Abstract
NADH:ubiquinone oxidoreductase (complex I) is a major source of reactive oxygen species in mitochondria and a significant contributor to cellular oxidative stress. Here, we describe the kinetic and molecular mechanism of superoxide production by complex I isolated from bovine heart mitochondria and confirm that it produces predominantly superoxide, not hydrogen peroxide. Redox titrations and electron paramagnetic resonance spectroscopy exclude the iron-sulfur clusters and flavin radical as the source of superoxide, and, in the absence of a proton motive force, superoxide formation is not enhanced during turnover. Therefore, superoxide is formed by the transfer of one electron from fully reduced flavin to O2. The resulting flavin radical is unstable, so the remaining electron is probably redistributed to the iron-sulfur centers. The rate of superoxide production is determined by a bimolecular reaction between O2 and reduced flavin in an empty active site. The proportion of the flavin that is thus competent for reaction is set by a preequilibrium, determined by the dissociation constants of NADH and NAD+, and the reduction potentials of the flavin and NAD+. Consequently, the ratio and concentrations of NADH and NAD+ determine the rate of superoxide formation. This result clearly links our mechanism for the isolated enzyme to studies on intact mitochondria, in which superoxide production is enhanced when the NAD+ pool is reduced. Therefore, our mechanism forms a foundation for formulating causative connections between complex I defects and pathological effects.
Keywords: flavin, iron-sulfur cluster, semiquinone, oxidative stress
The production of reactive oxygen species, such as superoxide (O2•−), by mitochondria is a major cause of cellular oxidative stress. It contributes to many pathological conditions such as Parkinson’s and other neurodegenerative diseases, ischemia reperfusion injury, atherosclerosis, and aging (1–3). In mammalian mitochondria, most of the superoxide originates from NADH:ubiquinone oxidoreductase (complex I) and ubiquinol: cytochrome c oxidoreductase (complex III) of the electron transport chain, but there is increasing evidence that superoxide production by complex I, into the mitochondrial matrix, is predominant (4, 5). Indeed, complex I deficiencies have been identified across a wide spectrum of pathologies and linked to enhanced superoxide production as well as to deficiencies in energy production (6–10). Therefore, it is imperative to define how, why, and when superoxide is produced by complex I to formulate causative connections with pathological effects and rational proposals for how defects may be addressed.
Many previous studies have addressed the question of how superoxide is produced by complex I (11–18). Most of these studies examined intact mitochondria or submitochondrial particles, in which it is difficult to correlate observations directly to complex I, or to define and control the conditions precisely (NADH, NAD+, ubiquinone, and ubiquinol concentrations, redox status, proton motive force, and pH). In addition, complex I is a highly complicated, membrane-bound enzyme with (in the human and bovine enzymes) 46 different subunits (19) and 9 redox cofactors [a flavin mononucleotide (FMN) and 8 iron-sulfur (FeS) clusters (20, 21)]. Consequently, despite recognition of the importance of the relationship between complex I and superoxide, no consensus on the mechanism, or the site in the enzyme at which superoxide is produced, has been reached. The strategy adopted here is to study pure complex I, isolated from bovine heart mitochondria with catalytic activity equal to that of the membrane-bound enzyme (22). Therefore, our system is simple, and we exercise stringent control over all experimental conditions. However, we are unable to impose a proton motive force. Extensive control experiments were carried out to establish that the purified enzyme accurately reflects the membrane-bound enzyme, to define the relative amounts of superoxide and hydrogen peroxide (H2O2) formed, and to avoid artifacts in their quantification. Consequently, we propose a comprehensive kinetic and molecular mechanism for superoxide production by complex I, compare our conclusions with known mechanisms for O2 reduction by related enzymes, and discuss how they are relevant to understanding the behavior of complex I in intact mitochondria.
Results
NADH Oxidation Provides Electrons for the Reduction of Oxygen by Complex I.
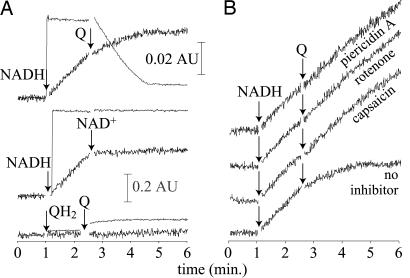
Fig. 1A displays the effects of the substrates and products of NADH:decylubiquinone oxidoreductase activity on superoxide production by complex I in aerobic buffer, detected by the reduction of acetylated cytochrome c (Cyt c). Addition of NADH, but not decylubiquinol, leads to superoxide production. The NADH-dependent activity is quickly stopped by excess NAD+. Although superoxide production is not affected directly by decylubiquinone, it is slowed, then stopped, upon its addition because catalytic turnover consumes NADH and generates NAD+. Importantly, superoxide production is not stimulated during turnover, indicating that, at least in the absence of a proton motive force, it is not mediated by a short-lived catalytic intermediate. Fig. 1B shows that addition of decylubiquinone in the presence of NADH and inhibitors that block ubiquinone reduction [rotenone, piericidin A, and capsaicin represent the different classes of complex I Q-site inhibitor (23)] does not affect superoxide production. The significance of these results for complex I in mitochondria is described in Discussion.
Fig. 1.
