Abstract
During infection with Chlamydia trachomatis, CD8+ T cells are primed, even though the bacteria remain confined to a host cell vacuole throughout their developmental cycle. Because CD8+ T cells recognize antigens processed from cytosolic proteins, the Chlamydia antigens recognized by these CD8+ T cells very likely have access to the host cell cytoplasm during infection. The identity of these C. trachomatis proteins has remained elusive, even though their localization suggests they may play important roles in the biology of the organism. Here we use a retroviral expression system to identify Cap1, a 31-kDa protein from C. trachomatis recognized by protective CD8+ T cells. Cap1 contains no strong homology to any known protein. Immunofluorescence microscopy by using Cap1-specific antibody demonstrates that this protein is localized to the vacuolar membrane. Cap1 is virtually identical among the human C. trachomatis serovars, suggesting that a vaccine incorporating Cap1 might enable the vaccine to protect against all C. trachomatis serovars. The identification of proteins such as Cap1 that associate with the inclusion membrane will be required to fully understand the interaction of C. trachomatis with its host cell.
Chlamydiae are obligate intracellular bacterial pathogens that replicate within membrane-bound vacuoles and are responsible for several major human diseases. Chlamydia trachomatis is the most common cause of bacterial sexually transmitted disease in the United States, as emphasized recently in a report showing an incidence of infection approaching 30% among teenage girls (1). Left untreated, the organism can cause pelvic inflammatory disease, the sequelae of which are a leading cause of preventable infertility. C. trachomatis is also responsible for the blinding disease trachoma. In addition, members of this genus have been suggested as contributing factors in atherosclerosis and other chronic human diseases (2–4).
Adaptive immune protection against C. trachomatis can be demonstrated by adoptive transfer of either Chlamydia-specific CD4+ or CD8+ T cells into infected mice (5–8), and both subsets are recruited to sites of infection in primate models (9). Experiments designed to define the relative contribution of CD4+ or CD8+ T-cell subpopulations in immunity to Chlamydia are extremely difficult. This is because cell-mediated and humoral elements of the immune system act in concert, and multiple redundancies exist in recognition and effector function, e.g., protective Th1-type cytokine production from both CD4+ and CD8+ T cells (10, 11). Multiple studies show a clear and important role for CD4+ T cells in immunity to C. trachomatis (5, 7, 12), and specific antigens recognized by CD4+ T cells during murine infection have been described (13). However, studies to investigate more fully the role of CD8+ T cells have been hampered by the absence of known murine CD8+ T-cell antigens.
The observed CD8+ T-cell response to C. trachomatis is intriguing, as this pathogen remains confined throughout its replication cycle to a host-cell vacuole known as an inclusion (14). Typically, CD8+ T cells recognize antigens derived from the cytosol. Thus, the response of these CD8+ T cells suggests that during infection, some C. trachomatis products access the cytosol of infected cells where they encounter the MHC class I processing machinery. These MHC class I accessible proteins are candidates for inclusion in Chlamydia vaccines and may also be critical factors required for growth within the inclusion.
Although viral and tumor CD8+ T-cell antigens have been identified by expression cloning in eukaryotic cells, these methods have not, until now, been successfully applied to bacterial pathogens. We report here the identification, by expression cloning, of a protein antigen from C. trachomatis that is localized to the inclusion membrane and stimulates CD8+ T cells. This antigen is encoded within an ORF initially defined as CT529 during genome sequencing as a 298-aa C. trachomatis hypothetical protein (15). We refer to this predicted protein as class I accessible protein-1, or Cap1. Cap1 has little homology to any known proteins, nor does it contain recognized domains predicted from known functional motifs. This lack of homology suggests a function of Cap1 that may be unique to this vacuolar parasite.
Methods
Mice.
Female BALB/cByJ mice (H-2d) were obtained from the Jackson Laboratory and were used for T-cell stimulations and challenge studies.
Tissue Culture.
The amphotropic retroviral helper packaging cell line, Phoenix-Ampho (17), was maintained in DMEM supplemented with 10% FCS and antibiotics. All other cells were maintained in RP-10, consisting of RPMI-1640 supplemented with 10% FCS and 50 μM 2-mercaptoethanol. C. trachomatis-specific CD8+ T-cell line 69 was derived from a BALB/cByJ mouse (H-2d) infected with C. trachomatis L2, as described (8). P815, an H-2d mastocytoma derived from DBA/2 mice, and J774, an H-2d BALB/c-derived monocyte/macrophage tumor, were obtained from the American Type Culture Collection (ATCC).
Growth of C. trachomatis.
Elementary bodies of C. trachomatis serovar L2 434/Bu were propagated within HeLa 229 cell monolayers in RP-10 and were purified by density gradient centrifugation, as described (18). Aliquots of C. trachomatis were stored at −70°C in a buffer containing sucrose, phosphate, and glutamate (SPG) (19). C. trachomatis serovar Ba (Apache-2), E (BOUR), G (UW57/Cx), I (UW12/Ur), K (UW31/Cx), L1 (440), L3 (404), and the mouse biovar of C. trachomatis (MoPn, NiggII) were obtained from ATCC. C. trachomatis serovar E (MTW447), F (P0670), and Ia (P0483) were obtained from W. Stamm (University of Washington, Seattle, WA). C. trachomatis serovar F (NI1) was obtained from J. Orfila (University of Picardie, Amiens, France). These strains were propagated on McCoy murine fibroblast monolayers in Eagle's minimal essential medium, and bacteria were released with glass beads, harvested into SPG at a concentration of 107 inclusion forming units (IFU)/ml, and stored at −70°C.
