Abstract
During mammalian aging, cellular proteins become increasingly damaged: for example, by carbonylation and formation of advanced glycation end products (AGEs). The means to ensure that offspring are born without such damage are unknown. Unexpectedly, we found that undifferentiated mouse ES cells contain high levels of both carbonyls and AGEs. The damaged proteins, identified as chaperones and proteins of the cytoskeleton, are the main targets for protein oxidation in aged tissues. However, the mouse ES cells rid themselves of such damage upon differentiation in vitro. This elimination of damaged proteins coincides with a considerably elevated activity of the 20S proteasome. Moreover, damaged proteins were primarily observed in the inner cell mass of blastocysts, whereas the cells that had embarked on differentiation into the trophectoderm displayed drastically reduced levels of protein damage. Thus, the elimination of protein damage occurs also during normal embryonic development in vivo. This clear-out of damaged proteins may be a part of a previously unknown rejuvenation process at the protein level that occurs at a distinct stage during early embryonic development.
Keywords: advanced glycation end products, aging, embryogenesis, proteasome, protein carbonylation
Organismal aging is associated with progressively increased damage to DNA, but genetic reassortment and recombination during meiosis ensures that such corruptions in nuclear genetic information are not transmitted to the offspring (e.g., refs. 1 and 2). In addition to age-related damage to DNA, proteins are damaged by a large number of reactions involving reactive oxygen species, and such oxidatively damaged proteins accumulate with age (3, 4). Carbonylation is an example of one such important oxidative modification of proteins and is associated with a large number of age-related disorders, including Parkinson’s disease, Alzheimer’s disease, and diabetes (e.g., refs. 3 and 5–9). Carbonyl derivatives are formed by a direct metal-catalyzed oxidative attack on the amino acid side chains of proline, arginine, lysine, and threonine and also on lysine, cysteine, and histidine by secondary reactions with reactive carbonyl compounds (8). Compared with other oxidative modifications, carbonyls are relatively difficult to induce, and in contrast to, for example, methionine sulfoxide and cysteine disulfide bond formation, carbonylation is an irreversible oxidative process (7).
Advanced glycation end product (AGE) formation is another protein modification that increases during aging (e.g., ref. 10). AGE is the collective name for the end products resulting from a complex set of nonenzymatic reactions between reduced sugars and proteins. AGEs accumulate in aging tissues, are enhanced by hyperglycemia, and have been associated with many diabetes complications (11). In addition, AGEs have been identified in protein deposits of Alzheimer’s amyloid plaques and in Lewy bodies of Parkinson’s disease (11, 12).
Modifications such as protein carbonyls and AGEs are suggested to cause cellular degeneration by two different means. The first denotes an age-dependent wear and tear of specific enzymes, such as aconitase and the nucleotide translocator ANT (e.g., refs. 13 and 14), causing a gradual loss of enzymatic activities. The second problem with oxidatively damaged proteins is that they tend to form large aggregates that not only escape degradation but clog up the proteasomes (6). The latter mechanism constitutes a negative feed-back loop that could explain why damaged and potentially protease-susceptible substrates accumulate with time during aging (6). Young specimens of aging mammals are essentially free of oxidative damage to proteins (4), but the underlying mechanism behind this “rejuvenation” phenomenon is unknown. Because germ-line cells are kept free of DNA damage, it is tempting to speculate that the same is true for protein damage, but no data support this view at present. Although it has been shown that undifferentiated murine ES cells exhibit a superior capacity, compared with differentiated cells, to cope with external oxidative stress (15), there are no data available on the carbonyl and AGE content of ES cells or the early differentiating embryo.
We have analyzed the levels of carbonyl and AGE modifications to proteins during differentiation of ES cells in vitro and in early embryonic development in vivo and present data that suggest that early embryonic development includes a mechanism of efficient damage elimination, which is associated with elevated 20S proteasomal activity.
Results
Detection and Identification of Carbonylated Proteins in ES Cells.
