Abstract
Protein allostery provides mechanisms for regulation of biological function at the molecular level. We present here an investigation of global, ligand-induced allosteric transition in a protein by time-resolved x-ray diffraction. The study provides a view of structural changes in single crystals of Scapharca dimeric hemoglobin as they proceed in real time, from 5 ns to 80 μs after ligand photodissociation. A tertiary intermediate structure forms rapidly (<5 ns) as the protein responds to the presence of an unliganded heme within each R-state protein subunit, with key structural changes observed in the heme groups, neighboring residues, and interface water molecules. This intermediate lays a foundation for the concerted tertiary and quaternary structural changes that occur on a microsecond time scale and are associated with the transition to a low-affinity T-state structure. Reversal of these changes shows a considerable lag as a T-like structure persists well after ligand rebinding, suggesting a slow T-to-R transition.
Keywords: allosteric protein transitions, intersubunit communication, kinetics
A fundamental goal in biology is to understand how organisms respond to environmental signals. Central to this process are allosteric proteins that undergo structural transitions between alternate states in response to stimuli such as ligand binding. Although static structures of alternate states are available for a number of allosteric proteins, information about the kinetic pathway between such functionally important states is limited. Time-resolved crystallographic analysis (1–9) provides the unique opportunity to obtain direct, time-dependent structural information at high resolution on the entire protein molecule as it undergoes structural change.
Much of our understanding of allosteric protein function has come from investigations into mammalian hemoglobins. Evidence of substantial structural changes accompanying ligand binding dates back to the 1930s with the observation that unliganded, high-salt, hemoglobin crystals exposed to oxygen shatter (10), foreshadowing the discovery of large quaternary changes that are linked to oxygen binding (11). Unliganded crystals of human hemoglobin grown under certain low-salt conditions can maintain their integrity upon oxygen binding; however, oxygen binding in this case is noncooperative (12). Although the ability to follow allosteric transitions in the crystalline state is limited when such large quaternary structural changes accompany cooperative ligand binding, spectroscopic investigations of hemoglobin solutions have revealed some aspects of the kinetic pathway of allosteric structural transitions (13–15). Results to date suggest a multistep pathway, with key tertiary structural transitions taking place within a few microseconds and quaternary rearrangements occurring in the range of tens of microseconds. In addition to providing structural information on high-affinity R and low-affinity T quaternary forms at the atomic level (16–18), crystallographic experiments have also succeeded in illuminating ligand-linked structural tertiary transitions within the constraints of R or T forms (19).
In contrast to mammalian hemoglobins, the dimeric hemoglobin (HbI) found in the mollusk Scapharca inaequivalvis is particularly well suited for the investigation of allosteric mechanism by time-resolved crystallography. Binding of oxygen to crystals of HbI is fully cooperative (20), while associated structural transitions are relatively localized and compatible with a well ordered crystal lattice (21). Despite limited structural differences, the two allosteric states show dramatic functional changes with an ≈300-fold difference in oxygen affinity between the high-affinity R-state and the low-affinity T-state (22, 23). Three important tertiary structural rearrangements contribute to the differences in oxygen affinity between the two states: movement of Phe-97 (F4) from the subunit interface into the proximal heme pocket (23, 24), a redistribution of interfacial water molecules (22), and movement of the heme groups toward the subunit interface (25) (see Movie 1, which is published as supporting information on the PNAS web site). In addition, a small subunit rotation (≈3.3°) is required to fully accommodate the tertiary changes (21).
Solution studies of structural transitions in HbI have been much more limited than those in mammalian hemoglobins. One important investigation by nanosecond time-resolved resonance Raman spectroscopy (26) revealed several significant changes upon ligand release in the low-frequency vibrational modes. The modes that undergo a change include those associated with motions of peripheral substituents on the heme, such as the propionates, and the 756-cm−1 mode, assigned as a pyrrole ring deformation mode of the heme, sensitive to steric interaction between the heme and amino acid residues within van der Waals contact. Based on time evolution of these heme-associated vibrational modes, it was proposed that tertiary structural changes, such as relaxation of the heme propionate groups toward their T-state and packing of F4 Phe in contact with the heme, occur with a half-life of 1–2 μs.
We report here a time-resolved crystallographic study following the allosteric transition of Scapharca HbI. The experiments were designed to address several important questions. What is the cascade of structural events upon ligand release that leads from state R to state T? Do functionally important structural transitions occur in a concerted or sequential fashion? Do structural intermediates facilitate the R-to-T transition?