Derivation of Retroviral Expression Vector.
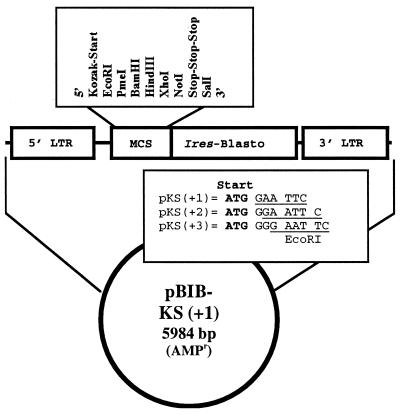
To allow eukaryotic expression of proteins from random bacterial genomic DNA fragments, a retroviral expression vector was modified to contain a multiple cloning site (16) (MCS) flanked by a Kozak eukaryotic translation initiation site with start codon and stop codons (in each of the three frames). Briefly, the pBIB-KS1,2,3 vectors were constructed from the retroviral vector pBIB by replacement of the pBIB MCS with GGATCTGCC GCCACCATG(+1,G+2,GG+3)GAATTCGTTTAAACGGATCCAAGCTTGAGCTCGAGCGCGGCCGCTAATTAGCTGAGTCGAC oligos, respectively. The parental pBIB vector was derived in several steps from pBabeMN (20), a vector nearly identical to pMX (21). First, a 1.0-kb SalI–NheI restriction fragment containing the intraribosomal entry site (IRES)-bsr sequences from pWZL-Blast (kindly provided by T. Kinsella, Stanford University, Stanford, CA) was ligated into SalI–NheI cut pBabeMN to derive pBMN-I-Blasto(H+). This creates a bicistronic mRNA containing the IRES (22) of the encephalomyocarditis virus driving bsr gene (23) translation, downstream of the cDNA insert (MCS), allowing for blastocidin-S selection of DNA-expressing transductants (24). The HindIII site at position 5919 was removed by HindIII digestion, nucleotide fill-in with the Klenow fragment of DNA polymerase, and religation, to derive pBMN-I-B. The MCS of pBMN-I-B was then replaced by a BglII/SalI PCR fragment containing the MCS from pcDNA3.1+ to derive pBIB.
Construction of a Chlamydia DNA Expression Library.
C. trachomatis L2 genomic DNA was prepared from elementary bodies propagated as described above. Briefly, DNA was purified by phenol/chloroform extraction of 5 × 109 IFU elementary bodies previously treated with proteinase K. For expression library construction, 3 μg of purified DNA was fragmented by incomplete restriction enzyme digestion by using BamHI and MboI. The resulting DNA fragments were ligated into the unique BamHI site of pBIB-KS and transformed into Escherichia coli XL-1 blue. Analyses of a subset of recombinants revealed an insert frequency of 60%, average insert size in the library of 0.5–1.0 kb, and a complexity of 18,000 independent clones containing inserts. DNA from pools of 80 clones/pool was prepared and used for transfection and preparation of retrovirus.
Transfection of Retroviral Packaging Line Phoenix-Ampho.
Amphotropic retroviral particles representing the C. trachomatis DNA fragment expression library were produced by transient transfection of the high-titer retroviral helper packaging line, Phoenix-Ampho (17), provided by G. Nolan (Stanford University, Stanford, CA). For transfections, 5 μg DNA from each plasmid pool was precipitated by the addition of 250 mM CaCl2 and Hanks' buffered saline solution in a final volume of 100 μl. For each pool, 70 μl of DNA solution was added drop wise to 1 well of a 24-well plate containing 0.3 ml Phoenix-Ampho cells that were seeded 18 h earlier in DMEM + 10% FCS at a density of 0.5 × 106/ml. After 6 h at 37°C, medium was replaced with fresh DMEM + 10% FCS. After 24 h, transfected cells were placed at 32°C for 18 h, after which supernatants containing retrovirus were collected and stored at −80°C.
Retroviral Transductions.
For library screening, P815 cells (H-2d) were transduced in 96-well plates prepared by coating V-bottom plates (Costar) with 50 μl/well of 10 μg/ml retronectin (BioWhitaker) in PBS. After a 2-h incubation, retronectin was removed, and the plates were blocked with 50 μl/well of 2% BSA in PBS. After 30 min, the BSA was removed, and the wells were washed twice with PBS. For transductions, exponentially growing P815 cells (0.25 × 106/ml) were then added at 50 μl/well to each well of these pretreated plates, and retroviral supernatants of individual library pools, or control supernatants, were added (50 μl/well) in the presence of 5 μg/ml DEAE dextran. Plates were centrifuged at 500 × g for 30 min at 4°C, incubated at 32°C for 18 h, and then moved to 37°C for 48 h. Seventy-two hours after transduction, the enhanced green fluorescent protein control wells were analyzed by FACS to determine transduction efficiency. Blastocidin-S (Calbiochem) was added at 10 μg/ml to select transduced P815 cells.
Stimulation and Maintenance of C. trachomatis-Specific CD8+ T Cells.