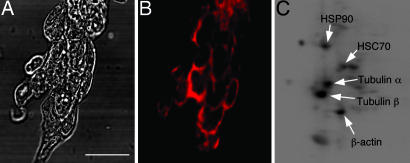
By modifying an in situ detection method (see Materials and Methods) that has previously been used for detection of carbonylated proteins in bacteria and yeast (16, 17), we were able to detect carbonylated proteins in undifferentiated murine ES cells (Fig. 1A and B). This method is based on the reaction of 2,4-dinitrophenyl-hydrazine (DNPH) with carbonyl groups forming 2,4-dinitrophenyl-hydrazone that can be immunochemically detected (9, 18). After fixation of cells in ice-cold EtOH (95%) and permeabilization by using 0.25% Triton X-100, carbonylated proteins were derivatized with DNPH in situ followed by immunodetection under the microscope. The unexpectedly high signal surrounding the nucleus (Fig. 1B) that was detected in the undifferentiated ES cells enabled us to estimate how the carbonyl levels corresponded to those of differentiated tissues of adult mice. We prepared protein extracts from 6-month-old brain and liver and found that the carbonyl levels in ES cells were in the same range as these tissues (data not shown).
Fig. 1.
Protein carbonyls in ES cells. (A and B) Bright-field image of fixed ES cells (A) and the corresponding image of carbonyls (B) visualized immunohistochemically. (Scale bar, 25 μm.) (C) Detection of the major targets for carbonylation in undifferentiated ES cells by two-dimensional Western blot analysis. The targets identified included the chaperones HSP90, HSC70, and GR75, as well as the α- and β-chains of tubulin and β-actin (GR75 was not observed in TCA-precipitated proteins samples).
A proteomics approach based on two-dimensional immunodetection and mass spectrometry of protein carbonyls established that a discreet number of proteins carry the main load of carbonylation in undifferentiated ES cells (Fig. 1C). These proteins included the chaperones HSP90, GR75, and HSC70; the α- and β-chains of tubulin; and β-actin (Fig. 1C). Interestingly, several of these proteins have been shown to be highly carbonylated in aged organisms ranging from bacteria and plants to mammals (18–22).
Protein Carbonyls Are Eliminated During ES Cell Differentiation.
We next initiated differentiation of ES cells by removing the cytokine leukemia inhibitory factor (LIF) from the culture. In situ detection of protein carbonyls demonstrated that cells that had embarked on differentiation [stage-specific embryonic antigen 1 (SSEA-1)-negative cells; Fig. 2] displayed significantly lower levels of carbonyls than undifferentiated ES cells (SSEA-1-positive cells; Fig. 2). This elimination of protein carbonyls was confirmed by analysis of protein extracts obtained before and after the removal of LIF (Fig. 3A). The analysis demonstrated a drastic drop in carbonyl levels between 2 and 5 days after the removal of LIF and that the elimination of protein carbonyls affected the targeted proteins in a general fashion (Fig. 3A Inset). This drop in protein carbonyls cannot be explained simply by a dilution of damaged proteins, because the undifferentiated murine ES cells (E14.1) reproduce with a shorter generation time than their differentiated counterparts. Yet, protein carbonyls are not eliminated during repeated passages of the undifferentiated and dividing ES cells. To determine whether LIF itself may act as an oxidant, ES cells were allowed to go through spontaneous differentiation in adherent cultures on a gelatin-coated tissue culture plate for 5 and 8 days in the presence of LIF. The cells that were generated displayed the same remarkable reduction in carbonyl levels (Fig. 3B), excluding LIF as a contributing oxidant. In addition, differentiation of ES cells into embryonic bodies (EBs) and retinoic acid (RA)-induced differentiation into neuronal and/or primitive endoderm cells resulted in the same characteristic clear-out of carbonylated proteins in the absence and presence of LIF (Fig. 3B).
Fig. 2.
In situ immunohistochemical detection of carbonylated proteins in undifferentiated (SSEA-1 positive) and differentiated (SSEA-1 negative) murine ES cells. (A) DAPI staining for localization of DNA/nucleus. (B) Immunodetection of SSEA-1 to score for differentiated and undifferentiated cells. (C) Detection of protein carbonyls. (D) Overlay of DAPI and protein carbonyl signals. (E) Overlay of DAPI, SSEA-1, and protein carbonyl signals. In A–E, the arrows indicate a cell conglomerate that has embarked on differentiation. (Scale bar, 25 μm.) (F) Expanded view of the boxed area in E showing localization of protein carbonyls between the cell surface (SSEA-1 signal) and the nucleus (DAPI signal) in undifferentiated ES cells. Representative images are shown.
Fig. 3.
Levels of protein carbonyls in protein extracts of ES cells upon differentiation. (A) Protein carbonyl levels in undifferentiated ES cells and in cells in which differentiation has been induced by withdrawing LIF. The values are related to those of undifferentiated cells (day 0), which were assigned a value of 1.0. Inset shows Western-blot carbonylation patterns after 0 (lane 1), 0.5 (lane 2), 2 (lane 3), 5 (lane 4), and 8 (lane 5) days of differentiation. (B) Carbonyl levels during differentiation of ES cells, in the absence and presence of LIF, into EBs and RA-induced differentiation into neuronal and/or visceral cells. (C) The overall concentration of the proteins identified as main carbonyl targets in differentiated and undifferentiated ES cells. The concentration is expressed as the ratio of protein levels in undifferentiated cells to those of differentiated ones. Error bars represent standard error of at least three measurements.
