Abstract
Using an appropriately designed and replicated study of a latitudinal influence on rates of evolution, we test the prediction by K. Rohde [(1992) Oikos 65, 514–527] that the tempo of molecular evolution in the tropics is greater than at higher latitudes. Consistent with this prediction we found tropical plant species had more than twice the rate of molecular evolution as closely related temperate congeners. Rohde’s climate-speciation hypothesis constitutes one explanation for the cause of that relationship. This hypothesis suggests that mutagenesis occurs more frequently as productivity and metabolic rates increase toward the equator. More rapid mutagenesis was then proposed as the mechanism that increases evolutionary tempo and rates of speciation. A second possible explanation is that faster rates of molecular evolution result from higher tropical speciation rates [e.g., Bromham, L. & Cardillo, M. (2003) J. Evol. Biol. 16, 200–207]. However, we found the relationship continued to hold for genera with the same number of, or more, species in temperate latitudes. This finding suggests that greater rates of speciation in the tropics do not cause higher rates of molecular evolution. A third explanation is that more rapid genetic drift might have occurred in smaller tropical species populations [Stevens, G. C. (1989) Am. Nat. 133, 240–256]. However, we targeted common species to limit the influence of genetic drift, and many of the tropical species we used, despite occurring in abundant populations, had much higher rates of molecular evolution. Nonetheless, this issue is not completely resolved by that precaution and requires further examination.
Keywords: latitude, metabolic rate, molecular evolution, mutagenesis, speciation
The decrease in species richness along the continuum from tropical to polar latitudes is perhaps one of the most widely recognized patterns in nature, yet there is no consensus as to a causal mechanism (1, 2). Indeed, since Hutchinson (3) explored a range of possibilities in his 1959 address, “homage to Santa Rosalia or why are there so many kinds of animals?”, a plethora of competing ideas that attempt to explain the pattern have emerged (4, 5). In 1808, von Humboldt (6) first suggested that energy was key in generating this relationship, and there are now several theories invoking energy as a determinant of diversity (5). These include the climate-speciation hypothesis of Rohde (7). The central element of that hypothesis is the idea that in warmer, more productive environments metabolic rates are higher and that because the rate of mutagenesis is thought to be positively correlated with metabolic rate (8) the tempo of both evolution and speciation is also greater (7). If rate heterogeneity in molecular evolution is controlled by climate it would also suggest that evolution is a spatially ordered phenomenon as was first proposed by Darwin (9). Here, we test the prediction made by Rhode (7) that rates of molecular evolution are faster in the tropics than at higher latitudes. Alternative hypotheses that might also explain faster molecular evolution in the tropics include: first, the concept under nearly neutral theory that faster rates of genetic drift occur in generally smaller tropical populations (10, 11), and second, the concept of more rapid tropical speciation rates themselves produce faster rates of molecular evolution at low latitudes (12).
Nucleotide substitution rates have been found to correlate positively with body temperature where phylogenetically disparate animal taxa are compared (endotherms and ectotherms) (8, 13), and Bleiweiss (14) found slower rates of molecular evolution at higher elevation within a single group of organisms, namely, hummingbirds. By contrast, Bromham and Cardillo (12), using congeneric pairings of birds, did not find a significant difference in rates of molecular evolution between high- and low-latitude taxa. However, the power of that study was affected by the inclusion of species pairs with comprehensively overlapping distributions (up to 100%) when seasonal migration is taken into account. High-latitude summer ranges were used as the latitudinal designates for species that experience warmer low-latitude climates during winter. Species that experience essentially tropical or subtropical winter climates cannot be defined as temperate in terms of their temperature regime on the basis of summer incursions into higher latitudes.
In this study with plants, using the internal transcribed spacer (ITS) region of rRNA-encoding DNA, we have selected 45 phylogenetically independent congeneric, or conspecific, pairs to examine the relationship between latitude and rates of molecular evolution. One taxon of each pairing occurs in the lowland/lower montane tropics, with the other being from the highest latitude and elevation possible while remaining within the same (rainforest) biome. None of these pairings have overlapping latitudinal distributions. There are two problems with overlapping latitudinal distributions: first, there is a high risk of gene flow between species where such overlap occurs in common geographic space; second, nucleotide substitutions can occur anywhere within a species population, and therefore any latitudinally affected environmental prompt for mutagenesis is equal for individuals from different congeners in overlapping sectors of their distributions.