C. trachomatis-specific H-2d-restricted CD8+ T-cell line 69 (8) was maintained and expanded by weekly stimulation on C. trachomatis-infected J774 (H-2d) cells, as described (8).
Stimulation of Cap1-specific T cells from infected mice was carried out as follows. BALB/c mice were infected i.p. with 108 IFU of C. trachomatis serovar L2. Control mice were given an injection of PBS. After 14 days, splenocytes from infected and control mice were washed once in RP-10 and then cultured with stimulator cells. Cells used as stimulators were naive, irradiated (2,000 rad), syngeneic splenocytes treated for 1 h with 10 μM sterile Cap1139–147 peptide. Cultures contained 4 × 107 stimulator cells and 4 × 107 splenocytes from either immunized or control mice. These were incubated upright for 5 days in a T-25 flask at 37°C in 7% CO2 in a total volume of 20 ml of RP-10.
Library Screening.
IFN-γ ELISPOT assays were used to screen the C. trachomatis DNA expression library for activation of T cells. For screening, 3 × 104 P815 cells from transduced and blastocidin-S-selected microcultures were added to nitrocellulose ELISPOT plates (Millipore) coated previously with anti-mouse IFN-γ mAb (no. 18181D; PharMingen), washed three times with PBS, and blocked with cell culture medium 1 h before cell addition. T cells (line 69) were added (105/well) to each well and cocultured in the presence of human IL-2 (3 ng/ml) and murine IL-12 (3 ng/ml). After 48-h incubation, the plates were washed, and the cultures were analyzed for IFN-γ production by the addition of a second biotinylated anti-mouse IFN-γ mAb (no. 18112D; PharMingen). After a 2-h incubation, plates were washed in PBS containing 0.1% Tween-20 and then incubated with streptavidin-horseradish peroxidase (SA-HRP). After 1 h, excess SA-HRP was removed by washing. Plates were then incubated with HRP Color development reagent (Bio-Rad) for 5 min, washed with distilled H2O, and air dried.
Cytotoxicity Assays.
Standard chromium release assays were used to determine the percent specific lysis of peptide-pulsed or C. trachomatis-infected (14 h) target cells, as previously described (8).
PCR and DNA Sequencing.
Transgenic DNAs from immunoreactive transduced P815 clones were amplified by using primers specific for pBIB-KS retroviral vector sequences flanking the MCS. The 5′ primer sequence was 5′-CCTTACACAGTCCTGCTGAC, and the reverse strand primer was 3′-GTTTCCGGGCCCTCACATTG. PCR reactions were performed by using Pwo Polymerase (Roche Diagnostics) with genomic DNA as template. The insert from 2C7.8 contained the sequence ATCTTTGTGTGTCTCATAAGCGCAGAGCGGCTGCGGCTGTCTGTA GCTTCATCGGAGGAATTACCTACCTCGCGACATTCGGAGCTATCCGTCCGATTCTGTTTGTCAACAAAATGCTGGCGCAACCGTTTCTTTCTTCCCAAATTAAAGCAAATATGGG. Full-length CT529 from serovar L2 was obtained by PCR amplification by using 5′-TTTTGA AGCAGGTAGGTGAATATG (forward) and 5′-TTAAGAAATTTA AAAAATCCCTTA (reverse) primers, by using purified C. trachomatis L2 genomic DNA as template. This PCR product was gel purified and cloned into pCRBlunt (Invitrogen) for sequencing. Full-length DNA coding for CT529 from serovars Ba, E (BOUR), E (MTW447), F (NI1), G, I, Ia, K, L1, L3, and MoPn were amplified from bacterial lysates containing 105 IFU (25) by using primers specific for serovar L2 DNA (external to the ORF). Primers sequences were 5′-GGTA TAATATCTCTCTAAATTTTG (forward) and 5′-AGATAAAAAAGGCTGTTTC (reverse) except for MoPn, which required 5′-TTTTGAAGCAGGTAGGTGAATATG (forward) and 5′-TTTACAAT AAGAAAAGCTAAGCACTTTGT (reverse). PCR-amplified DNA was cloned into pCR2.1 (Invitrogen) for sequencing.
Generation of Recombinant Vaccinia Virus.
A DNA fragment containing a modified Kozak sequence and base pairs 319–530 of cap1 was amplified from C. trachomatis L2 genomic DNA by using PCR and ligated into pSC11ss (26) DNA digested with SalI and StuI. The resulting plasmid was used to transfect CV-1 cells that were subsequently infected with wild-type vaccinia virus. Homologous recombination between the wild-type virus and plasmid DNA generated recombinant vaccinia viruses that were selected on the basis of both β-galactosidase expression and the inactivation of thymidine kinase, as described previously (27). Recombinant virus was plaque purified three times and titered after growth in human TK-143B cells. Virus preparations were treated with equal volume of 0.25 mg/ml trypsin for 30 min at 37°C and diluted in PBS before immunization of mice.
Quantitatation of C. trachomatis After Mouse Challenge.
Groups of five mice were used for all experimental and control groups. The data are representative of three independent experiments. Number of IFU per spleen was determined as previously described (8). Significance of these results was calculated by Wilcoxon's Rank Sum analysis.
Antibodies.