Superoxide production by isolated complex I in the presence and absence of catalytic substrates and products (A) and inhibitors (B). Superoxide production was monitored via Cyt c reduction [black traces, 0.02 absorption units (AU) scale marker, 550–541 nm]. NADH oxidation was monitored directly (gray traces, 0.2 AU scale marker, 340–541 nm). Complex I and phospholipids (and inhibitors when added) were present at the start of each measurement. For the inhibitor assays (B), the results were identical if the order of addition was Q − inhibitor − NADH, showing that prior occupancy of the binding site does not affect the results. Conditions were as follows: pH 7.5, 32°C, 30 μM NADH, 300 μM NAD+, 50 μM Cyt c, 100 μM Q (decylubiquinone), 100 μM QH2 (decylubiquinol), 0.4 mg ml−1 bovine heart polar phospholipids, 0.5 μM piericidin A, 2.5 μM rotenone, and 300 μM capsaicin.
Stoichiometric Detection of Superoxide Produced by Reduced Complex I and O2.
Three independent measurements established that, in the absence of additional electron acceptors, the electrons from NADH oxidation are conserved predominantly in superoxide formation (Table 1). The decrease in NADH concentration was compared with the rates of O2•− and H2O2 generation, by using the Cyt c and Amplex Red assays (Fig. 2). The specificity of the two detection systems was confirmed by using superoxide dismutase (SOD) and catalase (CAT), and the stoichiometry of 1 NADH:2 O2•− or 1 NADH:1 H2O2 suggests that all of the electrons from NADH oxidation are used to form O2•−. However, in competition assays (reaction of superoxide with Cyt c vs. dismutation; Fig. 3), a small amount of H2O2 (≈1.8 nmol min−1 mg−1; <10%) is not susceptible to Cyt c. Therefore, it is produced either directly or by dismutation in the active site before the superoxide becomes accessible to Cyt c.
Table 1.
The rates of NADH oxidation, superoxide formation, and hydrogen peroxide formation by complex I (nmol min−1 mg−1)
| Measurement | NADH oxidation | Cyt c reduction | Resorufin formation |
|---|---|---|---|
| NADH only | 20.9 ± 0.8 | 42.1 ± 2.8 | 21.1 ± 2.7 |
| NADH + SOD | 22.7 ± 2.7 | 3.9 ± 0.8 | 21.1 ± 2.9 |
| NADH + CAT | 20.5 ± 1.5 | 39.9 ± 2.8 | 1.9 ± 0.8 |
| % of e− detected (row 1) | 100.0 ± 3.6 | 100.6 ± 3.6 | 100.5 ± 9.3 |
| Background (− complex I) | 0.035 ± 0.014 | 0.011 ± 0.016 | 0.003 ± 0.002 |
The electrons from NADH oxidation are detected quantitatively, and >90% of them are conserved in superoxide formation. Rates were calculated using linear regression over 2 min, and background rates (nmol min−1 ml−1) were subtracted. Conditions were as follows: pH 7.5 and 32°C.
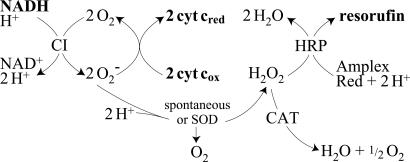
Fig. 2.
The generation, interconversion, and detection of superoxide and hydrogen peroxide in complex I assays. CI, complex I; HRP, horseradish peroxidase; CAT, catalase; SOD, superoxide dismutase.
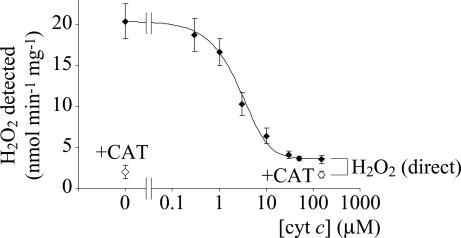
Fig. 3.
Competition assay showing that most of the electrons are conserved in superoxide formation, but that a small fraction of them form hydrogen peroxide directly. In the absence of Cyt c, all of the electrons form H2O2. In 150 μM Cyt c superoxide detection is stoichiometric (it outcompetes dismutation). However, ≈10% of the H2O2 is not susceptible to Cyt c. Amplex Red oxidation was measured at 557–620 nm (557 nm is an isosbestic point for Cyt c reduction). The curve was calculated by using constant O2•− production and simple bimolecular reactions and is illustrative only. CAT, catalase. Conditions were as follows: pH 7.5 and 32°C.
Comparison of the rates of O2 (≈40 nmol e− min−1 mg−1) and decylubiquinone reduction (typically 4.2 μmol e− min−1 mg−1) showed that, under our experimental conditions, ≈1% of the electrons are diverted from decylubiquinone to O2. However, the observed percentage is strongly dependent on both the NADH concentration and the O2 concentration, which is much higher (≈250 μM) than under physiological conditions (24).
Qualitatively, NADH oxidation and H2O2 generation by isolated complex I and mitochondrial membranes responded identically to catalytic substrates and inhibitors, provided that stigmatellin was included in the membrane assays to inhibit ubiquinol:cytochrome c oxidoreductase (complex III). Superoxide could not be detected directly because of direct reduction of Cyt c by the respiratory enzymes. Table 2 compares the rates of NADH oxidation and H2O2 generation under a range of conditions. In all cases, the specific activity of the membranes was ≈17% that of isolated complex I, in agreement with reported values for the ratio of complex I to the other respiratory complexes in bovine heart mitochondrial membranes (20 ± 4% by mass) (25). Therefore, Table 2 confirms that the catalytic properties of the isolated enzyme are representative of the membrane-bound enzyme and indicates that it is a valid experimental system for investigation of the mechanism of superoxide production.
Table 2.