Differentiation is accompanied by alterations in cell morphology and the cellular proteome. Therefore, it is possible that the reduction in protein carbonyls could be due to a reduction in the concentration of specific proteins rather than the damaged form of the proteins. The identity of the damaged proteins (Fig. 1C) allowed us to approach this question. As shown in Fig. 3C, Western blot analysis demonstrated that the overall concentration of the affected proteins that were analyzed (β-actin, HSP90, HSP70, α-tubulin, and β-tubulin) did not change appreciably during differentiation. In other words, the carbonylation load of the affected proteins is reduced upon differentiation, not the levels of the proteins per se.
The Levels of AGEs Are Diminished During Differentiation.
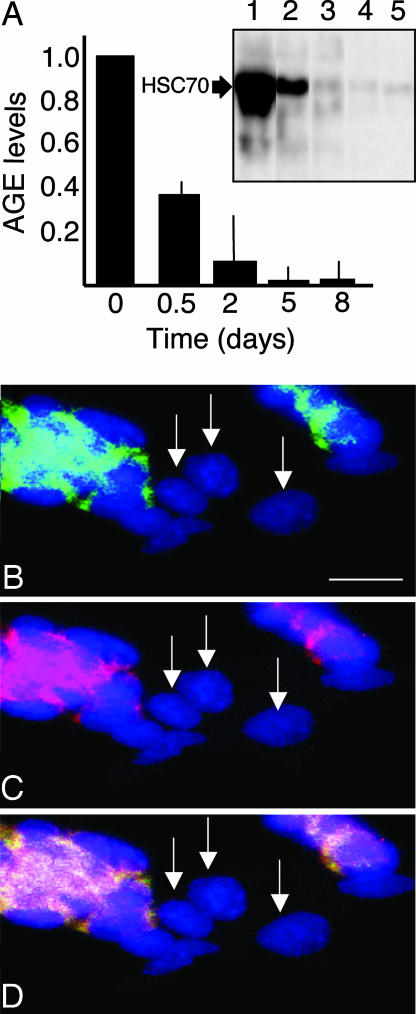
Next, we asked whether ES cells rid themselves of other types of protein damage (e.g., AGEs) upon differentiation. Protein extracts made from undifferentiated cells and cells differentiated as described above were analyzed for the content of AGEs. The antibody used exhibits immunospecificity to a common structure among AGE-structures, Nε-carboxymethyllysine (23). Undifferentiated ES cells displayed AGE modification of one major protein, HSC70 (Fig. 4A Inset). Similar to protein carbonyls, the AGE modification of this protein was efficiently eliminated upon differentiation as a consequence of LIF withdrawal (Fig. 3C). All other ways to induce differentiation (see Fig. 3B) also resulted in the same riddance of AGE modifications (not shown), indicating a general removal of damaged proteins rather than a specific elimination of carbonylated proteins.
Fig. 4.
AGE modification in undifferentiated and differentiated ES cells. (A) AGE levels in undifferentiated ES cells (day 0) and cells that have been triggered to differentiate by the removal of LIF. Inset shows a Western blot demonstrating an almost exclusive AGE modification of one single protein, which was identified as HSC70. AGE modification of HSC70 is shown after 0 (lane 1), 0.5 (lane 2), 2 (lane 3), 5 (lane 4), and 8 (lane 5) days of differentiation. Error bars represent standard error of at least three measurements. (B) Overlay of AGE immunodetection (green) and DAPI (blue) signals. (C) Overlay of carbonyl (red) and DAPI signals. (D) Overlay of AGE, carbonyl, and DAPI signals. In B–D, arrows indicate cells that are negative for both carbonyl and AGE staining. Representative images are shown. (Scale bar, 25 μm.)
Interference between the SSEA-1 and AGE immunodetection prevented simultaneous in situ detection of AGE and SSEA-1. However, we noticed that the AGE signal overlapped with the carbonyl signal and that the cells with low or no carbonyls also exhibited low AGE signal (Fig. 4 B–D).
Differentiation of ES Cells Results in a Boost in 20S Proteasomal Activity.