Anti-Cap1 polyclonal antibody was obtained by immunization of rabbits with a recombinant polypeptide (Cap11–125) encompassing the N-terminal portion of Cap1 from C. trachomatis serovar E. The immunizations were given in 3 doses of 100 μg (in incomplete Freund's adjuvant) at 3-wk intervals, and serum was harvested 5 wk after injection of the final dose. Cap11–125 protein was obtained from E. coli transformed with a pET expression plasmid containing the gene fragment encoding Cap11–125. Recombinant protein was purified by using a Ni2+-charged resin, as described by the manufacturer (Novagen).
Immunocytochemistry.
McCoy cell monolayers grown on glass coverslips were inoculated with either C. trachomatis serovar L2 or Chlamydia psittaci strain 6BC, at concentrations of 106 IFU per milliliter. After 2 h, medium was aspirated and replaced with fresh RP-10 supplemented with cycloheximide (1.0 μg/ml). Infected cells were then incubated at 37°C in 7% CO2 for 24 h. The cells were then rinsed once with PBS and fixed with methanol for 5 min. For antigen staining, fixed cell monolayers were washed with PBS and incubated at 37°C for 2 h with 1:100 dilutions of specific or control antisera. Cells were rinsed with PBS and incubated for 1 h with FITC-labeled anti-rabbit IgG (Kirkegaard and Perry Laboratories) and were stained with Evans blue (0.05%) in PBS. Coverslips were rinsed, removed, and inverted onto microscope slides in a drop of mounting fluid. Fluorescence was observed with a ×100 objective (Zeiss epifluorescence microscope) and photographed.
Results
Identification of a Chlamydia-Specific CD8+ T-Cell Antigen.
A genomic DNA fragment expression library from C. trachomatis serovar L2 was constructed in the retroviral vectors pBIB-KS1, KS2, and KS3 (Fig. 1). These vectors contain a Kozak eukaryotic translation initiation consensus sequence (16) upstream of the MCS to allow expression of polypeptides encoded by short prokaryotic chromosomal DNA fragments. The library was packaged and used to transduce the H-2d mastocytoma P815.
Figure 1.
Design of the pBIB-KS retroviral expression vectors used for eukaryotic expression of prokaryotic chromosomal DNA.
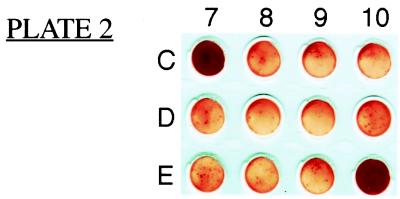
Thirty-thousand cells from each transduction pool were transferred into wells of 96-well nitrocellulose plates for screening by the Chlamydia-specific CD8+ T-cell line 69 (8). IFN-γ ELISPOT analysis was used to screen for pools containing transduced P815 cells recognized by the T cells. Two of the assay wells, 2C7 and 2E10, were strongly positive in the assay, showing confluent spots within the well (Fig. 2). To identify individual antigenic clones within these pools, transductants of P815 cells from positive pools 2C7 and 2E10 were cloned by limiting dilution. These clones were then screened individually for activation of T cells by using the IFN-γ ELISPOT assay.
Figure 2.
IFN-γ ELISPOT analysis to identify transduced cell pools recognized by Chlamydia-specific T cells. Twelve wells are shown, including positive pools 2C7 and 2E10. Positive wells contained confluent layers of ELISPOTs, precluding numerical quantitation.
A positive clone from the 2C7 pool was identified and designated 2C7.8. The C. trachomatis DNA fragment from clone 2C7.8 was amplified by using PCR with vector-specific primers. Sequencing revealed the fragment to be a short (160-bp) fragment of DNA with extremely high homology to a previously uncharacterized predicted ORF designated CT529 in the published genome sequence of C. trachomatis (15). The published sequence is from C. trachomatis serovar D. The full CT529 ORFs from serovar L2 and serovar D share 94% amino acid identity. Likewise, sequencing of DNA encoding the CT529 ORFs from several human serovars (Ba, E, F, G, IA, L1, and L3) reveals they share no less than 94% amino acid identity [see Fig. 7, published as supplemental data (www.pnas.org)].
We have designated the predicted 31-kDa gene product of CT529 class I accessible protein-1, Cap1. The C. trachomatis gene expressed by the 2E10 clones is different from that expressed by 2C7.8, and we have yet to identify the gene fragment in 2E10.
The Cap1139–147 Epitope Is Recognized by C. trachomatis-Specific T-Cells in the Context of H-2Kd.
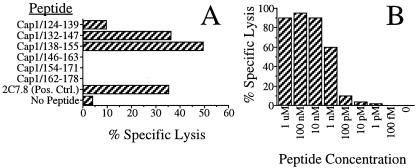
To identify the peptide epitope recognized by the T cells, we synthesized overlapping 18-mer peptides corresponding to the translation of the cap1 fragment within 2C7.8. These peptides were loaded onto surface MHC class I molecules of P815 cells and were tested for recognition by the Chlamydia-specific T cells. As shown in Fig. 3A, two peptides, Cap1132–147 and Cap1138–155, were recognized by the T-cell line. The 10 amino acids (Cap1138–147) representing the overlap of the two peptides are CSFIGGITYL. To confirm these results and to further map the epitope, truncated peptides within the Cap1138–147 sequence were synthesized and were tested for recognition by the T cells. Truncations that eliminate either Ser-139 (Cap1140–147) or Leu-147 (Cap1138–146) abrogate T-cell recognition (data not shown). These results identified the 9-mer peptide, Cap1139–147 (SFIGGITYL), as the minimal epitope recognized by the Chlamydia-specific T cells. Cap1139–147 is able to target cells for lysis at concentrations as low as 1 nM (Fig. 3B). The homologous serovar D peptide is SIIGGITYL. Chlamydia-specific T cells recognize the serovar D peptide at a minimum concentration of 10 nM. Although reproducible, this difference is likely to be insignificant. For all other experiments in this report, we used SFIGGITYL as the Cap1139–147 peptide.