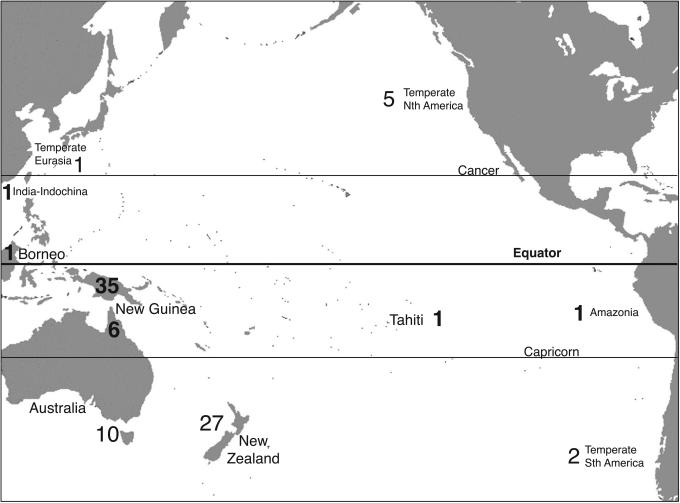
The study involves a phylogenetically diverse range of woody rainforest plants [conifers, magnoliids, basal eudicots, and core eudicots (rosids and asterids)]. Because they are sedentary ectotherms, plants are more appropriate organisms for exploring the molecular effects of latitudinally varying climates. The selection criteria for candidate taxa included that each of the species within a given paired comparison had to occur in wet forest, where water is unlikely to be strongly limiting. Therefore, the latitudinal difference in thermal environment between species pairs is likely to represent a variable closely linked to productivity potential (5). Because of the potential effect of generation time and body mass on rates of molecular evolution (7, 13, 15, 16) and of vegetation stratum on light flux and productivity (17, 18), we ensured that the contrasted taxa were from the same forest stratum and had the same body plan and size. Last, we targeted common species to avoid as far as possible influences from greater rates of genetic drift caused by small population sizes (10). Tropical samples were taken from species inhabiting New Guinea, northeast Australia, Borneo, India, Tahiti, and South America (Fig. 1). Temperate samples were taken from species inhabiting North America, southern Australia, New Zealand, Eurasia, and South America (Fig. 1). This study uses a well replicated data set with a robust spatial design to test for rate heterogeneity in molecular evolution between high and low latitudes.
Fig. 1.
Geographical distribution of sampled species. Numbers for tropical species are in bold face, and numbers for temperate species are in light face.
Results and Discussion
The rate of nucleotide substitution in the ITS region of rRNA-encoding DNA for tropical taxa was found to be more than twice that in the taxa from temperate latitudes [mean ratio = 2.1, n = 45, Wilcoxon statistic (W) = 194, P = 0.0001] (Table 1).
Table 1.
Ingroup pairs, outgroup taxa, and nucleotide substitution ratios (1 − shorter branch length/longer branch length)
| Tropical ingroup | Temperate ingroup | Outgroup | Ratio |
|---|---|---|---|
| Agathis borneensis | Agathis australis | Araucaria araucana | 0.92672 |
| Metrosideros salomonensis | Metrosideros umbellata | Cloezia floribunda | 0.90795 |
| Podocarpus archboldii | Podocarpus cunninghamii | Nageia nagi | 0.88851 |
| Lithocarpus rufovillosus | Lithocarpus densiflorus | Quercus suber | 0.86659 |
| Sophora tomentosa | Sophora tetraptera | Calpurnia aurea | 0.83811 |
| Nothofagus grandis | Nothofagus antarctica | Betula pendula | 0.78243 |
| Elmerrillia tsiampacca | Magnolia virginiana | Liriodendron tulipifera | 0.74319 |
| Eucalyptus deglupta | Eucalyptus coccifera | Angophora costata | 0.73571 |
| Geniostoma rupestre | Geniostoma rupestre | Mitreola petiolata | 0.72605 |
| Pennantia cunninghamii | Pennantia corymbosa | Corynocarpus cribbianus Griselinia lucida | 0.71942 |
| Clematis javana | Clematis paniculata | Naravelia laurifolia | 0.69887 |
| Schefflera macrostachya | Schefflera digitata | Polyscias ledermanii | 0.69509 |
| Kunzea graniticola | Kunzea ericoides | Agonis flexuosa | 0.68131 |
| Pittosporum ramiflorum | Pittosporum tenuifolium | Sollya heterophylla | 0.67852 |
| Opocunonia nymanii | Caldcluvia paniculosa | Ceratopetalum apetalum | 0.66781 |
| Araucaria hunsteinii | Araucaria araucana | Agathis australis | 0.66353 |
| Metrosideros whiteana | Metrosideros parkinsonii | Cloezia floribunda | 0.66026 |
| Rapanea leucantha | Rapanea howittiana | Myrsine oliveri | 0.61595 |
| Dysoxylum arborescens | Dysoxylum spectabile | Guarea glabra | 0.61172 |