Results and Discussion
Experiments were conducted on crystals of an HbI mutant in which the methionine at helix position B10 is substituted with a valine (M37V). This mutant has an enlarged distal pocket and shows lower geminate rebinding of oxygen in solution but maintains cooperative ligand binding and undergoes the same ligand-linked structural transitions as wild-type HbI (unpublished work). We collected Laue data in a pump-probe type diffraction experiment. A 7-ns laser pulse was used to photodissociate the CO ligands, and an x-ray pulse of appropriate duration was used to probe structural changes initiated by the ligand release. Data were obtained at 16 time delays between the laser and x-ray pulses, from 5 ns to 80 μs. Diffraction data before laser exposure were also collected for direct comparison with the data from photolyzed crystals. Difference electron density (Δρ) maps along with atomic models based on difference refinement against data at various time delays were examined to reveal structural changes. The analysis of structural changes in HbI has the considerable advantage of two subunits per asymmetric unit providing crystallographically independent views of the events.
Ligand Photodissociation.
Strong negative density at 5 ns at the position of the bound CO ligand (labeled CO in Fig. 1) results from significant initial photolysis of the ligand, estimated at ≈40% (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). This density decays to ≈10% by 1 μs (Fig. 2A) indicating a large amplitude of the geminate ligand rebinding. Positive density in the distal pocket adjacent to Val B10 (labeled CO* in Fig. 1) at 5 ns corresponds to a large fraction of the photodissociated CO molecules (≈50%) that has docked in the enlarged distal pocket and remains at this site for hundreds of nanoseconds. The decay of the CO* density follows closely that of the CO density (Fig. 2A).
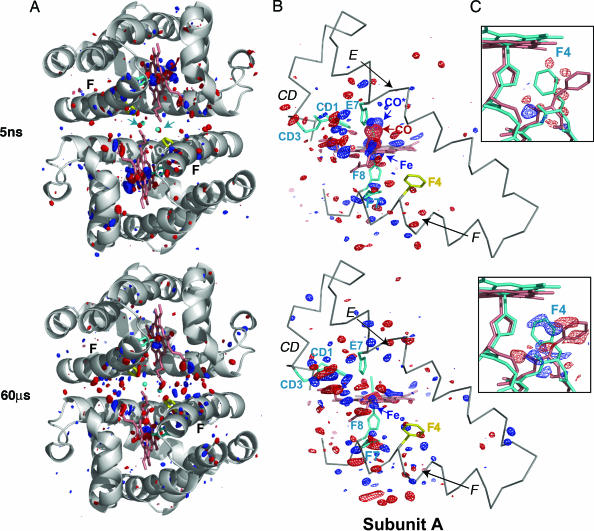
Fig. 1.
Difference Fourier map HbI* (photoproduct) minus HbI-CO at time delays of 5 ns and 60 μs is shown for the entire dimer (A); CD, E, F, and heme regions of subunit A (B); and the Phe F4 of subunit A (C). Fig. 5, which is published as supporting information on the PNAS web site, provides equivalent views for subunit B [the figure was produced with pymol (38)]. (A) A ribbon diagram of the HbI-CO dimer (gray) with side chains for His F8 (cyan), Phe F4 (yellow), and key interface water molecules (small cyan spheres) are shown along with the difference Fourier map. The maps are contoured at ±3.5σ (blue and red, respectively) for both A and B. Note the concentration of difference density mainly in the immediate heme region and along the F helix at 5 ns. The density distributes toward the interface by 60 μs. Arrows (in cyan) point out the position of two key R-state water molecules in the 5-ns map that show clear negative density as they rapidly respond to the loss of ligand. Removal of these two water molecules is required for the subsequent movement of the heme groups toward the subunit interface. (B) An α-carbon trace (gray) for the CD region and E and F helices along with the heme group (salmon), side chains for CD1, CD3, E7, F7, and F8, (cyan) and F4 (yellow) are shown. The photolysis signal at the bound CO position (labeled CO) is highly significant at 5 ns: −14σ and −17σ for the A and B subunits, respectively. The strong positive feature indicating the iron displacement (labeled Fe) is at +12σ and +14σ, for the A and B subunits, respectively. Note the extensive structural rearrangement involving the heme group at 5 ns, along with that of the CD region and F helix. (C) Difference electron density is shown for the region around F4 Phe at ±2.5σ in blue and red, respectively, along with the atomic model for the liganded (salmon) and unliganded (cyan) structures. Phe F4 undergoes the largest ligand-linked side-chain rearrangement during the R-to-T transition. As the density maps show, this movement has not occurred at 5 ns but is completed by 60 μs after the ligand release.
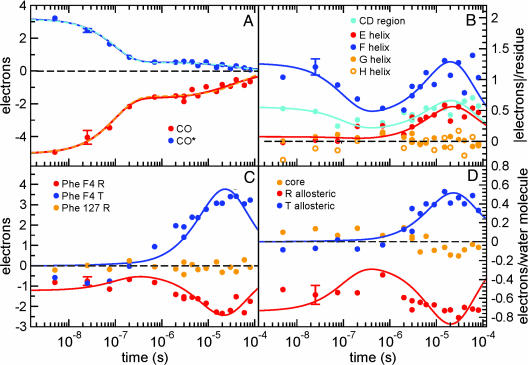
Fig. 2.