Figure 3.
Assays to identify peptides able to target P815 cells for lysis by Chlamydia-specific T cells. In A, 51Cr-labeled P815 cells were pulsed with the indicated peptides, washed, and incubated with the Chlamydia-specific T-cell line 69 at an effector to target ratio (E/T) of 10:1. Targeting was evaluated by measuring the amount of 51Cr release. In B, Cap1139–147 was chemically synthesized and tested at the dilutions shown for its ability to target P815 cells for lysis by the C. trachomatis-specific T cells. The E/T ratio used in the assay was 8:1.
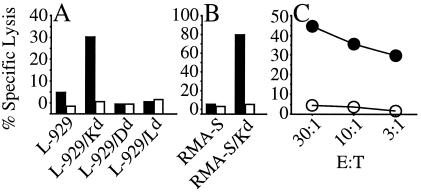
We determined the MHC restriction for T-cell recognition of Cap1139–147 by testing Cap1139–147 -specific T cells for their ability to recognize the Cap1139–147 peptide presented by a panel of L-929 cells (H-2k) transfected individually with the genes encoding the Dd, Kd, and Ld molecules. The Cap1-specific T cells were generated by stimulating T-cell line 69 for 2 consecutive weeks by using 2C7.8 as antigen-presenting cells. This enriched for only those T cells that respond to an epitope within Cap1. As shown in Fig. 4A, only the L-929 cells transfected with the Kd molecule were recognized by Cap1139–147-specific T cells. The Cap1139–147-specific T cells were also able to recognize Cap1139–147-pulsed RMA-S cells (H-2b) that had been transfected with Kd but were not able to recognize pulsed RMA-S cells that had not been transfected (Fig. 4B). Taken together, these data suggest the Cap1139–147-specific T cells recognize Cap1139–147 in the context of the Kd classical MHC class I molecule.
Figure 4.
Cap1139–147 peptide was tested for the ability to target
cells for lysis by Cap1139–147-specific T cells in a
51Cr release assay. In A and
B, the cells were tested with (solid bars) and without
(open bars) Cap1139–147 peptide. The target cells shown in
A are L-929 (H-2k) cells individually
transfected with the genes encoding the Dd, Kd,
or Ld molecules. L-929 cells alone are shown as a negative
control. In B, the target cells are RMA-S cells
(H-2b) and RMA-S cells transfected with the gene encoding
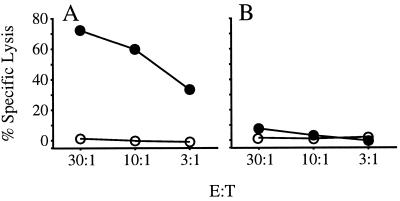
Kd. The E/T ratio used in the assay was 10:1. In
C, the target cells were C.
trachomatis-infected
( )
or uninfected
(
)
or uninfected
( )
Balb/3T3 cells.
)
Balb/3T3 cells.
The Cap1139–147-specific T cells were also able to recognize infected cells, confirming that the Cap1139–147/Kd complex is presented on the surface of cells infected with C. trachomatis (Fig. 4C).
Cap1139–147-Specific T Cells Are Primed During Murine Infection with C. trachomatis.
To confirm that murine infection with C. trachomatis primes a Cap1139–147-specific T-cell response, mice were infected with C. trachomatis and allowed to recover for 2 wk. The mice were then killed, and splenic T cells were stimulated on irradiated syngeneic spleen cells pulsed with Cap1139–147 peptide. After 5 days of stimulation, the cultures were used in a cytotoxicity assay to determine whether Cap1139–147-specific T cells were present in the culture. As shown in Fig. 5, these cultures contained Cap1139–147-specific cytolytic T cells, supporting our hypothesis that Cap1139–147-specific T cells are primed during murine infection with C. trachomatis.
Figure 5.
Chromium release assays showing recognition of Cap1139–147
by T cells primed during murine infection with C.
trachomatis. Mice were immunized i.p. with 108 IFU
of C. trachomatis serovar L2. Two weeks after infection,
spleen cells from an immunized mouse (A) or a control
mouse injected with PBS (B) were cultured for 5 days
with Cap1139–147-coated syngeneic spleen cells. Target
cells were Cap1139–147-coated
( )
or untreated
(
)
or untreated
( )
P815 cells. Graphs are representative of three experimental and three
control mice.
)
P815 cells. Graphs are representative of three experimental and three
control mice.
Immunization with a Recombinant Vaccinia Virus Expressing Cap1 Reduces the Level of C. trachomatis Infection After Challenge.