| Alectryon connatus | Alectryon excelsus | Pappea capensis | 0.60566 |
| Albizia procera | Albizia julibrissin | Inga edulis | 0.59658 |
| Elaeocarpus sphaericus | Elaeocarpus hookerianus | Aristotelia serrata | 0.58196 |
| Coprosma nadeandii | Coprosma tenuifolia | Nertera dichondrifolia | 0.56816 |
| Piper methysticum | Macropiper excelsum | Sarcorhachis sydowii | 0.55044 |
| Diospyros ferrea | Diospyros virginiana | Euclea crispa | 0.52719 |
| Streblus glaber | Streblus heterophylla | Morus wittiorum | 0.47233 |
| Passiflora foetida | Passiflora tetrandra | Adenia heterophylla | 0.46786 |
| Celtis latifolia | Celtis occidentalis | Trema orientalis | 0.42441 |
| Polyscias ledermanii | Polyscias sambucifolia | Schefflera digitata | 0.41911 |
| Litsea globosa | Litsea calicaris | Phoebe formosana | 0.40927 |
| Rhus taitensis | Rhus typhina | Searsia quartiniana | 0.40531 |
| Phyllanthus cicciodes | Phyllanthus gunnii | Sauropus granulosus | 0.38169 |
| Carpodetus arboreus | Carpodetus serratus | Cuttsia viburnea | 0.35508 |
| Grevillea papuana | Grevillea robusta | Buckinghamia celsissima Opisthiolepis heterophylla | 0.35455 |
| Acacia aulacocarpa | Acacia frigescens | Parkia timoriana | 0.34539 |
| Melicope cf. crassiramus | Melicope simplex | Haplophyllum bastetanum | 0.32393 |
| Pouteria macropoda | Pouteria costata | Xantolis siamensis | 0.32046 |
| Tasmannia insipida | Tasmannia lanceolata | Takhtajania perrieri | 0.30354 |
| Vitex cofassus | Vitex lucens | Callicarpa dichotoma | 0.29509 |
| Acmenosperma claviflorum | Syzygium maire | Thaleropia queenslandica | 0.24377 |
| Ceratopetalum succirubrum | Ceratopetalum apetalum | Caldcluvia paniculosa | 0.23931 |
| Corynocarpus cribbianus | Corynocarpus laevigatus | Griselinia lucida Pennantia corymbosa | 0.17074 |
| Banksia dentata | Banksia marginata | Austromuellera trinervia | 0.14173 |
| Dodonaea viscosa | Dodonaea viscosa | Harpullia cupanioides | 0.01769 |
| Weinmannia fraxinea | Weinmannia racemosa | Ackama rosaefolia | 0.00058 |
n = 45. Longer temperate branches are in bold face.
There are several broad hypotheses that might explain higher rates of molecular evolution for tropical plants. These include, first, the idea that tropical populations are generally smaller than temperate populations because of greater species richness at low latitudes (11) and therefore the rate of genetic drift has been more rapid in the tropics (10). However, for this sample we targeted species that were common so that the species in the data set are unlikely to have experienced more rapid drift than their temperate counterparts because of small population sizes. Indeed, within this data set, species such as Araucaria hunsteinii, Dysoxylum arborescens, Elaeocarpus sphaericus, Eucalyptus deglupta, Lithocarpus rufovillosus, Nothofagus grandis, Pittosporum ramiflorum, Podocarpus archboldii, and Schefflera macrostachya are among the most widespread and abundant species in the tropical rainforest communities in which they are found, yet all show much higher rates of molecular evolution than their temperate counterparts. It is nevertheless possible that some of the tropical species have gone through population declines in the past, which, however, is also possible for their temperate counterparts. It is unlikely that population declines have been systematically more prevalent in the tropical species across a data set of 45 paired comparisons in which common species have been specifically targeted. Nonetheless, we cannot preclude the possibility that more rapid genetic drift caused by small tropical population sizes may have contributed to the results.