Integrated difference electron density values, Δρ, as a function of time for several structural moieties. The values are averaged over subunits A and B (see Supporting Materials and Methods for error estimates). (A) Integrated Δρ(t) values for the loss of bound CO (red circles) and the photodissociated CO at the distal pocket docking site (CO*; blue circles). Both features exhibit similar biphasic behavior and are well described by either a sum of an exponential geminate ligand rebinding phase and a bimolecular rebinding phase (solid lines) or a sum of two exponential geminate phases (dashed lines; see Materials and Methods). (B) Integrated absolute values of difference electron densities, |Δρ(t)|, for helices E–H and the CD-loop region (residues 51–63). The nanosecond signal in the F-helix and CD regions corresponds to the early intermediate. Both helices E and F as well as the CD loop exhibit an increase in the signal in the microsecond time domain, simultaneous with the signal associated with a rearrangement of Phe F4 (C) and allosteric water molecules (D). Simultaneous fits of data in B–D by a common exponential function in the microsecond time region are shown as solid lines (see Materials and Methods). The fits take into account ligand rebinding and assume rapid reversal of structural changes upon ligand rebinding. The deviation of the fit from the data in the 30- to 80-μs time region indicates that the reversal of allosteric changes lags behind ligand rebinding. (C) Integrated Δρ(t) values for the Phe F4 ring at the R- and T-state atomic coordinates. Values for the Phe GH3 ring (R-state) are shown for comparison (no signal is observed). (D) Integrated Δρ(t) values for three distinct classes of water molecules at the dimer interface: 9 allosteric R-state, 12 allosteric T-state, and 5 core water molecules. An early (nanosecond) negative density at the allosteric R-state water molecules results from the disruption of this network. The formation of the T-state network of water molecules occurs on the microsecond time scale.
Progression of Structural Transitions.
The difference electron density maps reveal that photorelease of the ligand triggers a two-phase process involving the key structural moieties that mediate cooperative ligand binding in HbI: the early intermediate phase (5 ns to 1 μs) lays a foundation for the allosteric phase (1 μs to 80 μs). These two phases are readily distinguished by the positions of the heme iron atoms, as is evident from both difference maps and difference refinement (Fig. 3; and see Movies 2 and 3 and Fig. 4, which are published as supporting information on the PNAS web site).
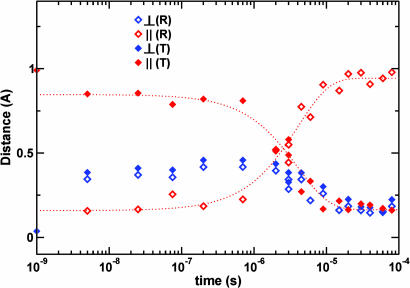
Fig. 3.
Time-dependent change in the heme iron position. This shift is broken down into components that are perpendicular to the heme plane (blue symbols) and components that are parallel to the heme plane (red symbols). The change is measured as the difference in position relative to the starting R-state position (open diamonds) and ending T-state position (filled diamonds). Flash photolysis causes the heme iron to move 0.4 Å perpendicular to the heme plane while shifting by only 0.15 Å parallel to the plane, away from its starting R-state position and toward the T-state position. The heme iron stays in this vicinity during the nanosecond time domain and moves toward its ending T-state position in the microsecond time domain, synchronously with other structural changes involved in the allosteric transition shown in Fig. 2.
Early Intermediate.
An early intermediate has formed by 5 ns after ligand photorelease and persists until ≈1 μs. Rapid displacements of the electron-dense heme iron atoms result in particularly strong positive peaks in Δρ maps (labeled Fe in Fig. 1). The irons are displaced toward the proximal histidine, in a direction perpendicular to the heme plane, by ≈0.4 Å from their starting positions in the R-state. They remain in the vicinity of this intermediate position, with only a small change (<0.2 Å) throughout the early intermediate time domain. The intermediate iron positions are 0.9–1.0 Å away from their T-state positions (Fig. 3 and Movie 3). The early intermediate thus remains highly populated during this time domain, with no significant motion of the hemes toward their T-state location.
Heme buckling, simultaneous with the iron out-of-plane displacement, results in strong negative and positive electron density features observed below and above the hemes (Fig. 1). It involves mainly B, C, and D pyrrole rings, while the A ring is least affected. Shift of the B ring opens additional space on the proximal side that facilitates the subsequent allosteric Phe F4 transition. The tilt of ring D results in the shift of the heme D propionate toward the Arg CD3, while positive and negative density features for Arg CD3 (±4σ) indicate movement toward its T-state location (Fig. 1). In response to changes at the heme, residues Phe CD1 and Leu E11 move deeper into the heme pocket, and His E7 moves toward the space vacated by the released ligand.
