Abstract
The new Golgi in the protozoan parasite Trypanosoma brucei grows near to the old and adjacent to the growing new endoplasmic reticulum exit site. Growth is now shown to be at least a two-stage process, in which a representative matrix marker (GRASP) and enzyme (GntB) are delivered to the site of assembly, followed ≈10 min later by a COPI component (ε-COP) and a trans-Golgi network (TGN) marker (GRIP70). A secretory cargo marker (signal sequence-YFP) appeared early near the new endoplasmic reticulum exit site but did not enter the Golgi until the second stage. Together these data suggest that structural and enzymatic components of the new Golgi stack are laid down first, followed by those needed to move and sort the cargo passing through it.
Keywords: COP, Golgi matrix, organelle biogenesis, trans-Golgi network, transport
The Golgi apparatus is an essential component of the eukaryotic secretory system. It is often composed of stacks of flattened cisternae that function in the modification and sorting of newly synthesized proteins made in the endoplasmic reticulum (ER). Exiting from the ER exit site, newly synthesized proteins are packaged into COPII-coated vesicles and ferried to the cis face of the Golgi stack. Several models have been put forward to explain transport through the Golgi, but in all cases COPI vesicles play a key role in mixing the transiting cargoes with resident enzymes in an ordered manner so that posttranslational modifications can be performed. Cargoes are then sorted in the late Golgi compartment, the trans-Golgi network (TGN), before onward delivery to their final destinations (1, 2).
During the cell cycle, the Golgi undergoes biogenesis to help ensure propagation through successive generations (3). Biogenesis involves duplication followed by partitioning between daughter cells. Partitioning has been most studied, particularly in mammalian cells. The present weight of evidence suggests that the Golgi ribbon helps mediate entry into mitosis, whereupon it undergoes fragmentation, vesiculation, and dispersal throughout the mitotic cell cytoplasm, followed by reassembly during telophase (refs. 4–8; but see ref. 9).
Golgi duplication has been less studied, in part because there are several hundred copies of the Golgi stack in mammalian cells subsumed within the Golgi ribbon (10), precluding direct observations on the assembly of new copies. Studies have, therefore, been restricted to analysis of Golgi reassembly after treatment with drugs such as Brefeldin A (11–14), although it is not clear to what extent this is a suitable model for biogenesis.
The difficulty of studying duplication in mammalian cells has led to the search for simpler systems, with fewer Golgi. Protozoan parasites such as Toxoplasma gondii and Trypanosoma brucei have a single Golgi stack, whereas the budding yeast Pichia pastoris has up to six (15–20). Fewer Golgi make it much easier to follow the duplication process in live cells using video fluorescence imaging technology. In T.gondii, the Golgi stack grows by a process of lateral extension followed by medial fission, suggesting that the old Golgi acts as a template for the assembly of the new (19). In contrast, In P.pastoris, new Golgi assemble de novo, suggesting that the old plays no role in the construction of the new. Instead, the suggestion has been that the new ER exit sites determine the site of new Golgi assembly (21). Similar results have been obtained In T.brucei, in which the single new Golgi stack appears at about the same time next to the new ER exit site (17), at a position that is determined, at least in part, by a structure containing TbCentrin2 (22). However, photobleaching experiments have shown that at least one putative Golgi enzyme is transferred from the old Golgi to the new during this assembly process (17). This transfer in turn suggests that the new Golgi may be the result of the combined actions of the old Golgi and the new ER exit site.
These data raise the issue of coordinating the outputs of the old Golgi and the new ER exit site to ensure correct assembly of the new Golgi. They also raise the question as to whether there is an order of assembly. At one extreme, one could imagine that all of the Golgi components are delivered together and are then sorted out. Alternatively, one might imagine that these components are delivered in a specific sequence and that Golgi assembly is an ordered process. We now show that the latter is correct, using additional markers tagged with variants of GFP and stably expressed In T.brucei cell lines. At least two distinct stages in the process have been identified that seem to represent the conversion of an inactive Golgi to one that can function in secretion.
Results and Discussion
Characterization of Additional Marker Proteins.