Comparison of isolated and membrane-bound complex I
| NADH oxidation (nmol min−1 mg−1) |
|||
|---|---|---|---|
| Complex I | Membranes | Ratio (%) | |
| NADH + Q | 1796 ± 193 | 308 ± 31 | 17.1 ± 2.5 |
| NADH + Q + NAD+ | 1666 ± 133 | 286 ± 32 | 17.2 ± 2.4 |
| NADH + Q + piericidin A | 28 ± 29 | 3 ± 2 | (10.7 ± 13.2) |
| Amplex Red (nmol min−1 mg−1) |
|||
|---|---|---|---|
| Complex I | Membranes | Ratio (%) | |
| NADH | 25.7 ± 1.9 | 4.1 ± 0.1 | 15.9 ± 1.3 |
| NADH + Q + piericidin A | 24.6 ± 1.7 | 4.3 ± 0.4 | 17.4 ± 1.3 |
| NADH + Q + NAD+ | 3.6 ± 0.3 | 0.7 ± 0.0 | 17.9 ± 1.6 |
Stigmatellin (0.1 μM) was used to inhibit complex III in membrane assays and piericidin A (0.5 μM) to inhibit complex I. Conditions were as follows: 30 μM NADH, 200 μM Q (decylubiquinone), 1 mM NAD+, 32°C, and pH 7.5.
The Kinetic Mechanism of Superoxide Production.
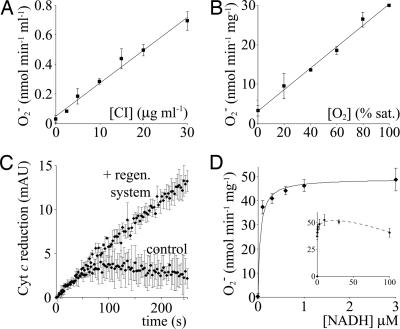
NADH oxidation transfers two electrons to complex I. They may then reduce ubiquinone, O2, or an artificial oxidant such as [Fe(CN)6]3− or [Ru(NH3)6]3+ (26). In air-saturated solution [O2 ≈250 μM (24)], complex I generates ≈40 nmol O2•− min−1 mg−1 (Table 1), so reduction of O2 is much slower than reduction of 1 mM [Fe(CN)6]3− (180,000 nmol min−1 mg−1) or [Ru(NH3)6]3+ (46,000 nmol min−1 mg−1). Therefore, the reaction between complex I and NADH, common to all of the assays, is >2,000 times faster than the reaction of reduced complex I with O2. Fig. 4A and B show that the rate of O2•− production is directly proportional to [complex I] and [O2], but only very low NADH concentrations are required for the rate to become independent of [NADH] (Fig. 4D). At >1 μM NADH, the rate-limiting step is the bimolecular reaction between reduced complex I and O2.
Fig. 4.
The rate of superoxide production by isolated complex I as a function of complex I, O2, and NADH concentration. The rate depends linearly on complex I (A) and O2 concentrations (B) (% saturated in air), 23°C. (C) Superoxide production in the presence of 300 nM NADH (average and standard deviations of three experiments). In the presence of the NADH regenerating system, [NADH] is maintained, and the assay is linear; in its absence, NADH is rapidly oxidized, and the rate drops to zero (control). (D) Dependence of O2•− production on [NADH], maintained by the regenerating system: KM(app) = 0.05 μM, Vmax = 49.1 nmol O2•− min−1 mg−1 (Inset, data recorded at high [NADH]). General conditions were as follows: pH 7.5 and 32°C.
Superoxide generation by complex I is strongly inhibited by NAD+ (Fig. 1A), so the data in Fig. 4 A, B, and D were recorded by using a regenerating system to maintain [NADH] and minimize [NAD+]. Otherwise, NADH is converted to NAD+ at ≈0.2 μM min−1, leading to significant curvature in the assay traces, particularly at low [NADH] (Fig. 4C). For O2•− production KM(app) for NADH is 0.05 μM. Importantly, NAD+ does not exert its inhibitory effect by causing NADH oxidation by complex I to become rate limiting. For example, 30 μM NAD+, which decreases O2•− formation by ≈50%, does not affect the much faster [Fe(CN)6]3−, [Ru(NH3)6]3+, or decylubiquinone reductions. Our observations suggest strongly that a preequilibrium is established between NADH, NAD+, and different states of complex I (oxidized, reduced, nucleotide free, or nucleotide bound), to determine how much of the complex is able to reduce O2 and to thus determine the rate of O2•− formation.
The Molecular Mechanism of Superoxide Formation.
Superoxide formation by purified complex I is not affected by decylubiquinone directly or amplified during catalysis (Fig. 1). Therefore, the FMN or one of the FeS clusters is the cofactor that controls O2 reduction, cofactor X. If cofactor X is an FeS cluster, then NAD+ acts, with NADH, to set its equilibrium level of reduction, according to the Nernst equation. If cofactor X is the flavin, then active site occupancy exerts an additional effect on the preequilibrium and is likely to affect the rate of the bimolecular reaction also, as bound nucleotide may preclude or hinder O2 access.
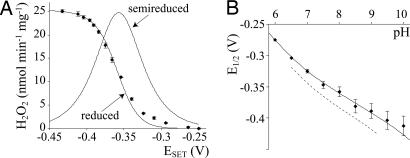
Fig. 5A shows the effect of varying the [NADH]/[NAD+] ratio on O2•− production, plotted as a redox titration of cofactor X (Eq. 1).
Fig. 5.