To evaluate whether an immunization strategy designed to stimulate Cap1-specific T cells would protect against C. trachomatis challenge, we constructed a recombinant vaccinia virus expressing amino acids 107–176 of Cap1. When used to immunize mice, this recombinant virus primes Cap1-specific CD8+ T cells (data not shown). A group of mice was immunized with 106 plaque-forming units (PFU) of the recombinant vaccinia i.p. and allowed to recover for 3 wk. Negative control groups were immunized with either buffer alone or with 106 PFU of wild-type vaccinia. As a positive control, a group of mice was infected i.v. with 106 IFU of C. trachomatis. The number of organisms given to the positive control group previously has been shown to be cleared within 2 wk.
After 3 wk, animals in each of the groups were challenged i.v. with 106 IFU of C. trachomatis. Three days after challenge, the mice were killed, and the number of IFU per spleen was determined. The mean number of organisms found in the spleens of animals immunized with the vaccinia virus expressing Cap1 (7.1 × 104) was 2.6-fold fewer (P < 0.01) than in animals in the control groups immunized with either buffer (1.8 × 105) or wild-type vaccinia (1.9 × 105). Animals in the positive control group had 77-fold fewer organisms (2.4 × 103) per spleen than animals in the negative control groups (P < 0.01). These data suggest that immunization with only a fragment of Cap1 can afford a modest, yet significant, level of protection against C. trachomatis.
Cap1 Localizes to the Inclusion Membrane of C. trachomatis-Infected Cells.
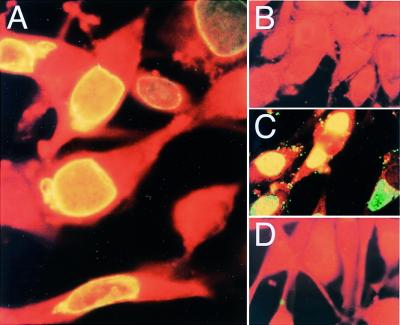
The CD8+ T-cell response to C. trachomatis suggests that part of the Cap1 protein has access to the host cell cytosol during bacterial replication. To determine the intracellular location of Cap1, a rabbit polyclonal antibody directed against a recombinant polypeptide encompassing the N-terminal 126 amino acids of Cap1 was used to stain McCoy cells infected with C. trachomatis (Fig. 6). Cap1-specific antibody localizes to the inclusion membranes of C. trachomatis-infected cells rather than the organisms contained within these inclusions (Fig. 6A). Conversely, rabbit antibody raised against whole infectious C. trachomatis particles clearly labeled bacteria within the inclusion as well as organisms bound to the cell surface (Fig. 6C). The anti-Cap1 antibody does not stain C. psittaci-infected cells (Fig. 6D), nor does preimmune sera stain cells infected with C. trachomatis (Fig. 6B). These results show that Cap1 is secreted from the bacteria and becomes associated, either directly or indirectly, with the inclusion membrane.
Figure 6.
Fluorescence immunocytochemistry of McCoy cells infected with C. trachomatis (A–C) or C. psittaci (D), all fixed 24 h postinfection. In A and D, cells were stained with rabbit anti-Cap1 antiserum. In B, cells were stained with preimmune serum. In C, cells were labeled with an antiserum directed against C. trachomatis elementary bodies.
Discussion
By using an expression cloning strategy, we have identified a gene from C. trachomatis that encodes an antigen, designated Cap1, capable of stimulating MHC class I restricted CD8+ T cells. No proteins have previously been described for this gene, although during sequencing of the C. trachomatis genome (15) the CT529 ORF was predicted to encode a hypothetical protein of 31 kDa. The ability to identify epitopes by expression cloning is particularly useful when applied to organisms such as C. trachomatis where large numbers of organisms are difficult to obtain, and where techniques for genetic manipulation of the organism are unavailable. However, one difficulty in identifying CD8+ T-cell antigens contained in libraries of bacterial DNA stems from inefficient expression of prokaryotic genes in eukaryotic cells. This is primarily because of differing translational requirements between prokaryotic and eukaryotic cells, including incompatible mRNA translation initiation sequences and alternative codon usage. The latter may result in limited prokaryotic polypeptide synthesis within a eukaryotic cell, where pools of specific rare tRNAs may become depleted. This problem is alleviated by using the approach described here, where only one to two cDNAs integrate after transduction.
Identification of Cap1 and epitopes such as Cap1139–147 will allow us to examine the extent to which experimental vaccines that stimulate CD8+ T cells can protect against C. trachomatis. For Cap1 to stimulate CD8+ T cells and be presented by MHC class I, it must access the cytosol of infected cells during the Chlamydia developmental cycle for processing and presentation by MHC class I. It is unlikely that an alternative processing pathway is solely responsible for presentation of the Cap1 epitope. Alternative processing has not been shown to occur in nonprofessional APCs such as Balb-3T3 cells, which we show can process and present this antigen. However, in vivo, some C. trachomatis antigens that remain confined to the vacuole may be presented via an alternative pathway. It is possible that human CD8+ T cells specific for the C. trachomatis major outer membrane protein, recently reported by Kim et al. (28), arise by such a mechanism.