Rapid structural changes along the F helix, particularly in the main-chain region near F4, are evident by positive and negative features along the helix observed at 5 ns (Fig. 1). We assessed the magnitude of these changes as a function of time by integrating |Δρ(t)| values along the helix. Similar integration for the CD-loop region and helices E, G, and H (Fig. 2B) clearly singles out the F helix and the CD loop as regions with the most structural changes at early time delays. Notably, these regions also show the largest differences between static R and T structures of HbI (27). Rapid F-helix displacement has also been observed in time-resolved studies of myoglobin (7) and appears to be a characteristic response of this class of proteins to deligation and subsequent heme iron movement.
The observed early movement of the F helix is in the direction of the subunit interface. It results in disruption of water molecules unique to the R-state, as evident by immediate negative density in difference maps at locations of these molecules (Fig. 2D) and by no difference density around those water molecules common to both R and T states. Most significantly affected are two R-state waters near the center of the dimer interface, with negative density evident at 5 ns (Fig. 1). These molecules are located near F4 and F7 side chains and are therefore susceptible to disruption by the observed F-helix shift. They are also hydrogen-bonded to the heme A propionates; their disordering is likely to facilitate a subsequent rearrangement of these propionates, which is in turn required for the shift of the hemes toward the interface.
The observed early intermediate does not lie on a direct trajectory between the R and T states, as is evident both in the heme position (Fig. 3) and in the quaternary structural arrangement. Difference refinement indicates that the subunits are rotated slightly (0.3°) and consistently away from the T-state arrangement during the early intermediate, before rotating toward the T-state quaternary arrangement during the allosteric phase.
Allosteric Phase.
During the second, allosteric phase (1–80 μs), unliganded HbI commences its approach toward the T-state. The tertiary T-state (i.e., unliganded T-state subunits in an R quaternary subunit arrangement) forms and reaches the maximum population by ≈10 μs (Fig. 2). Our results suggest a somewhat slower (≈5-fold) transition to the tertiary T-state conformation than is proposed based on resonance Raman solution studies (26), although the progression of concerted changes, involving heme propionates and Phe F4 as well as other groups identified here, is in full agreement with the solution results. Because none of the residues involved in the allosteric transition participate in lattice contacts, the different time constants probably reflect an overall effect of the crystal lattice. Formation of the tertiary T-state is followed by a slower subunit rotation in the final approach to the quaternary T-state, as is indicated by rigid body refinement of subunit orientations. A rotation of 0.6° from the R-state is observed by 80 μs, forming a “T-like” state, as compared with the expected rotation of 3.3° based on static R and T structures. Because the entire R-to-T rearrangement can be accommodated in this crystal lattice (21), we conclude that the full R-to-T subunit rotation has not been completed by 80 μs.
Transition to the tertiary T-state proceeds via rearrangements of several key local and global structural moieties (Fig. 2 B–D). The integrated Δρ(t) or |Δρ(t)| values associated with these moieties display remarkably similar time courses during the allosteric phase that can be described by an exponential function, with a common rate coefficient of 6.5 × 104 s−1. Electron density at the T-state position for Phe F4 begins to accumulate at ≈1 μs (Fig. 2C). Simultaneously, an increase in difference electron densities is observed for the E and F helices and the CD region, as well as for both R- and T-allosteric water molecules (Fig. 2 B and D). Populations of T-state Phe F4 reach a maximum at ≈10–30 μs. Based on the integrated Δρ for the T-state Phe F4 ring, we estimate ≈8% peak population of the T-state F4, consistent with the deligated fraction of hemes in this time region (Fig. 2A). This finding suggests that Phe F4 has switched from its R-to-T conformation in all deliganded subunits.
The kinetics of the T-state water molecules indicates a close coupling between their movements to T-state locations with that of Phe F4. This is expected because several T-state waters fill the volume vacated at the R-state Phe F4 position. An increase in the negative integrated Δρ values for the R-state waters is also observed, indicating their further displacement. The absence of difference density for the core of five ordered interface water molecules, present in both R and T states, confirms their stable conformation during both the early intermediate and the allosteric transition.
Is there a leading, trigger event that initiates the transition to the tertiary-T state after very rapid events associated with the early intermediate? To address this question we examined more closely the locations of heme irons, and thus the heme groups, by difference refinement. The observed heme shift toward the dimeric interface, itself facilitated by the early disruption of R-state water molecules at the interface and coupled with the F-helix displacement via the His F8 linkage, might reasonably be expected to precede the Phe F4 motion. It enlarges the proximal heme cavity, and structural changes in the pyrrole A ring and the A propionate are likely to reduce a steric barrier for transition of the Phe F4 to the T-state. In fact, the heme shift from the early intermediate toward the dimeric interface starts at the same time as accumulation of the positive density at the Phe F4 T-state location, at ≈1 μs (Figs. 3 and 2C). Populating the heme tertiary T-state therefore coincides with populating the Phe F4 T-state. Because the two events clearly follow each other very closely, either they cannot be resolved given the present data or the second event occurs essentially instantaneously (submicrosecond) after the first, rate-limiting step.