The rate and potential of superoxide production by isolated complex I. (A) The rate of H2O2 generation measured by using Amplex Red plotted against the NADH:NAD+ potential (♦). NADH was repurified anaerobically, and the lowest potential points were checked by using the NADH-regenerating system. The two curves are from Eq. 2 and the potentials from EPR, shifted by + 18 mV. The sigmoidal curve describes the fully reduced flavin, and the peak-shaped curve refers to the flavin radical. (B) pH dependence of E1/2 measured by using Cyt c (♦) and calculated from the pH dependence of the individual flavin potentials (pKS = 8.1, pKR = 6.8, solid line; dashed line from ref. 30). Assay conditions were as follows: pH 7.5, 32°C, 50 μM Cyt c, 10 μM Amplex Red, and 2 units ml−1 HRP.
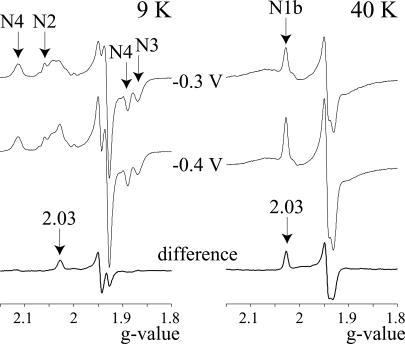
Fig. 5A shows that X is mainly reduced at ESET < −0.4 V, and mainly oxidized at ESET > −0.3 V. Fig. 6 shows electron paramagnetic resonance (EPR) spectra of complex I prepared at these two potentials, normalized to the signal from the high potential cluster N2 (27), to determine whether any of the FeS clusters change their oxidation state over the same potential range as cofactor X. The reduced clusters N2, N3, N4, N5, and N1b are evident at both potentials (20). The signal intensities of (N2), N3, N4, and N5 do not change, whereas the signal from N1b (g = 2.03 at 40 K) is larger at −0.4 V (N1b is ≈60% reduced at −0.3 V). Therefore, none of these clusters is cofactor X. Cluster N1a, the [2Fe-2S] cluster adjacent to the FMN (21), is not cofactor X either, because there is no signal from reduced N1a in either sample (see Table 3, which is published as supporting information on the PNAS web site), consistent with its low potential (28). Two further clusters are present in bovine complex I, in subunit TYKY (19–21), but they have not been observed by EPR, perhaps because their potentials are outside the accessible range, they have higher spin states, or because of spin-spin interactions. It is possible that the additional signal in the 9 K difference spectrum results partially from one of these clusters, because N1b is not expected to be observed at low temperature. In any case, Fig. 6 does not exclude the two TYKY clusters from being cofactor X. However, they are excluded by the flavoprotein (Fp) subcomplex of complex I, which contains only the 51 and 24 kDa subunits (the FMN and clusters N1a and N3) and not subunit TYKY (29) and produces H2O2 at approximately the same rate as intact complex I (211 ± 33 nmol min−1 mg−1 = 16.1 ± 2.6 min−1 in 30 μM NADH; O2•− production cannot be measured because Fp reduces Cyt c directly). We conclude that, in isolated complex I, superoxide is produced by the flavin, because it is not produced by any of the FeS clusters.
Fig. 6.
EPR spectra from complex I at high and low potential. The signals from clusters N2, N3, and N4 are present at 9 K in equal intensities at both potentials. Cluster N5 is observed in equal intensity but only at lower temperature (data not shown). Only the signal from cluster N1b is increased at −0.4 V, and there is no evidence for the signal from cluster N1a [gz = 2.00 (28)]. The difference signal at 9 K has not been assigned. Conditions were as follows: Complex I (pH 7.5 (0°C), 10 mg ml−1) was reduced with 1 mM NADH (≈−0.4 V) or with 1 mM NADH, 10 mM NAD+ (≈−0.3 V). The spectra were normalized by using the signal from cluster N2 (27). Microwave frequency, 9.39 GHz; microwave power, 1 mW; modulation amplitude, 5 G; time constant, 81.92 ms; conversion time, 20.48 ms.
Fig. 5A shows that E1/2, the midpoint potential of cofactor X, is −0.359 ± 0.005 V (n = 22), close to the reported two-electron potential of the flavin in complex I (−0.38 V at pH 7.5) (30). Previously, Kushnareva et al. (16) measured H2O2 generation as a function of the NAD+/NADH ratio in intact mitochondria and reported E1/2 = −0.39 V (pH 7.4), close to our value. Here, redox titrations measuring both O2•− and H2O2 production, and using a range of NADH concentrations (30–200 μM), all showed the same sigmoidal curve. Therefore, the O2•−:H2O2 ratio does not vary with potential, indicating that the mechanism of O2•− production is conserved over all potentials. In contrast, the flavin in fumarate reductase switches from mainly O2•− to mainly H2O2 as the potential is decreased, because of reduction of the proximal [2Fe−2S] cluster (31). Finally, although E1/2 measurements varied very little, the “n-value” of the titration curve, defining the number of electrons transferred, was difficult to define unambiguously, and so is not considered further.
Superoxide may be produced by either the fully reduced flavin, FMNH−/FMNH2, or by the semireduced flavin, FMN−/FMNH•; the two species are distinguished by their potential dependence (see Eq. 2, where EO/S is the reduction potential for converting oxidized and semireduced flavin and ES/R is the reduction potential for converting semireduced and reduced flavin) (32).
The fully reduced state is formed progressively as the potential decreases, but the semireduced state is present only at intermediate potentials, with a maximum at EAV (average of EO/S and ES/R). Fig. 5A shows that the fully reduced flavin is the source of superoxide by comparing our data with the calculated potential dependencies of the two states. The calculated lines use EO/S and ES/R as determined by EPR (30), with only a slight adjustment (+18 mV) to both potentials.