A second hypothesis in explanation of faster molecular evolution in the tropics is that because tropical assemblages are typically more speciose, higher rates of speciation (12) might positively affect nucleotide substitution rates by increasing the rate of natural selection (7) and/or increasing the potential for genetic drift (10, 19, 20). To test for an influence on molecular evolution caused by latitudinally determined rates of speciation, we isolated a subset of the data in which the genus has greater, or equivalent, species richness within the cooler landmass of a given paired geographic contrast. More rapid molecular evolution for the tropical species in this subset (mean ratio = 3.0, n = 18, W = 12, P = 0.0003) suggests that speciation is unlikely to be the cause of the observed rate heterogeneity favoring the tropics. It is possible that speciation and extinction rates have been greater among the tropical species (21) in this data subset and compensating extinction rates have thereby masked a speciation effect. However, most of these pairings involve genera with more species in higher latitudes. Therefore, for this relationship to be valid, the tropical extinction rates would need to be even greater relative to putatively elevated tropical speciation rates. This scenario of generally elevated speciation rates being overcompensated by even higher extinction rates in the tropics is, we think, less likely. If this effect was widespread it would tend to depress species richness in the tropics. In our view this could still be an issue across a small data set of just a few paired comparisons, but it is unlikely to be one across the 18 genera in this analysis.
Third, it has been proposed by Rohde (7) that climate determines the rate of molecular evolution and speciation. Rohde suggested that where there is greater biologically available energy and increased productivity in the tropics (22) resultant increases in metabolic rate and mutagenesis (7, 8, 13) and/or decreases in generation time (15, 16) might produce a positive effect on evolution for organisms inhabiting those latitudes. Martin and Palumbi (8) suggested that greater metabolic rates might influence substitution rates by increasing the rate of oxygen radical induced damage to DNA and the DNA replication rate. Generation time variability may influence the rate of molecular evolution by influencing the rate of genetic drift (13, 16) or the rate of natural selection (7, 23).
Given that many of the temperate comparator taxa that contributed to the data set are from geographically isolated New Zealand, we also tested for a unique geographic effect that might have been produced by that sampling emphasis. We divided the data into two subsets, one of pairings with temperate comparator taxa from New Zealand (n = 27) and one of pairings with temperate comparator taxa from outside of New Zealand (n = 18). Both data subsets had significantly greater rates of molecular evolution in tropical taxa (mean ratio = 2.5, W = 61, P = 0.001 and mean ratio = 1.5, W = 39, P = 0.02, respectively). This finding indicates that the results are not caused by the geographic influence of a sampling emphasis on New Zealand. That sampling emphasis arises from the fact that New Zealand carries into high latitudes a large number of woody plant genera that also occur in lowland tropical rainforest assemblages. We likewise tested for a bias caused by a large representation of New Guinean species in the sample by isolating a subset of data that excluded pairs with New Guinean species. Despite the small sample size (n = 10) for this comparison, a result with the same trend remains marginally significant (mean ratio = 2.3, W = 11, P = 0.05).
The most comprehensive study to date (24), done on a large sample of angiosperms from many different families, has demonstrated a positive correlation between rates of neutral molecular evolution and rates of protein and morphological evolution and a positive correlation between rates of neutral molecular evolution and species richness. That study suggests that the finding here of more rapid tropical substitution rates for ITS may be indicative of a wider genetic effect.
Faster rates of molecular evolution in the tropics present a plausible explanation for the dominance of positive correlations between productivity and diversity (5, 25). This general pattern is most notably expressed in the latitudinal gradient of species richness that typically sees much greater tropical diversity relative to cooler habitats (26). Support for this concept also comes from evidence in fossil records where consistently greater rates of tropical speciation over geological time have been found (27). Correspondingly, the latitudinal location of the first appearance in fossil records of apomorphic characters favors the tropics, with poleward deflections observed for the occurrence of plesiomorphic life forms (28–31).