Lag in Reversal of Structural Transitions.
Finally, our results provide important information on how fast structural changes occurring during the allosteric phase reverse upon ligand rebinding. As mentioned above, at 10–30 μs Phe F4 in all unliganded subunits has switched from the R-state to the T-state conformation. By 80 μs, significant further ligand rebinding has occurred (Fig. 2A). However, the signal related to Phe F4, the E and F helices, CD region, and allosteric water molecules (Fig. 2 B–D) has actually leveled off rather than significantly decayed during the 30- to 80-μs time period. Solid lines in Fig. 2 B–D indicate the trend that is expected in this time period if reversal of structural changes is fast compared with ligand rebinding. Because the data do not follow such a trend, we conclude that the reversal of the allosteric structural changes is slow and extends beyond 80 μs. This conclusion is also supported by the refined iron positions (Fig. 3) and from Fig. 1, where a prominent iron-related density is present well above 3.5σ at 60 μs while very little density associated with the CO photo dissociation is present. Data at time delays longer than presented here are clearly needed to follow the evidently slower-than-expected reversal of allosteric changes from the liganded T-state to the R-state.
This application of nanosecond time-resolved crystallography to examine a full allosteric protein transition has elucidated a sequence of structural events that mediate cooperativity after the release of bound ligands. An intermediate structure is formed rapidly, within 5 ns, and it is characterized by buckling of the heme and resulting initial rearrangements of the neighboring side chains and the F-helix and CD-loop region. Disruption of the R-state-specific network of water molecules also occurs rapidly, suggesting an important role for water molecules in facilitating the shift between alternate states. These changes lay the foundation for a concerted transition that commences at ≈1 μs. An intriguing aspect of our results is the substantial lag in transition back to the R-state after ligand rebinding. Evidently, the T-to-R transition is much slower than that from R to T, a result that implies substantial stability of the T-state, even for the singly liganded species, which is consistent with earlier equilibrium and kinetic studies (22, 25, 28).
Materials and Methods
Sample Preparation.
The M37V mutant protein was cloned, expressed, purified, and crystallized as described for recombinant HbI (25, 29). Crystals were mounted in a glass capillary under a poly(vinyl) film (30) to prevent laser-induced movement of the crystal (31). The capillaries were purged under a stream of carbon monoxide before a drop of stabilizing solution with saturated sodium dithionite was added to remove any residual oxygen before sealing.
Time-Resolved Data Collection.
Time-resolved x-ray diffraction experiments were conducted at BioCARS beamline 14-ID at the Advanced Photon Source, Argonne National Laboratory. In pump-probe-type diffraction experiments, HbI-CO crystals at 5°C were illuminated from two opposite sides by laser pulses from a Nd:YAG pumped dye laser (Continuum). The laser provided 7-ns (full width at half maximum) pulses at 615 nm (rhodamine 610/640 dye) with a total laser pulse energy of ≈2.2 mJ and beam diameters at the sample of 0.6 and 0.8 mm for the two laser beams. We estimated that the number of absorbed photons per laser pulse was approximately equal to the number of molecules in the crystals. A single 100-ps x-ray pulse during the hybrid mode of the Advanced Photon Source storage ring was used as the probe pulse for short time delays (5 ns to 3 μs). For longer time delays (3 μs to 80 μs) the probe pulse was either a 500-ns superpulse during the hybrid mode or a 2-μs pulse train during the standard operating mode of the storage ring. The 2-μs length of the pulse train was defined by the minimum opening of the BioCARS fast x-ray chopper. Experimental details of the time-resolved set-up at the 14-ID beamline are described in ref. 32. The time delays referred to in this work are measured from center to center between x-ray and laser pulses.
Time-resolved data were collected by using the Laue diffraction technique in the following way. At a fixed angular setting of the crystal, x-ray diffraction images were recorded without laser illumination (dark state) and at a series of time delays after the laser pulse. For the first time series, images were collected at 5 ns, 25 ns, 75 ns, 200 ns, 700 ns, and 3 μs. The procedure was repeated for 21 crystal orientations, with a 9° increment between angular settings of the crystal. The same measurement was repeated on four crystals, and the data were merged to obtain complete difference data sets for each time delay (see below). This method of data collection minimizes systematic errors in the time domain due to crystal-to-crystal variation in reaction photoinitiation as compared with data collection where a complete data set is collected for each time delay on a different crystal. Two additional time series of data sets were collected in a similar manner: dark, 3 μs, 6 μs, 15 μs, 30 μs, and 60 μs (four crystals); and dark, 2 μs, 4.5 μs, 9 μs, 20 μs, 40 μs, and 80 μs (three crystals).