Fig. 5B compares experimental and calculated values for E1/2 over a range of pH. Superoxide production could be measured quantitatively by using 50 μM Cyt c between pHs 6 and 10, because the rates of Cyt c reduction and NADH oxidation were equivalent, and so E1/2 could be determined accurately. Below pH 6, dismutation outcompetes Cyt c reduction (24) (and the extinction coefficient of Amplex Red becomes too small), and spontaneous NADH degradation is significant; above pH 10, catalysis slows significantly, challenging our preequilibrium model. In Fig. 5B, calculated E1/2 values are the potential at which 50% of the flavin is fully reduced (Eq. 2). Values for EO/S and ES/R were calculated from their pH dependencies (32): The semireduced flavin may exist as FMN− or FMNH• (pKS), whereas the fully reduced flavin may exist as FMNH− or FMNH2 (pKR). Only minor changes to the published pKa values and potentials were required to optimize the fit: pKS = 8.1, pKR = 6.8, at pH 7 EO/S = −0.363 V, and ES/R = −0.286 V [values from EPR: 7.7, 7.1, −0.383, and −0.304, respectively (30)]. Therefore, the pH dependence of E1/2 is further confirmation for the participation of the fully reduced flavin in superoxide production. Finally, the pH dependence of the rate of superoxide generation (NADH only) defines the pH dependence of the rate-limiting step (assuming that active site occupancy does not vary significantly). The rate increases from ≈40 nmol min−1 mg−1 at pH 6–7 to ≈90 nmol min−1 mg−1 at pH 10, and there is no evidence (no pKa value) for the participation of an active site protonatable residue in superoxide formation.
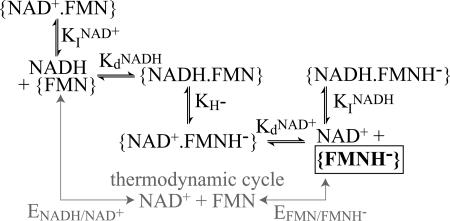
The data presented above are strong evidence for superoxide production by the fully reduced flavin, but simple interconversion of the flavin’s oxidation states cannot be considered separately from nucleotide binding. Our data do not define whether O2 can access the reduced flavin when nucleotide is bound, but all relevant structural models in the RCSB Protein Data Bank show a common configuration, with the nicotinamide moiety obscuring the isoalloxazine ring Re face. The structure of the hydrophilic domain of complex I from Thermus thermophilus does not contain bound nucleotide but suggests a similar configuration (21). Therefore, it is likely that only reduced flavin in an empty active site reacts with O2. Fig. 7, and Eq. 3 derived from it (32), show how NAD+ and NADH determine the equilibrium concentration of the free reduced flavin, via the flavin and NAD+ reduction potentials [ΔE = (EFMN − ENAD+)] and four binding constants, one for each oxidation state of nucleotide and flavin.
Fig. 7.
The preequilibrium of species that determines the rate of formation of superoxide by the reduced flavin. KdNADH and KdNAD+ are dissociation constants, and KINADH and KINAD+ are inhibition constants. The catalytically productive states interconvert by hydride transfer (KH−). Species enclosed by braces are enzyme bound, and the boxed species is that which produces O2•−. For simplicity, the flavin radical and the FeS clusters are not considered.
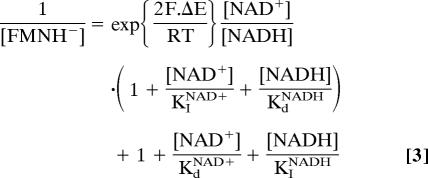 |
ENAD+ is well established [−0.335 V, pH 7.5 (32)]. EFMN has been measured by EPR (−0.375, pH 7.5) (30), but the values have not been revalidated. We found two reports for the four binding constants, both measured by varying the concentration of an artificial electron acceptor (NADH oxidation, assuming the flavin is reduced in steady state) or by reverse electron transport (NAD+ reduction, assuming the flavin is oxidized in steady state). Although the KI values are similar [KINADH = 80 μM (33) or 40 μM (34); KINAD+ = 1600 μM (33) or 2000 μM (34)], the Kd values are very different [KdNADH = 0.17 μM (33) or 40 μM (34); KdNAD+ ≈ 25 μM (33) or 0.9 μM (34)], and so calculated values of KH− are also very different (3.2 × 10−4 (33) or 2.1 (34), from the thermodynamic cycle in Fig. 7). Although predicted values of E1/2 for superoxide production (by using Eq. 3) are −0.44 V (33) and −0.38 V (34), close to the observed value of −0.36 V, the strong disagreement between the two sets of values prevents any meaningful interpretation. Consequently, accurately defining the binding constants and flavin potentials is an important, but challenging, future objective that is required for a quantitative understanding of the mechanism of superoxide production by isolated complex I.
Discussion
No consensus has yet been reached on the site of superoxide production in mitochondrial complex I. Proposals have included the flavin (11–13, 35), bound reduced nucleotide (14), FeS clusters N2 (15) and N1a (16), and a semiquinone radical (17, 18). Here, we demonstrate that fully reduced flavin, on the matrix side of the inner membrane, is a significant source of superoxide in isolated complex I. In comparison with other flavoenzymes that produce superoxide adventitiously, complex I (≈40 O2•− min−1) is unremarkable. For example, glutathione reductase, succinate dehydrogenase, and fumarate reductase produce 0.8, 13, and 1,600 O2•− min−1, respectively (36).