Several studies have shown that various types of cells are able to selectively degrade oxidatively damaged proteins and that mild oxidative stress correlates with increased intracellular proteolysis (24, 25). In mammalian cells, the 20S proteasome is the primary protease responsible for degradation of oxidatively damaged proteins in the cytosol and nuclei, whereas the ATP- and ubiquitin-dependent 26S proteasome is less important for the degradation of damaged proteins (e.g., ref. 25). To determine whether the reduction in protein damage upon differentiation was associated with altered 20S proteasome activity, an assay using the fluorogenic peptide succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin as a substrate for the proteasome was used (26). This analysis revealed that 20S proteasomal activity increased drastically upon differentiation of ES cells (by the removal of LIF) (Fig. 5). ATP-stimulated 26S proteasome was only marginally affected upon differentiation (not shown). Thus, differentiation of ES cells results in a considerable boost in the cells’ capacity to degrade damaged proteins, which coincides with the elimination of both carbonylated and AGE-modified proteins.
Fig. 5.
20S proteasome activity in undifferentiated ES cells and cells that have been triggered to differentiate by the removal of LIF. The chemotryptic activity of the proteasome was assayed by hydrolysis of the fluorogenic peptide succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin. The activities were all related to the activity obtained for the undifferentiated cells, which was assigned a value of 1.0. Day 0 denotes the undifferentiated ES cells, and “3” and “5” indicate 3 and 5 days of differentiation after the removal of LIF, respectively. Error bars represent standard deviation of three measurements in triplicate from three independent ES cell cultures.
Damaged Proteins Are Eliminated During Early Embryonic Development in Vivo.
If differentiation of ES cells were accompanied by elimination of damaged proteins during normal development in vivo, we would expect to observe differential carbonyl levels in cells within a blastocyst. Specifically, a higher load of damage would be expected in the inner cell mass, from where the pluripotent ES cells originate, than in the outer and differentiated cell layer (the trophectoderm), which was indeed the case (Fig. 6). We confirmed that protein carbonyls were predominantly associated with the pluripotent cells of the inner cell mass of the blastocyst by costaining with SSEA-1 (Fig. 6 A–D). Similar to carbonyl damage, AGE modification was primarily observed in the inner cell mass of blastocysts (Fig. 6 E–G). The data demonstrate that elimination of damaged proteins is an integral part of early embryonic development.
Fig. 6.
Protein damage in the undifferentiated cells of the inner cell mass in mouse blastocysts. (A–D) Series 1. (A) DAPI signal. (B) SSEA-1 signal. (C) Carbonyl signal. (D) Overlay of DAPI, SSEA-1, and carbonyl signals. (E–G) Series 2. (E) DAPI signal. (F) AGE immunodetection. (G) DAPI and AGE costaining. Arrows indicate the inner cell mass. A total of eight blastocysts were stained, and representative images are shown. (Scale bars, 25 μm.)
Discussion
Protein carbonylation is an indicator of severe oxidative damage, and the levels of protein carbonyls increase substantially during aging in animals (8, 4). In view of the fact that carbonylation is inhibiting the targeted proteins’ catalytic functions and may trigger formation of high-molecular-weight, potentially cytotoxic aggregates, this modification is likely to play havoc with cellular/tissue functions in the aging organism. In line with this, it has been demonstrated that the levels of carbonyl-damaged proteins are associated with the physiological age/performance or life expectancy of an organism rather than with its chronological age. For example, carbonyl levels are higher in crawlers (low life expectancy) than fliers in a cohort of houseflies of the same chronological age (27).
If oxidative protein damage is a genuine hazard to physiological performance and fitness, then the offspring should be kept free of such damage. For example, organisms producing reproductive organs at the closing stages of their development should have evolved means of keeping protein damage low throughout their life span or, alternatively, be equipped with systems that can clear out damage before reproduction. It appears that the plant Arabidopsis thaliana has evolved defense systems to do just that: Carbonylation first increases with the age of the plant, similar to animals, but drops abruptly before the vegetative-to-reproductive transition (22). In addition, unicellular yeast displays other means of keeping the young free of carbonylation damage. The asymmetrically dividing Saccharomyces cerevisiae has evolved a Sir2p-dependent system that retains carbonylated proteins in the mother cell compartment during mitotic cytokinesis (16). Thus, the progeny, exhibiting a full reproductive potential in contrast to the mother cell, starts out with a markedly reduced load of damage compared with the ancestor cell.