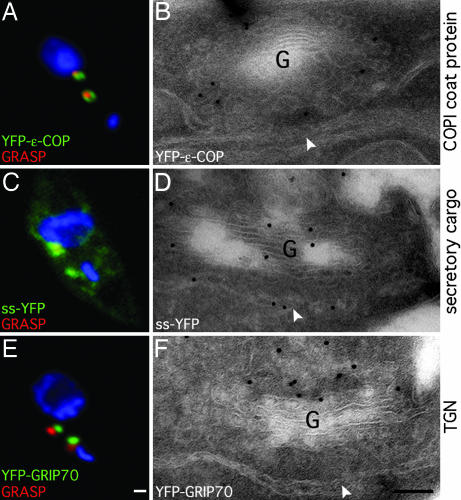
Table 1 summarizes the markers used in the present study. Earlier work characterized a representative Golgi matrix protein, GRASP, as well as GntB, a putative GlcNAc transferase. ER exit sites were marked by using Sec13p, a COPII coat component (17). In the present work, a marker for COPI coats was characterized. Coatomer comprises a hetero-polymer of seven subunits that, together with ARF, assemble the COPI coats that are needed for intra-Golgi transport (23). In mammalian cells, a CHO cell line has been described that is temperature-sensitive for the ε-COP subunit, and, at the nonpermissive temperature, the cells can be rescued by expression of a GFP-tagged version of ε-COP (24, 25). The T.brucei homolog of ε-COP was, therefore, chosen for tagging and stable expression. As shown in Fig. 1A, it localized to the Golgi, as a ring shape, surrounding the region marked by GRASP. Because earlier immuno-electron microscopy (immuno-EM) had shown that GRASP is present throughout the Golgi stack (17), these data suggested that ε-COP was present at the rims. This location was confirmed by immuno-EM, the labeling for ε-COP being found predominantly at the cisternal rims, from which COPI vesicles are known to bud (Fig. 1B) (26).
Table 1.
Summary of T. brucei markers
| Markers | Locations | Roles | References |
|---|---|---|---|
| Sec13p | ER exit site | COPII component: ER to Golgi transport | 42 |
| GRASP | Golgi stack | Golgi matrix: Golgi stacking and signaling | 4, 30 |
| GntB | Golgi stack | Golgi enzyme: Putative GlcNAc transferase | 31 |
| ε-COP | Golgi rim | COPI coatomer: Golgi/ER and intra-Golgi transport | 23, 25 |
| ss-YFP | Golgi and ER | Fluorescent secretory cargo | 29 |
| GRIP70 | TGN | Golgin: Endosome/TGN transport | 43, 44 |
Fig. 1.
Localization of stably expressed YFP-tagged markers. (A, C, and E) Fluorescence imaging of fixed cells expressing the indicated constructs (in green) labeled with antibodies to GRASP (red) and stained with DAPI (blue), marking the nucleus and the kinetoplast (the smaller structure). The images are of cells with one old and one new Golgi. The old Golgi is located near to the nucleus, whereas the new Golgi is located closer to the kinetoplast. (Scale bar: 1 μm.) (B, D, and F) Immuno-EM analysis of fixed cells expressing the indicated constructs labeled with antibodies to GFP followed by protein-A gold. G, Golgi stack; arrowheads, ER exit site. (Scale bar: 100 nm.) Note that the COPI coat protein, ε-COP, is mostly present at the cisternal rims by immuno-EM (B), and by fluorescence (A) forms a ring around the Golgi stack marked by the matrix protein GRASP. The secretory cargo, ss-YFP, is found in the Golgi and the ER (C and D). The TGN golgin, GRIP70, lies adjacent to the Golgi stack (E and F), on the side opposite to the ER exit site (F).
GRIP70 was used as a marker for the TGN. It is a coiled-coil protein that contains a conserved GRIP domain shown to localize to the TGN in many cell types (27, 28). Tagging the full-length GRIP70 with YFP at the N terminus and stable expression In T.brucei showed that the fluorescence was adjacent to the Golgi stack marked by GRASP (Fig. 1E). Immuno-EM localized it to a tubulo-reticular network adjacent to the side of the Golgi stack opposite the ER exit site, confirming its location to the TGN (Fig. 1F).
To determine when the new Golgi was functional for transport, a soluble secretory cargo was constructed. The signal sequence for procyclin, the major surface coat protein (29), was attached to the N terminus of YFP, and stable lines were generated. This construct, signal sequence-YFP (ss-YFP), was found predominantly in the Golgi, and in the ER, by fluorescence (Fig. 1C). There was no surface fluorescence because the construct was secreted (data not shown). Immuno-EM confirmed these locations, in that labeling was found over the Golgi stack and all elements of the ER (Fig. 1D).