Fully reduced flavin is a low potential electron donor capable of O2 reduction, it is in a hydrophilic domain (O2•− is anionic), and, in the substrate binding site, it is accessible to O2 [whereas the FeS clusters are buried beneath the solvent accessible surface (21)]. However, formation of O2•− from O2 [E0′ = −0.33 V, 1 atm (1 atm = 101.3 kPa) O2 (24)] is less favorable thermodynamically than formation of H2O2 [E0′ = +0.28 V, pH 7 (24)], questioning why a fully reduced flavin should produce O2•−. Typically, fully reduced flavins in dehydrogenases produce a mixture of O2•− and H2O2 (37), consistent with the small amount of H2O2 observed from complex I. Reduction of O2 to H2O2 by reduced flavin is a sequential process (37); in complex I, further reduction of the nascent O2•− does not compete effectively with its escape from the active site. In contrast, enzymes such as glucose oxidase produce only H2O2 (37). They stabilize O2•− electrostatically (38) and may discriminate even against N2 (39), whereas O2 and O2•− probably bind only weakly and nonspecifically in complex I. In addition, one of the steps preceding the second reduction may be slow [intersystem crossing (40) or protonation of O2•−] or sterically hindered [formation of the flavin hydroperoxide adduct (37)]. The properties of the flavin radical are also crucial: The electron may be retained and donated to a second O2 [for example, flavodoxin has a stable flavin radical and produces only O2•− (37)] or redistributed to another center. Electron redistribution is thought to control the O2•−/H2O2 ratio in xanthine oxidase (41, 42) and fumarate reductase, in which the proximal [2Fe−2S] cluster oxidizes the flavin radical (31). In complex I, the flavin radical is thermodynamically unstable (30), supporting redistribution, but it is not possible to identify a single FeS cluster to oxidize (or re-reduce) it. It is tempting to propose a specific role for cluster N1a, because it is close to the flavin yet has no obvious role in energy transduction (21). However, N1a is unlikely to be important during catalysis, when electrons are flowing from flavin to ubiquinone, and its potential is too low [−0.45 V (28)] for it to compete effectively with the flavin [−0.42 V (30)] for the extra electron. Finally, our results show clearly that the rate-limiting step in superoxide production by isolated complex I is a bimolecular reaction between “competent” enzyme and O2. The first electron transfer is rate limiting in H2O2 production by glucose oxidase (38), suggesting that it may also determine the bimolecular rate constant in complex I. Consequently, the rate increases with pH because the flavin potential decreases, increasing the thermodynamic driving force for electron transfer [“normal” Marcus behavior (43)].
Finally, how does the mechanism of superoxide production by isolated complex I relate to the enzyme under physiological conditions? Three observations have proved crucial in developing an understanding of superoxide production from complex I in intact mitochondria (13, 16, 17, 35, 44, 45). (i) Under “normal” conditions (respiration supported by malate/pyruvate), superoxide production is very low (<0.05 nmol min−1 mg−1). (ii) On addition of a complex I inhibitor (typically rotenone), the rate increases significantly (0.2–0.5 nmol min−1 mg−1). (iii) During reverse electron transport supported by succinate, the rate is high, although a wide range of values [0.3 (16)–2.7 (17) nmol min−1 mg−1) have been reported. Our mechanism for superoxide production by the reduced flavin in complex I, defining how it depends on ENAD+, is qualitatively consistent with all three observations: Under normal conditions ENAD+ is high ([NAD+] > [NADH]); when complex I is inhibited, NADH builds up and ENAD+ decreases; during reverse catalysis, complex I reduces NAD+ to NADH, and ENAD+ is again low. Furthermore, studies which explicitly considered the influence of the NAD+/NADH ratio on superoxide production in intact mitochondria found a close correlation, in agreement with our observations (13, 16, 35, 46). Importantly, however, our experiments are unable to address the effects of the proton motive force, which may exert an indirect influence on ENAD+, but which may also exert distinct and separate effects. In the presence of a proton motive force, reactive intermediates, formed only transiently in our experiments, may build up to significant levels. Such intermediates, most likely formed during quinone reduction, have been proposed to provide an additional source of electrons for superoxide production by complex I (17). Our mechanism for superoxide production by the fully reduced flavin provides a basis for the design and interpretation of future experiments to address the role of proton motive force and possible further sites of production, and so complete our understanding of superoxide production by mitochondrial complex I.
Materials and Methods
Enzyme Preparations.
Mitochondrial membranes were prepared from bovine hearts, and then complex I was purified by using asolectin and quantified by its FMN content (the properties of this preparation have been described in detail in ref. 22). The flavoprotein subcomplex was resolved by using sodium perchlorate (29) and purified chromatographically. Protein concentrations were determined by using the Pierce bicinchoninic acid (BCA) assay.
Kinetic Measurements.
Kinetic measurements were carried out at 32°C in 1-ml cuvettes [diode array spectrometer (Ocean Optics, Dunedin, FL) or the cuvette port of a microtiter plate reader (Molecular Devices)] or in 96-well plates (200 μl). The assay buffer comprised 20 mM Tris·Cl (pH 7.5) and 30 μM NADH (Sigma); at different pHs a mixed buffer system of 10 mM sodium acetate, Mes, Hepes, and N-[Tris(hydroxymethyl)methyl]-3-amino propanesulfonic acid (TAPS) was used. Complex I was added to 10 μg ml−1 from a 5 mg ml−1 stock in 20 mM Tris·Cl (pH 7.4 at 4°C), 150 mM NaCl, 10% glycerol, 1 mM EDTA, and 0.02% DDM (Anatrace, Maumee, OH). When required, bovine heart polar phospholipids (Avanti Polar Lipids) were added to 0.4 mg ml−1 from a 10 mg ml−1 stock solution (prepared anaerobically) in 20 mM Tris·Cl (pH 7.5), 2% wt/vol CHAPS {3-[(3-cholamidoproyl) dimethylammonio]-1-propane-sulfonate, Sigma}, and the enzyme and phospholipids were preincubated for 2 min in the assay buffer (22). Catalytic rates are reported in nmol min−1 mg−1 (1 nmol min−1 mg−1 ≈ 1 min−1, as the MR for complex I is 0.98 MDa).