In contrast to plants, the production of offspring in mammals coincides with the early to middle stages of the organism’s life cycle; i.e., it occurs at a time at which the overall oxidative damage in the organism is low. However, this fact alone cannot explain the low levels of oxidative damage in the offspring, and we show here that early embryonic development encompasses a drastic reduction of both carbonylated and AGE-modified proteins. This elimination of damaged proteins coincides with an elevated activity of the 20S proteasome, which has been shown previously to be essential for degradation of oxidatively damaged proteins (e.g., ref. 25).
Because AGE modifications are exceedingly slow reactions (11), it is likely that the AGE damage observed in the inner cell mass of the blastocyst (3.5–4.5 days old) is inherited from the germ cell [AGE modifications have previously been observed also in fetuses of rats (28)]. In addition, the targets of carbonylation in the undifferentiated ES cells (e.g., chaperones and proteins of the cytoskeleton) are similar to those found in aging organisms (17–22). Based on these results, we believe it is worth considering that the offspring of mammals may initially be free of protein damage because of an early developmental damage elimination rather than by a mechanism that keeps the germ-line cells free of deteriorated macromolecules. The degree of protein carbonylation in undifferentiated ES cells is on par with that of the liver and brain of fully developed, 6-month-old mice, which begs the questions: Why do aged tissues, in contrast to cells of the blastocyst, fail to rid themselves of protein carbonyls and AGE modifications, and how do ES cells accomplish this task? The fact that ES cells cultivated in vitro also displayed efficient removal of damaged proteins, and a concomitantly elevated 20S proteasome activity, upon differentiation makes it possible to approach this question by classical and chemical genetics as well as RNA interference technology. Elucidating these questions may not only shed light on the process of embryonic development but also help us understand the aging process, age-related disorders, and the reasons for the aged and differentiated cells’ shortcomings in counteracting the progressive accumulation of damaged proteins.
Materials and Methods
ES Cell Line and Culture Conditions.
The murine ES cell line E14.1 was used in all experiments (29, 30). Cells were cultured on gelatin-coated plates (except during EB formation; see below) in DMEM (GIBCO) supplemented with 15% FCS (GIBCO), 0.1 mM nonessential amino acids (GIBCO), 2 mM l-glutamine (GIBCO), 1 mM sodium pyruvate (GIBCO), 100 μM 2-mercaptoethanol (Sigma), 50 μg/ml penicillin (GIBCO), 50 μg/ml streptomycin (GIBCO), and, when appropriate, 1,000 units/ml LIF (LIF/ESGRO; Chemicon). A 37°C humidified incubator with 5% CO2 was used. Mouse embryonic fibroblasts (MEFs) treated with mitomycin C (10 μg/ml; Sigma) were used as feeders when ES cells were thawed to facilitate recovery of the ES cells. ES cells were routinely passaged every other day (at ≈80% confluence) by 0.25% trypsin-EDTA (3 min at 37°C; GIBCO) and cultivated in their undifferentiated state until the MEF feeder cells were diminished. Media were changed every day. The undifferentiated status of the ES cells was confirmed by Oct-4 (localized in the nucleus; ref. 31) and SSEA-1 (cell surface-localized; ref. 32) immunochemical staining.
ES Cell Differentiation.
From the time point when the undifferentiated ES cells were ready to be passaged, differentiation was induced in various ways. (i) Differentiation was induced by removal of LIF such that the cells were washed and cultured in cultivation media without LIF for 0.5, 2, 5, and 8 days. (ii) ES cells were allowed to embark on spontaneous differentiation in the presence of LIF by omitting continuous passage. The ES cells were kept in adherent culture on a gelatin-coated tissue culture plate under these conditions for 5 and 8 days in the presence of LIF. (iii) ES cells were triggered to differentiate by the addition of RA. The ES cells were washed and cultured for 7 days in media with and without LIF supplemented with 10−7 M RA. RA induces differentiation of ES cells into neural and visceral cell types (33–36). (iv) ES cells were allowed to differentiate into EBs such that undifferentiated ES cells were detached and dissociated into single cells and small clusters of ES cells by trypsin incubation. Cells were then cultured in suspension in the media (without LIF) for 3 days to form EBs. At day 3, the EBs were dissociated and plated onto gelatin-coated tissue culture plate with media with or without LIF and cultured for 2 and 5 days. (See ref. 35.)
Determination of Protein Levels and Protein Damage by One-Dimensional Western Blot Analysis.
Cells were lysed with a modified RIPA buffer (50 mM Na2HPO4/150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS/2 mM EDTA/1 mM pefablock) on ice. Cell debris was removed by centrifugation. Protein concentration was determined by using the BCA Protein Assay kit (Pierce).