We have previously shown that adoptive transfer of the CD8+ T-cell line used to identify Cap1 affords a modest but significant reduction in C. trachomatis after challenge (8). However, as noted above, the T-cell line transferred in the previous study appears to contain T cells with at least one additional specificity, and the adoptive transfer experiments used more pathogen-specific CD8+ T cells than might be stimulated normally by vaccination or infection. To determine whether immunization with Cap1139–147 alone afforded protection from C. trachomatis challenge, we vaccinated mice with recombinant vaccinia virus expressing a fragment of Cap1. We show here that immunization with this recombinant vaccinia reduced the number of organisms recovered from spleens after challenge. Mice that have recovered from previous C. trachomatis infection showed greater protection from challenge, although these convalescent mice are likely to have memory T cells against several C. trachomatis antigens as compared to mice vaccinated with a single antigen. As we use the procedures outlined here to identify additional antigens, we can incorporate them into vaccines designed to prime CD8+ T cells of several specificities. Full protection with a vaccine may require priming a CD8+ T-cell response to several epitopes, perhaps with boosting, to stimulate sufficient numbers of T cells. It also appears from work in several systems that during infection with intracellular bacteria such as C. trachomatis, CD8+ T cells act in concert with other elements of the immune system, including CD4+ T cells, to provide additional levels of adaptive immunity. Certainly an ideal vaccine for use in outbred populations such as humans would contain several antigens presented by MHC class I, as well as several antigens presented by MHC class II.
One difficulty in generating protective immunity to C. trachomatis is the serovar specificity of the response. Often this is a result of immune responses directed to the major outer membrane protein (MOMP), the primary determinant of a serovar. Although the antibody response to MOMP is dominant during natural infection, stimulation of other elements of the immune system such as the CD8+ T-cell response characterized here may significantly augment the efficacy of a vaccine. Additionally, Cap1 is virtually identical among human C. trachomatis serovars. Therefore, a vaccine incorporating Cap1 might enable the vaccine to protect against all C. trachomatis serovars.
An ortholog of Cap1 is present in the published sequence of C. pneumoniae (29). Residues 16–291 of Cap1 are 35% identical and 54% similar to residues 38–322 of CPn0648, a predicted Chlamydia-specific hypothetical protein from C. pneumoniae. If this predicted C. pneumoniae protein also has access to the cytosol of host cells, it may allow for its use in immunization strategies designed to reduce coronary heart disease and other conditions in which C. pneumoniae has been implicated.
In the study presented here, we use a human serovar of C. trachomatis, infecting mice systemically. Genital infection of mice with human strains of C. trachomatis does not yield the reproducible infections necessary for challenge studies. A mouse biovar of C. trachomatis, designated MoPn, can be used to infect the genital tract of mice and may serve as a model of mucosal infection and immunity. Interestingly, the Cap1 homologue in MoPn is less conserved (62% identical), raising the question of whether the protein functions in the same manner during replication of this biovar. We have tested the homologous 9 amino acids from MoPn, and Cap1-specific T cells do not recognize this peptide (data not shown). Additional experiments will be required to determine whether protective CD8+ T cells specific for the MoPn Cap1 homologue are stimulated after murine genital tract infection with MoPn.
The function of Cap1 is unknown and remains highly speculative, as no strong homologies have been observed when this 31-kDa protein is compared with proteins in available databases. Cap1 lacks the predicted characteristics of inclusion membrane proteins such as those described by Bannantine et al. (30, 31). However, as shown here, antibodies to Cap1 labeled the inclusion membranes of C. trachomatis-infected cells, demonstrating that Cap1 also associates with the inclusion membrane. Whether Cap1 is an integral membrane protein or indirectly localizes to the membrane by association with other membrane proteins remains to be determined. In either case, our data showing MHC class I presentation of a Cap1-derived peptide suggest that at least a portion, or domain, of the protein is accessible to the cytoplasmic face of the inclusion membrane. Indeed, Cap1 contains a COOH-terminal tripeptide sequence (“ARA” in several C. trachomatis serovars), which is known to function as a microbody-targeting signal for some proteins (32–34). Microbodies (glycosomes, glyoxisomes, peroxisomes, and lysosomes) reside in the cytosol.
As obligate intracellular pathogens, members of the Chlamydia genus are nutrient and energy scavengers, and little is known of how Chlamydia “samples” the cytosolic microenvironment. C. trachomatis proteins at the interface of the inclusion and the cytosol may have evolved specific critical roles in the Chlamydia life cycle such as scavenging nutrients, altering vesicular trafficking, or modifying other host-cell functions essential for the complex interaction of C. trachomatis with eukaryotic cells. If Cap1 has a critical role in the Chlamydia life cycle, it would also serve as a possible target for antibiotic development.
The T-cell expression cloning approach described here to identify Cap1 uses retroviral-meditated transexpression of polypeptides encoded by short chromosomal DNA fragments. This approach, unlike biochemical methods of peptide extraction that depend on the quantity of peptide present, can be used to identify both dominant and subdominant CD8+ T-cell epitopes. This method will be useful in the identification of additional epitopes from C. trachomatis and can be applied to the identification of antigens from other bacterial pathogens where there is evidence of a CD8+ T-cell response. It may be a particularly advantageous technique when applied to intracellular pathogens that are difficult or unsafe to grow in large quantities, such as Mycobacterium tuberculosis.