Each of these stable cell lines was also analyzed by Western blotting to show that each fusion protein was expressed as a single species of the correct molecular weight (Fig. 6, which is published as supporting information on the PNAS web site).
A Golgi Enzyme and Matrix Protein Arrive Simultaneously at the New Golgi.
To determine the arrival times of the different markers at the new Golgi, a reference protein was needed. GRASP was chosen as a well characterized and representative matrix marker (30). It also had the additional advantages that polyclonal antibodies useful for immunofluorescence microscopy had been successfully raised (17) and it could also be tagged with monomeric red fluorescent protein (mRFP) (see below). These properties facilitated the double-labeling experiments needed to determine the relative arrival times.
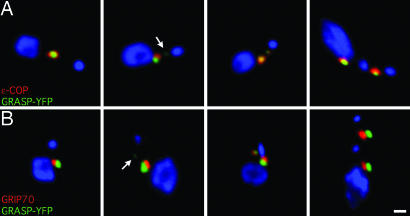
GntB was used as a representative Golgi enzyme, earlier characterized as a putative GlcNAc transferase by comparison with a related enzyme In Dictyostelium discoideum (31, 32). Photobleaching studies showed that this putative enzyme moves from the old Golgi to the new, but the timing relative to GRASP was not determined (17). To determine the relative timing, cells stably expressing YFP-tagged GntB were fixed and stained with antibodies against GRASP (Fig. 2A). They were then surveyed for new Golgi that had GRASP, but not GntB, or vice versa, which would indicate different times of arrival. We were unable to find any such examples. GntB colocalized with GRASP at all stages of the cell cycle. It was even found in the smallest new Golgi detectable by GRASP (Fig. 2A, arrows), suggesting concomitant arrival of the two.
Fig. 2.
Appearance of YFP-tagged markers at the new Golgi. (A–D) Fluorescence imaging of fixed cells expressing the indicated constructs (in green) labeled with antibodies to GRASP (red). Panels are arranged from left to right, as the cells progress through the cycle. Cells at left have a single (old) Golgi, cells in the middle have a growing new Golgi, and cells at right have more additional Golgi that appear just before cytokinesis. Arrows indicate first appearance of the new Golgi, and Insets show the separated green and red channels. Note that GntB colocalizes with GRASP at all stages of the cell cycle. In contrast, YFP-ε-COP, ss-YFP, and YFP-GRIP70 are not present at the earliest stage defined by the appearance of GRASP. Furthermore, ss-YFP is sometimes found immediately adjacent to the new Golgi at this early stage (arrowhead). (Scale bar: 5 μm.) (E) Quantitation of the total YFP fluorescence at the old Golgi (early in the cell cycle) in cells expressing the indicated constructs tagged with YFP. Results were expressed as the mean ± SEM (n = 6).
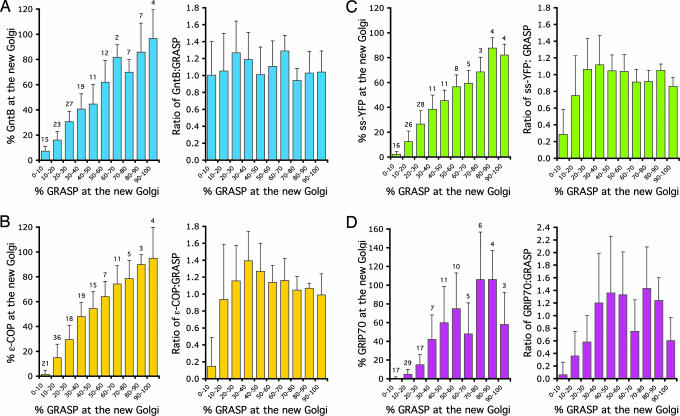
This conclusion was further strengthened by quantitating the fluorescence intensity of both markers during new Golgi growth. The new Golgi grew until it was the same size as the old. The percentage of GntB at the new Golgi (relative to the old) was plotted against the percentage of GRASP. The latter data points were binned for every 10% of GRASP at the new Golgi to simplify presentation (Fig. 3A Left). The clear proportionality was further emphasized by plotting the ratio of GntB to GRASP at the new Golgi (as a function of GRASP). As shown in Fig. 3ARight, the ratio was maintained at around 1 throughout new Golgi growth, strongly supporting the idea that GntB and GRASP both arrive simultaneously at the new Golgi assembly site.
Fig. 3.