For one-dimensional Western blot analysis of carbonylation, the carbonyl-derivatized samples were loaded directly onto the gels, according to the protocol of the Chemicon Oxyblot kit. Samples used for detection of AGE modification or quantification of specific proteins were prepared as described by Ballesteros et al. (37). Gel electrophoresis was followed by blotting the proteins onto a poly(vinylidene difluoride) membrane (Millipore). The rabbit polyclonal anti-2,4-dinitrophenyl and goat peroxidase-conjugated anti-rabbit IgG were from the Chemicon Oxyblot kit; the mouse peroxidase-conjugated monoclonal anti-AGE IgG1 6D12 was from TransGenic (Kumamoto, Japan); the rabbit polyclonal anti-HSP90, anti-HSC70, and anti-β-actin were from Biosite (San Diego); and the goat peroxidase-conjugated anti-rabbit IgG, the mouse monoclonal anti-α-tubulin and anti-β-tubulin, and the goat peroxidase-conjugated anti-mouse IgG were from Sigma. The levels of the antigens were determined by exposure in a charge-coupled device camera after the reaction between ECL Plus (Amersham Pharmacia Bioscience) and the peroxidase conjugated to the secondary antibodies.
Identification of Damaged Proteins by Two-Dimensional Western Blot Analysis.
The samples used for identification of carbonylated proteins were prepared for two-dimensional Western blot analysis gel electrophoresis as described by O’Farrell (38) and VanBogelen et al. (39) with the addition of a carbonyl derivatization step (according to the Chemicon Oxyblot kit protocol). Some samples were trichloroacetic acid (TCA)-precipitated after the derivatization with 10% TCA followed by 30-min centrifugation and acetone washes. The acetone was allowed to evaporate fully, and lysis buffer (39) was added to the protein pellet before loading. The samples for AGE detection were prepared with TCA precipitation without the carbonyl derivatization step. Immunochemical analysis was performed as described above for one-dimensional Western blot analysis. For every two-dimensional separation experiment, each sample was run in duplicate, of which one of the gels was stained with Coomassie brilliant blue, and the other was used for blotting and immunodetection. After immunodetection, the membrane was stained with Coomassie brilliant blue. The detected damaged protein spots were matched with the Coomassie spots on the membrane, and the corresponding protein spots on the two-dimensional gel were cut out and used for mass-spectrometric analysis. The samples were analyzed by using a MALDI-linear reflection mass spectrometer (Micromass, Manchester, U.K.) in reflectron mode. Tryptic digest (0.5 μl) was mixed with 0.5 μl of matrix solution [12 mg/ml α-cyano-4-OH-cinnamic acid in acetonitrile/water (1:1)/0.1% trifluoroacetic acid] directly on the MALDI probe and allowed to dry at ambient conditions. Monoisotopic mass values were used for peptide mass mapping with mascot against the nr database at the National Center for Biotechnology Information. All proteins that were identified had a score equivalent to a 95% confidence level or higher and were identified from at least two independent experiments.
Immunohistochemical Detection of Damage in ES Cells.
The cells were fixed in ice-cold EtOH (95%) for 15 min at −20°C and permeabilized at room temperature in 0.25% Triton X-100 for 20 min for detection of carbonyls and AGEs, permeabilized in 0.5% Triton X-100 for 10 min for detection of Oct4, and permeabilized in 0.5% Triton X-100 for 5 min for detection of SSEA-1. The carbonyl groups were derivatized by DNPH (Chemicon Oxyblot kit) for 15 min at room temperature, after which the reaction was stopped by the addition of neutralization solution (Chemicon Oxyblot kit). FCS (10% in PBS) was used for blocking (for 40 min at room temperature) when staining for carbonyls, the mouse-to-mouse blocking kit (Biosite) was used according to the manufacturer’s instructions when detecting AGEs, and normal donkey serum (Jackson ImmunoResearch) was used for blocking during SSEA-1 and Oct4 detection (for 1 h at room temperature). Cells were incubated with primary antibody overnight at 4°C and with secondary antibody for 60 min at room temperature. The antibodies used were rabbit anti-2,4-dinitrophenyl (1:62; Chemicon Oxyblot kit) with secondary IgG goat anti-rabbit Texas red (1:125; Jackson ImmunoResearch), mouse monoclonal anti-AGE (1:125; TransGenic) with secondary donkey anti-mouse IgG FITC (1:150; Jackson ImmunoResearch), Oct4 primary antibody (1:500; Santa Cruz Biotechnology) with secondary goat anti-mouse IgG2b Cy3 (1:500; Southern Biotechnology Associates), and SSEA-1 antibody (1:100; Developmental Studies Hybridoma Bank, Iowa City, IA) with secondary goat anti-mouse IgM FITC (1:150) or rabbit anti-mouse IgM Cy3 (both 1:500; Southern Biotechnology Associates). Carbonyl signal was not detected when the primary antibody was omitted from the protocol or when DNPH was replaced by the DNPH control solution. The protocol for carbonyl detection was completed before the SSEA-1 staining to avoid possible background carbonyls.