Laue data were processed by using the programs precognition (indexing and integration) and epinorm (scaling) (Zhong Ren; www.renzresearch.com). All data sets were integrated to 1.6 Å. Data reduction statistics are presented in Tables 1 and 2, which are published as supporting information on the PNAS web site. From the dark-state structure factor (SF) amplitudes |FD(hkl)| and corresponding time-dependent SF amplitudes |F(hkl, t)|, time-dependent difference SF amplitudes ΔF(hkl, t) = |F(hkl, t)| − |FD(hkl)| were calculated for each time point t and each crystal. Difference SF amplitudes ΔF(hkl, t) from multiple crystals were merged by using weighted averaging, where weights used for ΔF(hkl, t) for each crystal were calculated as described in Ren et al. (9). When averaging over multiple crystals, the weight for the mean value 〈ΔF(hkl, t)〉 was calculated as the sum of the weights of ΔF(hkl, t) that were averaged. Weighted difference electron density maps (Δρ) were then calculated for each time point by using merged difference SF amplitudes 〈ΔF(hkl, t)〉 and phases φhklD obtained from the known M37V HbI-CO structural model (unpublished work).
Difference Electron Density Integration.
Integration of difference electron densities in selected regions of Δρ(t) maps (Fig. 2) was performed by using the program promsk (2). The Δρ(t) or |Δρ(t)| values were summed within a specified mask and multiplied by a factor of two to account for the fact that the Δρ maps calculated as described above are on approximately half of the absolute scale (32). The integration mask was generated by supplying atomic coordinates of the structural region of interest of the R- or T-state and specifying a radius of integration around given coordinates (1.2 Å was used in all cases, except for helices where 2 Å was used for integration around R-state coordinates).
Ligand Rebinding and Kinetics of Structural Changes.
Decay of the CO* density follows closely that of the CO density (Fig. 2A) because both time courses exhibit two major phases with similar rates. A detailed description of the ligand migration pathways is not possible given that other docking sites were not detected but are required to account for all photodissociated CO molecules at early times. We therefore cannot determine unambiguously whether the second phase of the CO and CO* time courses is geminate or bimolecular. Fig. 2 shows two possible options that both describe data well. The CO and CO* decays are fit jointly by a sum of an exponential function, C1 × exp[−k1t], describing geminate ligand-rebinding and a bimolecular-rebinding function, C2/[1 + k2′ t] (solid lines). The common rate coefficients are k1 = 1.0 × 107 s−1 and k2′ = 3.2 × 104 s−1, and the amplitude of the geminate phase is 0.7 for CO and 0.8 for CO*. An alternative fit by two exponential functions describing two geminate phases (dashed lines) results in similar rates (k1 = 9.6 × 106 s−1 and k2 = 1.7 × 104 s−1) and identical amplitudes.
Possible causes for the unexpectedly large amplitude of the fast geminate phase in the crystal are the alteration of the ligand escape rates by the crystal lattice and the migration of dichloroethane used for crystal immobilization (31) into a cavity near the heme pocket, occupying a site suggested (25, 33) to be on the entry and exit pathway of the ligand (unpublished work).
Time courses shown in Fig. 2 B–D correspond to structural changes that involve two key players in the allosteric mechanism of HbI: Phe F4 and allosteric water molecules, as well as several larger structural moieties such as E and F helices and the CD-loop region. Whereas some time courses exhibit significant integrated Δρ(t) values in the nanosecond time domain (most notably those corresponding to allosteric R-state water molecules, F helix and CD loop region), they all exhibit an increase in the integrated Δρ(t) values in the microsecond time domain, with an apparently similar rate. They were therefore fitted jointly by a single exponential function corresponding to the R-to-T transition, C3 + C4[1 − exp(−k3t)], with a rate coefficient k3 = 6.5 × 104 s−1 (solid lines in Fig. 2 B–D). Constant C3 accounts for subnanosecond structural changes in some structural moieties. The difference electron density signal is expected to diminish as ligands rebind because it represents structural changes in a decaying number of unligated molecules. This is, however, true only if the reversal of structural changes upon ligand rebinding is fast compared with ligand rebinding. To assess whether the reversal is indeed fast, the above fit function describing the time courses was multiplied by a normalization term corresponding to the ligand rebinding, with parameters fixed to those derived from data in Fig. 2A (using the first fit discussed above). The deviation of the data from the fit observed in the 20- to 80-μs time period in Fig. 2 B–D, where data level off rather than decay in accordance with the ligand rebinding trend, suggests that the reversal of the allosteric structural changes is slow and not complete by 80 μs. The resetting of the structure to the R-state in the liganded tertiary T-state is lagging behind the ligand rebinding.
Difference Refinement.
