), tomato GRL1 (DQ372898), tomato GRL2 (DQ372899), potato GRL1 (DQ372900), potato GRL2 (SGN-U276841), A. thaliana RTE1 “AtRTE1” (NP_180177), Arabidopsis RTE1-HOMOLOG “AtRTH” (NP_190673), rice 1 (NP_916598), rice 2 (AAV59409), rice 3 (AAO37528), human (NP_115501), mouse (AAH37609), honey bee (XP_393764), fruit fly (NP_723362), Caenorhabditis elegans (AAF39886), dog (XP_852505), Xenopus (AAH87509), zebrafish (NP_001013334), pufferfish (CAG00266), Trypanosoma brucei (EAN77990), Giardia (XP_767284), Trypanosoma cruzi (XP_804751), Plasmodium (NP_703394), Aquilegia (TC10814), grape (TC40111), sorghum 1 (TC94016), sorghum 2 (TC97340), barley (TC132972), wheat (TC268045), oilseed rape (TC3567), poplar (TC31211), cotton (TC34712), and spruce (TC2575). Figure courtesy of Cornelius Barry (Boyce Thompson Institute for Plant Research, Ithaca, NY).
RTE1/GR are somewhat unusual with respect to their functions in ethylene signaling. In Arabidopsis, RTE1 appears to interact specifically with ETR1. The dominant etr1 allele used in the second-site suppressor screen exhibits reduced ethylene sensitivity. The rte1/etr1 mutant, in contrast, is more sensitive to ethylene than is wild type. The phenotype of the double mutant and single rte1 mutant is nearly identical to that of the etr1 loss-of-function mutant, suggesting that ETR1 function depends on a functional RTE1. Surprisingly, rte1 did not suppress the dominant ethylene insensitivity of either a much stronger etr1 allele or mutants in any of the other receptor genes tested. Thus, the data are consistent with a specific interaction between RTE1 and ETR1. Because ETR1 is the most prevalent of the ethylene receptors (11), the results could give ETR1 the appearance of being the main target of RTE1. This seeming specificity for ETR1 might also be the consequence of differential distribution of RTE1 and the receptors within or between cells.
The tomato results also indicate specificity in GR function. The Gr mutant was previously shown to be the consequence of a tissue-specific ethylene insensitivity (10). Whereas seedlings show a normal ethylene response, fruits do not respond to ethylene. Although tissue-specific ethylene responses are not unprecedented (2), they are unusual. What makes Gr unusual is that the ethylene-insensitive phenotype is the consequence of constitutive overexpression of the gene product. That is, the gene is aberrantly expressed in all tissues, but only a subset exhibit ethylene insensitivity. As further proof, a constitutively expressed CaMV35::GR transgene recreates the Gr mutant phenotype but does not lead to a global reduction in ethylene responsiveness. Together, these results suggest tissue-specific modulation of ethylene responses in tomato. Specific GR/receptor interactions, such as the Arabidopsis RTE1:ETR1 data suggest, could explain the tissue specificity of the tomato response. There are six ethylene receptor genes in tomato, and their expression varies greatly with different tissue types (12). One receptor, LeETR4, is highly expressed in flowers and fruits, and reduced expression of LeETR4 greatly increases fruit ethylene responses. Thus, GR could achieve the appearance of tissue-specific function if it interacts with specific receptors.
The presence of RTE1/GR in plant and animal genomes suggests an important conserved function. Because ethylene receptors are not found in animals, it seems likely that these proteins have functions beyond ethylene signal transduction. These proteins clearly have the capacity to alter ethylene signaling in vivo, but the actions of GR and RTE1 on the ethylene receptor(s) could be indirect. Because these proteins are integral to membranes, demonstration of direct physical interactions with receptors in vivo are challenging but clearly necessary. Regardless of mechanism, the RTE1/GR family of proteins likely interacts in some way with the receptor complex.
Conflict of interest statement: No conflicts declared.
References
- 1.Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 2.Roman G., Lubarsky B., Kieber J. J., Rothenberg M., Ecker J. R. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gamble R. L., Coonfield M. L, Schaller G. E. Proc. Natl. Acad. Sci. USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moussatche P., Klee H. J. J. Biol. Chem. 2004;279:48734–48741. doi: 10.1074/jbc.M403100200. [DOI] [PubMed] [Google Scholar]
- 5.Hua J., Meyerowitz E. M. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 6.Schaller G. E., Bleecker A. B. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- 7.Clark K. L., Larsen P. B., Wang X. X., Chang C. Proc. Natl. Acad. Sci. USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Resnick J. S., Wen C.-K., Shockey J. A., Chang C. Proc. Natl. Acad. Sci. USA. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry C. S., Giovannoni J. J. Proc. Natl. Acad. Sci. USA. 2006;103:7923–7928. doi: 10.1073/pnas.0602319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry C. S., McQuinn R. P., Thompson A. J., Seymour G. B., Grierson D., Giovannoni J. J. Plant Physiol. 2005;138:267–275. doi: 10.1104/pp.104.057745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Malley R. C., Rodriguez F. I., Esch J. J., Binder B. M., O'Donnell P., Klee H. J., Bleecker A. B. Plant J. 2005;41:651–659. doi: 10.1111/j.1365-313X.2004.02331.x. [DOI] [PubMed] [Google Scholar]
- 12.Tieman D., Taylor M., Ciardi J., Klee H. J. Proc. Natl. Acad. Sci. USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]