Quantitation of markers at the new Golgi. Cells stably expressing GntB-YFP (A), YFP-ε-COP (B), ss-YFP (C), and YFP-GRIP70 (D) were fixed and labeled with anti-GRASP antibodies as in Fig. 2, and the levels of YFP and GRASP fluorescence were quantitated and expressed as the percentage in the new Golgi relative to the old. (Left) Plots of the percentage of YFP-tagged protein against the percentage of GRASP, with the latter data points binned for every 10% of GRASP at the new Golgi. (Right) Plots of the ratio of YFP-tagged protein to GRASP against the binned data for GRASP. Each panel was constructed from three experiments and a total of 127 (A), 139 (B), 115 (C), and 109 (D) experimental points. Note that GntB and GRASP are present in similar amounts at the new Golgi at all sizes, and the ratio remains relatively constant at ≈1. In contrast, YFP-ε-COP, ss-YFP, and YFP-GRIP70 were mostly absent from the new Golgi containing up to 10% GRASP, and the ratio was <0.3, arguing that these are later arrivals. Results were expressed as the mean ± SEM. The number of experimental points for each binned data set is indicated above the error bars.
COPI Coatomer Arrives After the Golgi Matrix Protein.
The arrival of the COPI coatomer and GRASP at the new Golgi site was next compared by using the cell line stably expressing ε-COP fused to YFP. Fixed cells were double-labeled by using antibodies to GRASP, and fluorescence of the new Golgi was examined. At the earliest stages of growth, small Golgi defined by GRASP had little if any detectable ε-COP (Fig. 2B, arrows). The converse was never observed, that is, ε-COP fluorescence in the absence of GRASP. Quantitation confirmed these observations. New Golgi containing <10% GRASP had <2% of ε-COP (Fig. 3B Left) and a ratio of ε-COP to GRASP of 0.15 (Fig. 3B Right). Thereafter, ε-COP was added together with GRASP to the growing Golgi.
One concern is that these results might have been affected by the relative fluorescence intensity of the two markers. The delayed detection of ε-COP at the new Golgi might simply reflect a lower fluorescence signal when compared with GRASP. A lower signal seems unlikely because the protein (Fig. 6) and fluorescence (Fig. 2E) levels of ε-COP were similar to those of GntB, which was readily detectable in all GRASP-labeled Golgi. Furthermore, the reciprocal experiment gave the same result. Cells stably expressing GRASP-YFP were fixed and labeled with polyclonal antibodies to endogenous ε-COP. The smallest Golgi contained GRASP-YFP but not ε-COP (Fig. 4A, arrow).
Fig. 4.
Appearance of endogenous ε-COP and GRIP70 at the new Golgi. Fluorescence imaging of cells stably expressing GRASP-YFP (green), fixed and stained with antibodies against ε-COP (A) or GRIP70 (B) (red). The nucleus and kinetoplast were stained with DAPI (blue). Panels are arranged from early (Left) to late (Right) stages of the cell cycle. Note that the endogenous ε-COP and GRIP70 were not present at early stages in the new Golgi marked by GRASP (arrows). (Scale bar: 1 μm.)
Delayed Entry of Secretory Cargo into the New Golgi.
The absence of ε-COP in the small, new Golgi argued that it was not functional for transport. To assay transport, cells stably expressing ss-YFP were fixed and labeled for GRASP. ss-YFP was present throughout the ER and was concentrated in the Golgi. However, it could not be detected in the smallest new Golgi defined by GRASP (Fig. 2C, arrows). Quantitation confirmed this result, showing that new Golgi containing <10% GRASP had <2% ss-YFP (Fig. 3C Left) and a ratio of ss-YFP to GRASP of 0.29 (Fig. 3C Right). Thereafter, the ss-YFP appeared in the new Golgi in amounts proportional to those of GRASP. Again, there was a concern that the results might simply reflect low fluorescence levels of ss-YFP, but the protein levels (Fig. 6) were only slightly lower and the fluorescence (Fig. 2E) levels were similar to GntB that was readily seen in the smallest Golgi defined by GRASP (Fig. 2A, arrows).
Interestingly, whereas ss-YFP was excluded from the smallest Golgi, it was found to accumulate adjacent to it up until the new Golgi contained 10–25% of the final levels of GRASP (Fig. 2C, arrowhead). The precise site of accumulation is presently unclear although it does not seem to be the ER exit site, as marked by the COPII component, Sec13p (data not shown). Because Sec13p and GRASP arrive at the new site at the same time (17), one speculative possibility is that the cargo is present in COPII-derived vesicles, awaiting entry until the Golgi becomes functional. It will be beneficial to conduct studies using other markers to identify them.