To assess the structural changes as a function of time, a model of the photoproduct was obtained for each time delay according to the difference refinement approach of Terwilliger and Berendzen (34, 35). Refined structures, obtained as described in Supporting Materials and Methods, help assess the direction of motion for structural moieties and populations of the underlying intermediates. We emphasize that by this approach we are not suggesting that each time delay represents a single intermediate state and that an overall structural change proceeds via a large number of intermediate states. Quite to the contrary, our data (Fig. 2) suggest only one early intermediate that forms in the subnanosecond time domain and a concerted R-to-T transition of several structural moieties in the microsecond time domain to the tertiary T-state. In general, when the simple chemical kinetics mechanism holds, as seems to be the case here given that the microsecond allosteric transition from the early intermediate to the tertiary T-state is well described by an exponential function (32, 36, 37), well defined time-independent intermediate states exist and only their populations vary with time. At any time, a mixture of intermediate structures is likely to be present (32). The refinement of a single structural model at any individual time delay is therefore clearly inappropriate for extracting the structures of intermediate states when a mixture of states is present. At short time delays, before the microsecond transition, and at longer time delays, after the microsecond transition, the refined models provide estimates for the structures of the early intermediate and the tertiary T state. A full structural characterization of all intermediates involved in the R-to-T transition will require data more finely sampled in time and a global data analysis by the singular value decomposition method (32).
Supplementary Material
Acknowledgments
We thank members of Keith Moffat’s group for stimulating discussions on all aspects of time-resolved crystallography; Keith Moffat for his comments on an early version of the manuscript; Michael Wulff and his team at European Synchrotron Radiation Facility beamline ID09, along with Tsu-Yi Teng, Claude Pradervand, and Wilfried Schildkamp, for contributions to preliminary experiments on HbI; Marius Schmidt for the use of promsk; the BioCARS staff for their assistance; and Bill Kobertz for assistance with making the movies. This work was supported by National Institutes of Health Grant GM66756 (to W.E.R. and V.Š.) and was initiated with funds from National Institutes of Health Grants DK43323 (to W.E.R.) and GM36452 (to Keith Moffat) and Postdoctoral Fellowship 9920261T from the New England affiliate of the American Heart Association (to J.E.K.). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38. Use of BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under Grant RR07707.
Abbreviation
- HbI
dimeric hemoglobin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2GRF for M37V, 2GRH for M37V-CO, and 2GRZ for 5-ns photoproduct).
References
- 1.Ihee H., Rajagopal S., Šrajer V., Pahl R., Anderson S., Schmidt M., Schotte F., Anfinrud P. A., Wulff M., Moffat K. Proc. Natl. Acad. Sci. USA. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt M., Nienhaus K., Pahl R., Krasselt A., Anderson S., Parak F., Nienhaus G. U., Šrajer V. Proc. Natl. Acad. Sci. USA. 2005;102:11704–11709. doi: 10.1073/pnas.0504932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt M., Pahl R., Šrajer V., Anderson S., Ren Z., Ihee H., Rajagopal S., Moffat K. Proc. Natl. Acad. Sci. USA. 2004;101:4799–4804. doi: 10.1073/pnas.0305983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajagopal S., Anderson S., Šrajer V., Schmidt M., Pahl R., Moffat K. Structure (Cambridge, MA) 2005;13:55–63. doi: 10.1016/j.str.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Bourgeois D., Vallone B., Schotte F., Arcovito A., Miele A. E., Sciara G., Wulff M., Anfinrud P., Brunori M. Proc. Natl. Acad. Sci. USA. 2003;100:8704–8709. doi: 10.1073/pnas.1430900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schotte F., Lim M., Jackson T. A., Smirnov A. V., Soman J., Olson J. S., Phillips G. N., Jr., Wulff M., Anfinrud P. A. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 7.Šrajer V., Ren Z., Teng T. Y., Schmidt M., Ursby T., Bourgeois D., Pradervand C., Schildkamp W., Wulff M., Moffat K. Biochemistry. 2001;40:13802–13815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 8.Šrajer V., Teng T., Ursby T., Pradervand C., Ren Z., Adachi S., Schildkamp W., Bourgeois D., Wulff M., Moffat K. Science. 1996;274:1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- 9.Ren Z., Perman B., Šrajer V., Teng T. Y., Pradervand C., Bourgeois D., Schotte F., Ursby T., Kort R., Wulff M., Moffat K. Biochemistry. 2001;40:13788–13801. doi: 10.1021/bi0107142. [DOI] [PubMed] [Google Scholar]