NADH oxidation (ε = 6.22 mM−1 cm−1) was followed at 340–420 nm or at 340–541 nm in the presence of Cyt c. To measure the NADH:decylubiquinone oxidoreductase activity, decylubiquinone (Sigma) was added to 200 μM (10 mM stock solution in ethanol), and εNADH = 6.81 mM−1 cm−1 was used to account for the decylubiquinone absorbance. The NADH regenerating system (see Fig. 8, which is published as supporting information on the PNAS web site) comprised 2 mM fructose bisphosphate, 1 mM sodium arsenate, and 1 unit ml−1 of aldolase, triose isomerase, and glyceraldehyde 3-phosphate dehydrogenase (BDH). Piericidin A, rotenone, capsaicin, and stigmatellin were from Sigma.
Superoxide formation was quantified stoichiometrically by the reduction of 50 μM oxidized Cyt c (47, 48) (partially acetylated equine heart Cyt c; ε550–541 = 18.00 ± 0.6 mM−1 cm−1; Sigma). When required, superoxide dismutase (SOD) (Cu-Zn-SOD from bovine erythrocytes; Sigma) was added to 10 units ml−1. H2O2 was quantified by using the horseradish peroxidase (2 units ml−1; MP Biomedicals, Aurora, OH) dependent oxidation of Amplex Red (10 μM; Invitrogen) to resorufin (ε557–620 = 51.6 ± 2.5 mM−1 cm−1 at pH 7.5; Sigma) (16, 48). Note that [NADH] >30 μM interferes with the Amplex Red assay. When required, catalase (bovine liver; Sigma) was added to 1000 units ml−1.
Redox Titrations Using NADH and NAD+.
NADH was repurified in a glovebox (O2 < 2 ppm) by anion exchange chromatography (5-ml HiTrap Q-Sepharose column; Amersham Pharmacia Biosciences) (49) to remove contaminating NAD+. After experimentation, the integrity of the NADH stock solution was reevaluated (0.08 ± 0.04% NAD+ formed in 6 h). Typically, redox potentials were set by using 30 μM NADH and a varying amount of NAD+ (Sigma), and the low potential limit was checked by using the NADH regenerating system.
EPR.
Complex I (10 mg ml−1) was reduced anaerobically by 1 mM purified NADH or by dialysis against purified NADH (≈−0.4 V) or to ≈−0.3 V by using 1 mM NADH and 10 mM NAD+, and frozen immediately. Spectra were recorded on a Bruker EMX X-band spectrometer by using an ER 4119HS high-sensitivity cavity and a ESR900 continuous-flow liquid helium cryostat (Oxford Instruments, Oxford, U.K.).
Supplementary Material
Acknowledgments
We thank Chérise D. Barker (Medical Research Council, Cambridge, U.K.) for preparing the flavoprotein subcomplex. L.K. thanks Boehringer-Ingelheim for a one-year fellowship. This work was supported by the Medical Research Council.
Abbreviations
- complex I
NADH:ubiquinone oxidoreductase
- Cyt c
cytochrome c
- EPR
electron paramagnetic resonance
- EO/S
reduction potential for converting oxidized and semireduced flavin
- ES/R
reduction potential for converting semireduced and reduced flavin
- EAV
average of EO/S and ES/R
- FMN
flavin mononucleotide
- pKS
pKa of semireduced flavin
- pKR
pKa of reduced flavin
- Q
oxidized ubiquinone
- QH2
reduced ubiquinone.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Li Y., Huang T.-T., Carlson E. J., Melov S., Ursell P. C., Olson J. L., Noble L. J., Yoshimura M. P., Berger C., Chan P. H., et al. Nat. Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 2.Raha S., Robinson B. H. Trends Biochem. Sci. 2000;25:502–508. doi: 10.1016/s0968-0004(00)01674-1. [DOI] [PubMed] [Google Scholar]
- 3.Balaban R. S., Nemoto S., Finkel T. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Brand M. D., Affourtit C., Esteves T. C., Green K., Lambert A. J., Miwa S., Pakay J. L., Parker N. Free Radical Biol. Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 5.Turrens J. F. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smeitink J., van den Heuvel L. Am. J. Hum. Genet. 1999;64:1505–1510. doi: 10.1086/302432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitkänen S., Robinson B. H. J. Clin. Invest. 1996;98:345–351. doi: 10.1172/JCI118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orth M., Schapira A. H. V. Am. J. Med. Genet. 2001;106:27–36. doi: 10.1002/ajmg.1425. [DOI] [PubMed] [Google Scholar]
- 9.Betarbet R., Sherer T. B., MacKenzie G., Garcia-Osuna M., Panov A. V., Greenamyre J. T. Nat. Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 10.Wallace D. C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 11.Vinogradov A. D., Grivennikova V. G. Biochemistry (Moscow) 2005;70:120–127. doi: 10.1007/s10541-005-0090-7. [DOI] [PubMed] [Google Scholar]