The TGN GRIP Protein Arrives After the Golgi Matrix Protein.
The delayed arrival of ε-COP and ss-YFP at the Golgi stack defined by GRASP raised the question as to the arrival time of other parts of the Golgi apparatus. The arrival of the TGN was tested by using a stable line expressing YFP-GRIP70. In fixed cells labeled for GRASP, it was possible to find GRASP-labeled new Golgi that lacked GRIP70 but not vice versa (Fig. 2D, arrows). Quantitation provided confirmation, showing that new Golgi containing <10% GRASP had <1% GRIP70 (Fig. 3D Left) and a ratio of GRIP70 to GRASP of 0.06 (Fig. 3D Right). These results were not the consequence of low fluorescence levels because GRIP70 was 2- to 3-fold brighter than GntB (Fig. 2E) and was expressed at a similar level to GntB (Fig. 6). The result was also not an artifact of expressing a YFP-tagged GRIP70 because the same pattern was observed when cells expressing GRASP-YFP were fixed and stained with polyclonal antibodies to the endogenous GRIP70 (Fig. 4B, arrow). Together, these data suggest that the TGN is recruited to the Golgi region after the Golgi stack.
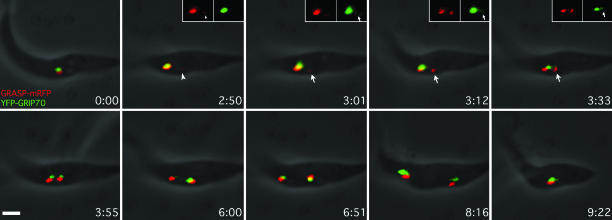
Interestingly, the percentage of GRIP70 at the new Golgi (and the ratio of GRIP70 to GRASP) varied considerably as the new Golgi grew (Fig. 3D). The reason for this variation became clear when TGN duplication was followed by using live-cell video microscopy. Stable cell lines expressing both GRASP-mRFP and YFP-GRIP70 were generated to mark the Golgi stack and the TGN, respectively, and duplication was followed through an entire cell cycle (≈9 h). Representative frames of the sequence are shown in Fig. 5, and the entire sequence is found in Movie 1, which is published as supporting information on the PNAS web site. YFP-GRIP70 arrived ≈10–15 min later than GRASP-mRFP (arrows). The levels of YFP-GRIP70 in the old and new then seemed to oscillate in opposite phases, providing an explanation for the wide variation in the percentage and ratio measurements of GRIP70 during later Golgi growth. This oscillation was not due to the Golgi moving in and out of the focal plane, because such behavior was not observed with GRASP-mRFP. GRIP70 is a peripheral membrane protein so exchange could be occurring through the cytoplasm. However, photobleaching experiments suggest that the half-time for recovery of GRIP70 is about five times longer than that for GRASP (a myristoylated, peripheral protein) (33) and more similar to that for GntB (a putative spanning protein). In other words, GRIP70 behaves more like an integral membrane protein than a peripheral one. The oscillations might, therefore, reflect the duplication process, or the en bloc consumption of the TGN as part of a cisternal maturation process. Nevertheless, such possibilities can only be investigated further once additional markers for the TGN are identified and characterized.
Fig. 5.
Arrival of GRIP70 at the new Golgi. Cells stably expressing GRASP-mRFP (red) and YFP-GRIP70 (green) were imaged during one cell cycle, and the fluorescence and phase images were merged. Selected images (and Insets) show the arrival of GRIP70 (arrows) ≈10–15 min after GRASP (arrowheads) at the new Golgi. The last frame shows one of the daughter cells initiating another round of the cell cycle. The complete image sequence is available in Movie 1. (Scale bar: 1 μm.) Numbers refer to hours:minutes of imaging.
A Two-Stage Model for Golgi Assembly.
The data suggest the following model for new Golgi assembly in the protozoan parasite T.brucei. The matrix marker, GRASP, and the putative enzyme, GntB, arrive at the new site at the same time as the ER exit site marker, Sec13p. A cargo marker, ss-YFP, also appears near to the ER exit site and the Golgi, perhaps arrested in COPII-derived vesicles. About 10 min later, after the arrival of coatomer (marked by ε-COP), transport of cargo through the Golgi occurs. The TGN is also added at this time, or perhaps a little later, allowing transport out of the Golgi. One could, therefore, argue that structural and enzymatic components of the Golgi are first laid down, followed by the addition of those components that are needed for cargo transport and sorting. Naturally, this conclusion is tentative and will require analysis of more types of markers and more examples of each marker. It will also be important to determine how each marker gets to the new site and what initiates and sustains this ordered delivery. Lastly, it will be crucial to carry out corresponding EM studies so as to identify and characterize the intermediates on this Golgi assembly pathway.