Proteasome Activity.
Cells were lysed by hypotonic suspension in 1 mM DTT in distilled water, shaking at 4°C for 1 h. Cell debris was removed by centrifugation, and protein concentration was determined by the Lowry DC Protein Assay (Bio-Rad).
The chemotryptic activity of the proteasome was assayed by hydrolysis of the fluorogenic peptide succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (suc-LLVY-AMC) (Bachem, Saffron Walden, U.K.) according to Reinheckel et al. (26). Approximately 10–30 μg of protein from each extract was incubated with 200 μM suc-LLVY-AMC in 50 mM Tris (pH 7.8) and 1 mM DTT in a total volume of 100 μl. Fluorescence was read on a spectrofluorometer using 390-nm excitation and 460-nm emission filters and using free AMC as a standard (Bachem). Proteasomal activity measurements were confirmed by lactacystin inhibition (Biomol, Plymouth Meeting, PA) at a concentration of 30 μM. This concentration of lactacystin inhibited the activity by 85–90%.
Immunohistochemical Detection of Carbonylated Proteins, AGEs, and SSEA-1 in Mouse Blastocysts.
Embryonic day 3.5 blastocysts were fixed in 95% ice-cold ethanol for 15 min at 4°C. Permeabilization, derivatization, blocking, and antibody incubation were performed as described above for carbonyl detection in ES cells. The above-described protocols for AGEs and SSEA-1 were also applied for the blastocysts. Cell nuclei were visualized by incubating for 4 min with DAPI (1:1,000; Sigma-Aldrich) in PBS at room temperature.
Acknowledgments
We thank Anne Bergeron for valuable methodological input regarding the 20S proteasome activity assay and Henrik Lindskog and Per Lindahl (both from Göteborg University) for vials of E14.1 mouse ES cells. The SSEA-1 primary antibody, developed by Davor Solter (Max-Planck-Institute, Freiburg, Germany), was obtained from the Developmental Studies Hybridoma Bank. This work was supported by a grant from the Swedish Research Council/Cell and Molecular Biology (to T.N.), the Göran Gustafsson Award in Molecular Biology (to T.N.), the Swedish Research Council/Medicine (H.S.), Juvenile Diabetes Research Foundation (H.S.), and the Swedish Diabetes Research Foundation (H.S.).
Abbreviations
- AGE
advanced glycation end product
- LIF
leukemia inhibitory factor
- RA
retinoic acid
- EB
embryonic body
- DNPH
2,4-dinitrophenyl-hydrazine
- SSEA-1
stage-specific embryonic antigen 1.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., Alt F. W. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Medvedev Z. A. Mech. Ageing Dev. 1981;4:331–359. doi: 10.1016/0047-6374(81)90052-x. [DOI] [PubMed] [Google Scholar]
- 3.Stadtman E. R. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 4.Levine R. L. Free Radical Biol. Med. 2002;32:790–796. doi: 10.1016/s0891-5849(02)00765-7. [DOI] [PubMed] [Google Scholar]
- 5.Bota D. A., Davies K. J. Nat. Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 6.Grune T., Jung T., Merker K., Davies K. J. A. Int. J. Biochem. Cell Biol. 2004;36:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I., Giustarini D., Colombo R., Rossi R., Milzani A. Trends Mol. Med. 2003;9:169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 8.Stadtman E. R., Levine R. L. Ann. N.Y. Acad. Sci. 2000;899:191–208. doi: 10.1111/j.1749-6632.2000.tb06187.x. [DOI] [PubMed] [Google Scholar]
- 9.Levine R. L., Williams J. A., Stadtman E. R., Shacter E. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 10.Baynes J. W. Exp. Gerontol. 2001;36:1527–1537. doi: 10.1016/s0531-5565(01)00138-3. [DOI] [PubMed] [Google Scholar]