- 10.Haurowitz F. Hoppe-Seyler’s Z. Physiol. Chem. 1938;254:266–274. [Google Scholar]
- 11.Perutz M. F. Nature. 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 12.Mozzarelli A., Rivetti C., Rossi G. L., Henry E. R., Eaton W. A. Nature. 1991;351:416–419. doi: 10.1038/351416a0. [DOI] [PubMed] [Google Scholar]
- 13.Hofrichter J., Sommer J. H., Henry E. R., Eaton W. A. Proc. Natl. Acad. Sci. USA. 1983;80:2235–2239. doi: 10.1073/pnas.80.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayaraman V., Rodgers K. R., Mukerji I., Spiro T. G. Science. 1995;269:1843–1848. doi: 10.1126/science.7569921. [DOI] [PubMed] [Google Scholar]
- 15.Goldbeck R. A., Esquerra R. M., Kliger D. S. J. Am. Chem. Soc. 2002;124:7646–7647. doi: 10.1021/ja025855l. [DOI] [PubMed] [Google Scholar]
- 16.Fermi G., Perutz M. F., Shaanan B., Fourme R. J. Mol. Biol. 1984;175:159–174. doi: 10.1016/0022-2836(84)90472-8. [DOI] [PubMed] [Google Scholar]
- 17.Shaanan B. J. Mol. Biol. 1983;171:31–59. doi: 10.1016/s0022-2836(83)80313-1. [DOI] [PubMed] [Google Scholar]
- 18.Silva M. M., Rogers P. H., Arnone A. J. Biol. Chem. 1992;267:17248–17256. [PubMed] [Google Scholar]
- 19.Adachi S., Park S. Y., Tame J. R., Shiro Y., Shibayama N. Proc. Natl. Acad. Sci. USA. 2003;100:7039–7044. doi: 10.1073/pnas.1230629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mozzarelli A., Bettati S., Rivetti C., Rossi G. L., Colotti G., Chiancone E. J. Biol. Chem. 1996;271:3627–3632. doi: 10.1074/jbc.271.7.3627. [DOI] [PubMed] [Google Scholar]
- 21.Knapp J. E., Royer W. E., Jr. Biochemistry. 2003;42:4640–4647. doi: 10.1021/bi027136p. [DOI] [PubMed] [Google Scholar]
- 22.Royer W. E., Jr., Pardanani A., Gibson Q. H., Peterson E. S., Friedman J. M. Proc. Natl. Acad. Sci. USA. 1996;93:14526–14531. doi: 10.1073/pnas.93.25.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knapp J. E., Bonham M. A., Gibson Q. H., Nichols J. C., Royer W. E., Jr. Biochemistry. 2005;44:14419–14430. doi: 10.1021/bi051052+. [DOI] [PubMed] [Google Scholar]
- 24.Pardanani A., Gibson Q. H., Colotti G., Royer W. E., Jr. J. Biol. Chem. 1997;272:13171–13179. doi: 10.1074/jbc.272.20.13171. [DOI] [PubMed] [Google Scholar]
- 25.Knapp J. E., Gibson Q. H., Cushing L., Royer W. E., Jr. Biochemistry. 2001;40:14795–14805. doi: 10.1021/bi011071t. [DOI] [PubMed] [Google Scholar]
- 26.Rousseau D. L., Song S., Friedman J. M., Boffi A., Chiancone E. J. Biol. Chem. 1993;268:5719–5723. [PubMed] [Google Scholar]
- 27.Royer W. E., Jr. J. Mol. Biol. 1994;235:657–681. doi: 10.1006/jmbi.1994.1019. [DOI] [PubMed] [Google Scholar]
- 28.Royer W. E., Jr., Fox R. A., Smith F. R., Zhu D., Braswell E. H. J. Biol. Chem. 1997;272:5689–5694. doi: 10.1074/jbc.272.9.5689. [DOI] [PubMed] [Google Scholar]
- 29.Summerford C. M., Pardanani A., Betts A. H., Poteete A. R., Colotti G., Royer W. E., Jr. Protein Eng. 1995;8:593–599. doi: 10.1093/protein/8.6.593. [DOI] [PubMed] [Google Scholar]
- 30.Rayment I., Johnson J. E., Suck D. J. Appl. Crystallogr. 1977;10:365–368. [Google Scholar]
- 31.Knapp J. E., Šrajer V., Pahl R., Royer W. E., Jr. Micron. 2004;35:107–108. doi: 10.1016/j.micron.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt M., Ihee H., Pahl R., Šrajer V. In: Protein-Ligand Interactions: Methods and Applications. Ulrich N. G., editor. Totawa, NJ: Humana; 2005. [DOI] [PubMed] [Google Scholar]
- 33.Chiancone E., Elber R., Royer W. E., Jr., Regan R., Gibson Q. H. J. Biol. Chem. 1993;268:5711–5718. [PubMed] [Google Scholar]
- 34.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1996;52:1004–1011. doi: 10.1107/S0907444996006725. [DOI] [PubMed] [Google Scholar]
- 35.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1995;51:609–618. doi: 10.1107/S0907444994013247. [DOI] [PubMed] [Google Scholar]
- 36.Moffat K. Chem. Rev. 2001;101:1569–1581. doi: 10.1021/cr990039q. [DOI] [PubMed] [Google Scholar]
- 37.Karplus M. Simplicity and Complexity in Proteins and Nucleic Acids. Berlin: Dahlem Univ. Press; 1999. p. 139. [Google Scholar]
- 38.DeLano W. L. The pymol Molecular Graphics System. San Carlos, CA: DeLano Sci.; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.