Whether Golgi duplication in mammals follows the same pattern has still to be determined. However, it is instructive to compare these results with those obtained studying Golgi reassembly after washout of drugs such as brefeldin A. Early work by Sandoval and colleagues (11) suggested a cis to trans assembly of the Golgi, consistent with what we interpret as assembly of the Golgi stack before the TGN. Later work by both the Linstedt (14) and Storrie (12) groups showed that certain matrix proteins appear at the reassembly site before other markers such as Golgi enzymes. This order differs from the present work for reasons that might simply reflect the use of different enzyme markers. It will, therefore, be important to look at more enzyme markers In T.brucei and other types of markers in mammalian cells, including cargo, to determine when transport is initiated in the reassembling Golgi. Only then will it be possible to determine the extent to which these processes are the same or different in the two organisms. If they are different, then drug washout might not be a valid model for Golgi duplication. Alternatively, the duplication mechanisms might be different in the two organisms. In this context, it is worth pointing out that the medial fission seen during Golgi duplication In T.gondii has no obvious counterpart In T.brucei orP.pastoris (17, 19, 21). Furthermore, in mammalian cells, Golgi partitioning during mitosis, involving fragmentation, vesiculation, and dispersal, is not universally observed. Plants, fungi, and protozoan parasites do not disassemble their Golgi during mitosis and cell division, nor do they arrest membrane traffic (3, 34). It will, therefore, become crucial to study Golgi duplication in animal cells directly before conclusions as to the generality of Golgi biogenetic mechanisms can be drawn.
Finally, the observation that the new Golgi reaches ≈10% of its final size before it functions is intriguing. EM reconstructions of serial thin sections show that the volume of the T.brucei Golgi is ≈0.06 μm3, equivalent to ≈100 COPI-sized vesicles (35). Because there are typically five compartments in the T.brucei Golgi [cis-Golgi-network (CGN), TGN, and three cisternae; J. Yelinek and G.W., unpublished data], each will comprise at most a few vesicles when the Golgi begins to work. Theoretically, one would need only one representative of each compartment to generate a functioning Golgi. The observed 10-min delay might, therefore, represent the time needed to deliver these representatives to the site where the new Golgi is assembled.
Materials and Methods
Cell Culture.
The procyclic T.brucei strain YTat1.1 was used throughout the study. Cells were grown and transfected as described (36).
DNA Constructs.
T.brucei ε-COP and GRIP70 have been identified previously by others (27, 37). Full-length coding sequences were amplified by PCR from genomic DNA of T.brucei strain 29.13 and cloned into a variant of the pXSGFPM3FUS expression vector (38, 39) where the GFP was replaced by either enhanced yellow fluorescent protein (EYFP) (Clontech) or mRFP (40). A soluble secretory fluorescent protein, ss-YFP, was constructed by fusing the signal sequence of procyclin (27 aa) (29) to the N terminus of YFP in the expression vector, pHD1034.
Antibodies and Western Blotting.
Antibodies against GRASP have been described (17). Polyclonal antibodies against ε-COP and GRIP70 were raised against the peptides CTKTDARKAEDIAAFHAEKE (amino acids 27–45) or CEKDEEASELLKEGRAPSQE (amino acids 123–141), respectively, and affinity-purified by using SulfoLink Coupling Gel (Pierce) according to the manufacturer's instructions. Western blotting analysis was performed by using 10% SDS polyacrylamide gels, and proteins were transferred onto nitrocellulose membranes. Antigens were detected by using the ECL system (Amersham Pharmacia Biosciences).
Immuno-EM.
T.brucei stably expressing YFP-ε-COP, YFP-GRIP70, or ss-YFP were fixed in 2% paraformaldehyde (PFA) and 0.2% glutaraldehyde for 1 h at room temperature, then further processed for cryosectioning. Sections were probed with an affinity-purified polyclonal anti-GFP antibody (41), followed by 10 nm of protein A-gold.
Immunofluorescence Assays.