- 11.Singh R., Barden A., Mori T., Beilin L. Diabetologia. 2001;44:129–146. doi: 10.1007/s001250051591. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki N., Fukatsu R., Tsuzuki K., Hayashi Y., Yoshida T., Fujii N., Koike T., Wakayama I., Yanagihara R., Garruto R., et al. Am. J. Pathol. 1998;153:1149–1155. doi: 10.1016/S0002-9440(10)65659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan L. J., Levine R. L., Sohal R. S. Proc. Natl. Acad. Sci. USA. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan L. J., Sohal R. S. Proc. Natl. Acad. Sci. USA. 1998;95:12896–12901. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saretzki G., Armstrong L., Leake A., Lako M., von Zglinickia T. Stem Cells. 2004;22:962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- 16.Aguilaniu H., Gustafsson L., Rigoulet M., Nyström T. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 17.Desnues B., Gregori G., Dukan S., Aguilaniu H., Nyström T. EMBO Rep. 2003;4:400–404. doi: 10.1038/sj.embor.embor799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dukan S., Nyström T. Genes Dev. 1998;12:3431–3441. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soreghan B. A., Yang F., Thomas S. N., Hsu J., Yang A. J. Pharm. Res. 2003;20:1713–1720. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- 20.Castegna A., Aksenov M., Thongboonkerd V., Klein J. B., Pierce W. M., Booze R., Markesbery W. R., Butterfield D. A. J. Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 21.Reverter-Branchat G., Cabiscol E., Tamarit J., Ros J. J. Biol. Chem. 2004;279:31983–31989. doi: 10.1074/jbc.M404849200. [DOI] [PubMed] [Google Scholar]
- 22.Johansson E., Olsson O., Nystrom T. J. Biol. Chem. 2004;279:22204–22208. doi: 10.1074/jbc.M402652200. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda K., Higashi T., Sano H., Jinnouchi Y., Yoshida M., Araki T., Ueda S., Horiuchi S. Biochemistry. 1996;35:8075–8083. doi: 10.1021/bi9530550. [DOI] [PubMed] [Google Scholar]
- 24.Rivet J. A. J. Biol. Chem. 1985;260:300–305. [PubMed] [Google Scholar]
- 25.Davies K. J. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 26.Reinheckel T., Grune T., Davies K. J. Methods Mol. Biol. 2000;99:49–60. doi: 10.1385/1-59259-054-3:49. [DOI] [PubMed] [Google Scholar]
- 27.Sohal R. S., Ku H. H., Agarwal S. Biochem. Biophys. Res. Commun. 1993;196:7–11. doi: 10.1006/bbrc.1993.2208. [DOI] [PubMed] [Google Scholar]
- 28.Ling X., Nagai R., Sakashita N., Takeya M., Horiushi S., Takahashi K. Lab. Invest. 2001;81:845–861. doi: 10.1038/labinvest.3780294. [DOI] [PubMed] [Google Scholar]
- 29.Hooper M., Hardy K., Handyside A., Hunter S., Monk M. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 30.Wakayama T., Rodriguez I., Perry A. C. F., Yanagimachi R., Mombaerts P. Proc. Natl. Acad. Sci. USA. 1999;96:14984–14989. doi: 10.1073/pnas.96.26.14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholer H. R., Hatzopoulos A. K., Balling R., Suzuki N., Gruss P. EMBO J. 1989;8:2543–2550. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solter D., Knowles B. B. Proc. Natl. Acad. Sci. USA. 1978;75:5565–5569. doi: 10.1073/pnas.75.11.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones-Villeneuve E. M., McBurney M. W., Rogers K. A., Kalnins V. I. J. Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pachernik J., Esner M., Bryja V., Dvorak P., Hampl A. Reprod. Nutr. Dev. 2002;42:317–326. [PubMed] [Google Scholar]
- 35.Okada Y., Shimazaki T., Sobue G., Okano H. Dev. Biol. 2004;275:124–142. doi: 10.1016/j.ydbio.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 36.Pachernik J., Bryja V., Esner M., Kubala L., Dvorak P., Hampl A. Phys. Res. 2005;54:115–122. doi: 10.33549/physiolres.930526. [DOI] [PubMed] [Google Scholar]
- 37.Ballesteros M., Fredriksson A., Henriksson J., Nystrom T. EMBO J. 2001;20:5280–5289. doi: 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Farrell P. H. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 39.VanBogelen R. A., Hutton M. E., Neidhardt F. C. Electrophoresis. 1990;11:1131–1166. doi: 10.1002/elps.1150111205. [DOI] [PubMed] [Google Scholar]