Supplementary Material
Acknowledgments
We thank K. Grabstein, M. Alderson, J. Mekalanos, D. Carter, and W. Loomis for reviewing the manuscript and for their helpful suggestions, A. Bhatia for providing L2 DNA, S. Engardt, S. Steen, and L. Ballweber for their excellent technical assistance, N. Hosken for ELISPOT development, and Y. Skeiky (Corixa Corporation, Seattle, WA) for production of Cap1-specific antiserum. This work was supported in part by National Institutes of Health Grants AI39558 (M.N.S.) and AI31448 (M.N.S. and M.F.L.) and by an award from the American Cancer Society (M.S.).
Abbreviations
- IFU
inclusion forming units
- MCS
multiple cloning site
References
- 1.Burstein G R, Gaydos C A, Diener-West M, Howell M R, Zenilman J M, Quinn T C. J Am Med Assoc. 1998;280:521–526. doi: 10.1001/jama.280.6.521. [DOI] [PubMed] [Google Scholar]
- 2.Ward M E. In: Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens R S, editor. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 171–210. [Google Scholar]
- 3.Wong Y K, Gallagher P J, Ward M E. Heart. 1999;81:232–238. doi: 10.1136/hrt.81.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shor A, Phillips J I. J Am Med Assoc. 1999;282:2071–2073. [Google Scholar]
- 5.Igietseme J U, Ramsey K H, Magee D M, Williams D M, Kincy T J, Rank R G. Reg Immunol. 1993;5:317–324. [PubMed] [Google Scholar]
- 6.Igietseme J U, Magee D M, Williams D M, Rank R G. Infect Immun. 1994;62:5195–5197. doi: 10.1128/iai.62.11.5195-5197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su H, Caldwell H D. Infect Immun. 1995;63:3302–3308. doi: 10.1128/iai.63.9.3302-3308.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starnbach M N, Bevan M J, Lampe M F. J Immunol. 1994;153:5183–5189. [PubMed] [Google Scholar]
- 9.Van Voorhis W C, Barrett L K, Sweeney Y T, Kuo C C, Patton D L. J Infect Dis. 1996;174:647–650. doi: 10.1093/infdis/174.3.647. [DOI] [PubMed] [Google Scholar]
- 10.Perry L L, Feilzer K, Caldwell H D. J Immunol. 1997;158:3344–3352. [PubMed] [Google Scholar]
- 11.Lampe M F, Wilson C B, Bevan M J, Starnbach M N. Infect Immun. 1998;66:5457–5461. doi: 10.1128/iai.66.11.5457-5461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison R P, Feilzer K, Tumas D B. Infect Immun. 1995;63:4661–4668. doi: 10.1128/iai.63.12.4661-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su H, Morrison R P, Watkins N G, Caldwell H D. J Exp Med. 1990;172:203–212. doi: 10.1084/jem.172.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moulder J W. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, et al. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pear W S, Scott M L, Nolan G P. In: Methods in Molecular Medicine: Gene Therapy Protocols. Robbins P, editor. Totowa, NJ: Humana; 1996. pp. 41–57. [Google Scholar]
- 18.Howard L, Orenstein N S, King N W. Appl Microbiol. 1974;27:102–106. doi: 10.1128/am.27.1.102-106.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson E B, Smadel J E. Am J Hyg. 1951;53:326–331. doi: 10.1093/oxfordjournals.aje.a119457. [DOI] [PubMed] [Google Scholar]
- 20.Kinsella T M, Nolan G P. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 21.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 22.Jackson R J, Howell M T, Kaminski A. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 23.Izumi M, Miyazawa H, Kamakura T, Yamaguchi I, Endo T, Hanaoka F. Exp Cell Res. 1991;197:229–233. doi: 10.1016/0014-4827(91)90427-v. [DOI] [PubMed] [Google Scholar]
- 24.Rees S, Coote J, Stables J, Goodson S, Harris S, Lee M G. BioTechniques. 1996;20:102–104. doi: 10.2144/96201st05. [DOI] [PubMed] [Google Scholar]
- 25.Denamur E, Sayada C, Souriau A, Orfila J, Rodolakis A, Elion J. J Gen Microbiol. 1991;137:2525–2530. doi: 10.1099/00221287-137-11-2525. [DOI] [PubMed] [Google Scholar]
- 26.Earl P L, Koenig S, Moss B. J Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarti S, Brechling K, Moss B. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S K, Angevine M, Demick K, Ortiz L, Rudersdorf R, Watkins D, DeMars R. J Immunol. 1999;162:6855–6866. [PubMed] [Google Scholar]
- 29.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman R W, Olinger L, Grimwood J, Davis R W, Stephens R S. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 30.Bannantine J P, Rockey D D, Hackstadt T. Mol Microbiol. 1998;28:1017–1026. doi: 10.1046/j.1365-2958.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- 31.Rockey D D, Lenart J, Stephens R. Infect Immun. 2000;68:5473–5479. doi: 10.1128/iai.68.10.5473-5479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blattner J, Swinkels B, Dorsam H, Prospero T, Subramani S, Clayton C. J Cell Biol. 1992;119:1129–1136. doi: 10.1083/jcb.119.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommer J M, Cheng Q L, Keller G A, Wang C C. Mol Biol Cell. 1992;3:749–759. doi: 10.1091/mbc.3.7.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih S, Hwang H Y, Carter D, Stenberg P, Ullman B. J Biol Chem. 1998;273:1534–1541. doi: 10.1074/jbc.273.3.1534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.