Cells expressing YFP-ε-COP were fixed with methanol for 10 min at −20°C, whereas all others were fixed with 4% paraformaldehyde and then permeabilized by 0.25% Triton X-100. BSA (3%) was then used for blocking before antibody staining. Anti-GRASP and anti-GRIP70 antibodies were used at 1:1,000. Anti-ε-COP antibodies were used at 1:4,000. Alexa Fluor-conjugated goat anti-rabbit IgG (Invitrogen) was used at 1:2,000. Fixed cells were visualized by using either a confocal microscope (Zeiss ConfoCor Lsm510) or an upright microscope (Axioplan2; Carl Zeiss MicroImaging, Inc.) equipped with a charge-coupled device (CCD) camera (Orca-II; Hamamatsu, Hamamatsu, Japan), and a Plan-Apochromat 100 × 1.4-numerical aperture differential interference contrast (DIC) microscopy objective. Confocal images were acquired by using lsm510 3.2 software (Carl Zeiss MicroImaging, Inc.) and processed by using Adobe photoshop 7.0 (Adobe Systems, San Jose, CA). Epifluorescence images were acquired and processed by using openlab software (Improvision, Lexington, MA).
Live Cell Imaging.
T.brucei stably expressing both YFP-GRIP70 and GRASP-mRFP were plated on the surface of a low-melting agarose gel made with conditioned medium and then air dried briefly to restrict cell motility. The gel was then placed onto an imaging dish (Delta TPG Dish; Bioptechs, Butler, PA) with the cell side facing the coverslip. Cells were imaged at 28°C by using an inverted microscope (Axiovert 100M; Carl Zeiss MicroImaging, Inc.) equipped with a CCD camera (Orca-100; Hamamatsu) and a Plan-Apochromat Ph3 100 × 1.4-numerical aperture objective. Phase, YFP, and mRFP images were acquired sequentially, every 10 min, and processed by using openlab software (Improvision).
Supplementary Material
Acknowledgments
We thank Elisabetta Ullu and the G.W./I. Mellman laboratories for helpful discussions. This work was supported by the National Institutes of Health.
Abbreviations
- ER
endoplasmic reticulum
- TGN
trans-Golgi network
- immuno-EM
immuno-electron microscopy
- mRFP
monomeric red fluorescent protein
- ss-YFP
signal sequence-YFP.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Bonifacino J. S., Glick B. S. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Gu F., Crump C. M., Thomas G. Cell. Mol. Life Sci. 2001;58:1067–1084. doi: 10.1007/PL00000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shorter J., Warren G. Annu. Rev. Cell Dev. Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- 4.Sutterlin C., Hsu P., Mallabiabarrena A., Malhotra V. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- 5.Yoshimura S., Yoshioka K., Barr F. A., Lowe M., Nakayama K., Ohkuma S., Nakamura N. J. Biol. Chem. 2005;280:23048–23056. doi: 10.1074/jbc.M502442200. [DOI] [PubMed] [Google Scholar]
- 6.Preisinger C., Korner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pecot M. Y., Malhotra V. Cell. 2004;116:99–107. doi: 10.1016/s0092-8674(03)01068-7. [DOI] [PubMed] [Google Scholar]
- 8.Axelsson M. A., Warren G. Mol. Biol. Cell. 2004;15:1843–1852. doi: 10.1091/mbc.E03-07-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altan-Bonnet N., Sougrat R., Liu W., Snapp E. L., Ward T., Lippincott-Schwartz J. Mol. Biol. Cell. 2006;17:990–1005. doi: 10.1091/mbc.E05-02-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rambourg A., Clermont Y., Hermo L., Segretain D. Biol. Cell. 1987;60:103–115. doi: 10.1111/j.1768-322x.1987.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 11.Alcalde J., Bonay P., Roa A., Vilaro S., Sandoval I. V. J. Cell Biol. 1992;116:69–83. doi: 10.1083/jcb.116.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasap M., Thomas S., Danaher E., Holton V., Jiang S., Storrie B. Traffic. 2004;5:595–605. doi: 10.1111/j.1398-9219.2004.00203.x. [DOI] [PubMed] [Google Scholar]