- 12.Galkin A., Brandt U. J. Biol. Chem. 2005;280:30129–30135. doi: 10.1074/jbc.M504709200. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y., Fiskum G., Schubert D. J. Neurochem. 2002;80:780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- 14.Krishnamoorthy G., Hinkle P. C. J. Biol. Chem. 1988;263:17566–17575. [PubMed] [Google Scholar]
- 15.Genova M. L., Ventura B., Giuliano G., Bovina C., Formiggini G., Castelli G. P., Lenaz G. FEBS Lett. 2001;505:364–368. doi: 10.1016/s0014-5793(01)02850-2. [DOI] [PubMed] [Google Scholar]
- 16.Kushnareva Y., Murphy A. N., Andreyev A. Biochem. J. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert A. J., Brand M. D. J. Biol. Chem. 2004;279:39414–39420. doi: 10.1074/jbc.M406576200. [DOI] [PubMed] [Google Scholar]
- 18.Ohnishi S. T., Ohnishi T., Muranaka S., Fujita H., Kimura H., Uemura K., Yoshida K.-I., Utsumi K. J. Bioenerg. Biomembr. 2005;37:1–15. doi: 10.1007/s10863-005-4117-y. [DOI] [PubMed] [Google Scholar]
- 19.Hirst J., Carroll J., Fearnley I. M., Shannon R. J., Walker J. E. Biochim. Biophys. Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi T. Biochim. Biophys. Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 21.Sazanov L. A., Hinchliffe P. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 22.Sharpley M. S., Shannon R. J., Draghi F., Hirst J. Biochemistry. 2006;45:241–248. doi: 10.1021/bi051809x. [DOI] [PubMed] [Google Scholar]
- 23.Degli Esposti M. Biochim. Biophys. Acta. 1998;1364:222–235. doi: 10.1016/s0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 24.Halliwell B., Gutteridge J. M. C. Free Radicals in Biology and Medicine. Oxford: Oxford Univ. Press; 1998. [Google Scholar]
- 25.Schägger H., Pfeiffer K. J. Biol. Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 26.Vinogradov A. D. Biochim. Biophys. Acta. 1998;1364:169–185. doi: 10.1016/s0005-2728(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 27.Ingledew W. J., Ohnishi T. Biochem. J. 1980;186:111–117. doi: 10.1042/bj1860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zu Y., di Bernardo S., Yagi T., Hirst J. Biochemistry. 2002;41:10056–10069. doi: 10.1021/bi026026f. [DOI] [PubMed] [Google Scholar]
- 29.Galante Y. M., Hatefi Y. Arch. Biochem. Biophys. 1979;192:559–568. doi: 10.1016/0003-9861(79)90126-7. [DOI] [PubMed] [Google Scholar]
- 30.Sled V. D., Rudnitzky N. I., Hatefi Y., Ohnishi T. Biochemistry. 1994;33:10069–10075. doi: 10.1021/bi00199a034. [DOI] [PubMed] [Google Scholar]
- 31.Messner K. R., Imlay J. A. J. Biol. Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- 32.Clark W. M. Oxidation-Reduction Potentials of Organic Systems. Baltimore: Williams & Wilkins; 1960. [Google Scholar]
- 33.Avraam R., Kotlyar A. B. Biochemistry. 1991;56:1181–1189. [Google Scholar]
- 34.Vinogradov A. D. J. Bioenerg. Biomembr. 1993;25:367–375. doi: 10.1007/BF00762462. [DOI] [PubMed] [Google Scholar]
- 35.Kudin A. P., Bimpong-Buta N. Y.-B., Vielhaber S., Elger C. E., Kunz W. S. J. Biol. Chem. 2004;279:4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 36.Imlay J. A. J. Biol. Chem. 1995;270:19767–19777. [PubMed] [Google Scholar]
- 37.Massey V. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 38.Roth J. P., Klinman J. P. Proc. Natl. Acad. Sci. USA. 2003;100:62–67. doi: 10.1073/pnas.252644599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goto Y., Klinman J. P. Biochemistry. 2002;41:13637–13643. doi: 10.1021/bi0204591. [DOI] [PubMed] [Google Scholar]
- 40.Prabhakar R., Siegbahn P. E. M., Minaev B. F., Ågren H. J. Phys. Chem. B. 2002;106:3742–3750. [Google Scholar]
- 41.Hille R., Massey V. J. Biol. Chem. 1981;256:9090–9095. [PubMed] [Google Scholar]
- 42.Porras A. G., Olson J. S., Palmer G. J. Biol. Chem. 1981;256:9096–9103. [Google Scholar]
- 43.Marcus R. A. Angew. Chem. Int. Ed. Engl. 1993;32:1111–1121. [Google Scholar]
- 44.Votyakova T. V., Reynolds I. J. J. Neurochem. 2001;79:266–277. doi: 10.1046/j.1471-4159.2001.00548.x. [DOI] [PubMed] [Google Scholar]
- 45.Han D., Canali R., Rettori D., Kaplowitz N. Mol. Pharmacol. 2003;64:1136–1144. doi: 10.1124/mol.64.5.1136. [DOI] [PubMed] [Google Scholar]
- 46.Starkov A. A., Fiskum G. J. Neurochem. 2003;86:1101–1107. doi: 10.1046/j.1471-4159.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- 47.Azzi A., Montecucco C., Richter C. Biochem. Biophys. Res. Commun. 1975;65:597–603. doi: 10.1016/s0006-291x(75)80188-4. [DOI] [PubMed] [Google Scholar]
- 48.Tarpey M. M., Fridovich I. Circ. Res. 2001;89:224–236. doi: 10.1161/hh1501.094365. [DOI] [PubMed] [Google Scholar]
- 49.Orr G. A., Blanchard J. S. Anal. Biochem. 1984;142:232–234. doi: 10.1016/0003-2697(84)90544-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.