- 13.Lippincott-Schwartz J., Yuan L. C., Bonifacino J. S., Klausner R. D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puri S., Linstedt A. D. Mol. Biol. Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duszenko M., Ivanov I. E., Ferguson M. A., Plesken H., Cross G. A. J. Cell Biol. 1988;106:77–86. doi: 10.1083/jcb.106.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field H., Sherwin T., Smith A. C., Gull K., Field M. C. Mol. Biochem. Parasitol. 2000;106:21–35. doi: 10.1016/s0166-6851(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 17.He C. Y., Ho H. H., Malsam J., Chalouni C., West C. M., Ullu E., Toomre D., Warren G. J. Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joiner K. A., Roos D. S. J. Cell Biol. 2002;157:557–563. doi: 10.1083/jcb.200112144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier L., Stern C. A., Pypaert M., Sheff D., Ngo H. M., Roper N., He C. Y., Hu K., Toomre D., Coppens I., et al. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- 20.Rossanese O. W., Soderholm J., Bevis B. J., Sears I. B., O'Connor J., Williamson E. K., Glick B. S. J. Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bevis B. J., Hammond A. T., Reinke C. A., Glick B. S. Nat. Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- 22.He C. Y., Pypaert M., Warren G. Science. 2005;310:1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
- 23.Nickel W., Brugger B., Wieland F. T. J. Cell Sci. 2002;115:3235–3240. doi: 10.1242/jcs.115.16.3235. [DOI] [PubMed] [Google Scholar]
- 24.Presley J. F., Ward T. H., Pfeifer A. C., Siggia E. D., Phair R. D., Lippincott-Schwartz J. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 25.Guo Q., Vasile E., Krieger M. J. Cell Biol. 1994;125:1213–1224. doi: 10.1083/jcb.125.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orci L., Glick B. S., Rothman J. E. Cell. 1986;46:171–184. doi: 10.1016/0092-8674(86)90734-8. [DOI] [PubMed] [Google Scholar]
- 27.McConville M. J., Ilgoutz S. C., Teasdale R. D., Foth B. J., Matthews A., Mullin K. A., Gleeson P. A. Eur. J. Cell Biol. 2002;81:485–495. doi: 10.1078/0171-9335-00268. [DOI] [PubMed] [Google Scholar]
- 28.Munro S. Biochem. Soc. Trans. 2005;33:601–605. doi: 10.1042/BST0330601. [DOI] [PubMed] [Google Scholar]
- 29.Clayton C. E., Mowatt M. R. J. Biol. Chem. 1989;264:15088–15093. [PubMed] [Google Scholar]
- 30.Wang Y., Satoh A., Warren G. J. Biol. Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West C. M., Van Der Wel H., Sassi S., Gaucher E. A. Biochim. Biophys. Acta. 2004;1673:29–44. doi: 10.1016/j.bbagen.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Van Der Wel H., Morris H. R., Panico M., Paxton T., Dell A., Kaplan L., West C. M. J. Biol. Chem. 2002;277:46328–46337. doi: 10.1074/jbc.M208024200. [DOI] [PubMed] [Google Scholar]
- 33.Barr F. A., Puype M., Vandekerckhove J., Warren G. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 34.Warren G. Annu. Rev. Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- 35.Malhotra V., Serafini T., Orci L., Shepherd J. C., Rothman J. E. Cell. 1989;58:329–336. doi: 10.1016/0092-8674(89)90847-7. [DOI] [PubMed] [Google Scholar]
- 36.Wirtz E., Leal S., Ochatt C., Cross G. A. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 37.Maier A. G., Webb H., Ding M., Bremser M., Carrington M., Clayton C. Mol. Biochem. Parasitol. 2001;115:55–61. doi: 10.1016/s0166-6851(01)00268-7. [DOI] [PubMed] [Google Scholar]
- 38.Marchetti M. A., Tschudi C., Kwon H., Wolin S. L., Ullu E. J. Cell Sci. 2000;113:899–906. doi: 10.1242/jcs.113.5.899. [DOI] [PubMed] [Google Scholar]
- 39.Bangs J. D., Brouch E. M., Ransom D. M., Roggy J. L. J. Biol. Chem. 1996;271:18387–18393. doi: 10.1074/jbc.271.31.18387. [DOI] [PubMed] [Google Scholar]
- 40.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seedorf M., Damelin M., Kahana J., Taura T., Silver P. A. Mol. Cell. Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 43.Short B., Haas A., Barr F. A. Biochim. Biophys. Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Yoshino A., Setty S. R., Poynton C., Whiteman E. L., Saint-Pol A., Burd C. G., Johannes L., Holzbaur E. L., Koval M., McCaffery J. M., Marks M. S. J. Cell Sci. 2005;118:2279–2293. doi: 10.1242/jcs.02358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.