Conclusions
The results of this study, showing a doubling in the rate of nucleotide substitution in the tropics, indicate that evolution may be a spatially ordered phenomenon with a distinct latitudinal polarity. We have also found that the relationship of more rapid molecular evolution in the tropics holds for a subset of the data with higher, or equivalent, temperate species richness. This result suggests that the typical pattern of more speciose tropical communities may be consequential rather than causal relative to faster molecular evolution in the tropics. Two other hypotheses may explain the results: first, genetic drift is more rapid in the tropics because of smaller population sizes; second, mutagenesis is increased in the tropics because of greater ambient temperatures and productivity. We suggest the former is less likely because we have targeted common species in this sample to avoid the influence of greater rates of genetic drift in small populations and some of the highest rates of molecular evolution we found were in widespread and abundant tropical species, many of which are dominant components of the communities where they occur. However, further research is required to distinguish between these two hypotheses.
Greater rates of molecular evolution in the tropics constitute a possible explanation for one of the most fundamental patterns in nature, namely higher tropical species richness. Beginning with von Humboldt (6) in the early 19th century, the idea that the driver for evolution and speciation might be energy has been repeatedly suggested but the identification of a causal mechanism has proved to be elusive. Consistent with this venerable idea, we present empirical evidence that the rate of molecular evolution is greater in the tropics than in temperate latitudes.
Materials and Methods
We selected a total of 45 tropical woody plant taxa with latitudinal/altitudinal ranges coinciding with the warmest sectors of the total climatic ranges of their respective genera. Each of these was paired with (mostly) a congeneric species from the coolest sector of the climatic range for a given genus. In two comparisons, contrasts were made between tropical and temperate conspecific populations within wide-ranging species entities (Dodonaea and Geniostoma). A subgeneric level paired contrast for each of two subgenera within the same genus was also made (Metrosideros). We targeted common species on the presumption that rare species might show substitution rates influenced by population size (10). We avoided herbaceous species because of the possibility of the confounding influence of generation time heterogeneity for annuals versus biennials versus short-period perennials versus long-period perennials (32, 33). We also ensured that the contrasted taxa were from the same forest stratum and had the same body plan and size to avoid an influence caused by mass and/or generation time on substitution rates (7, 13, 15, 16) and light flux on productivity (17, 18). Where practicable, species occupying the same subclade, within a given generic clade, were compared to avoid the potential confounding effect of greater phylogenetic distance. In all cases there is no latitudinal overlap in the natural ranges of each of the taxa forming a given contrasting pair. The selection criteria for candidate taxa included that each of the species within a given paired comparison had to occur in wet forest, where water is unlikely to be strongly limiting. We therefore avoided plants growing in productivity regimes severely limited by water deficits. The combined stringency of these criteria meant that the majority of the ingroup samples had to be collected and sequenced by us, with only a small part of the total data set being derived from the public domain (GenBank). In all instances where species with different generic nomenclatures have been compared, we have ensured that these comparisons are between phylogenetically proximate lineages (congeneric level) on the basis of recently published DNA-based phylogenies. Only complete ITS data of nuclear rRNA-encoding DNA (ITS1–5.8S-ITS2) has been used for all ingroup accessions.
Leaf tissue was collected fresh and stored at 4°C, dried over silica gel immediately after collection and stored at room temperature, or sampled from Herbarium material. For most plant species, DNA was isolated by using the Sigma Extract-N-Amp Kit (XNAP2). For some species the biochemistry of the kit proved to be incompatible, and in these cases DNA was isolated by using the Qiagen (Valencia, CA) DNeasy Plant Kit (69104). Preliminary trials showed that the phylogenetically diverse range of plant genera differed with respect to optimal PCR conditions, in particular the annealing temperature. Rather than attempting to optimize for each species in this study, a standard touchdown PCR protocol was developed that gave adequate amplification for most plant species. The touchdown protocol was a 2-min hold at 94°C followed by 10 “touchdown” cycles in which the annealing temperature was reduced by 1°C in each cycle, 94°C for 30 s, 65–56°C for 30 s, 72°C for 1 min followed by 35–40 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min followed by a 7-min hold at 72°C. For ITS amplification the 18S (forward) primer used was CY1 (TACCGATTGAATGATCCGGTGAAG) and the 28S (reverse) primer was CY3 (CGCCGTTACTAGGGGAATCCTTGT) (C. G. Yong, personal communication). For amplification of members of Araucariaceae and Podocarpaceae, where ITS PCR products could exceed 2.2 kb in length, the 72°C extension times were increased to 3 min. For some plant species, the presence of an unusually long ITS1 (Araucariaceae and Podocarpaceae), or of indels, caused sequencing difficulties. In these cases the use of internal primers, located in the 5.8S gene, helped to complete double-stranded sequencing. The forward and reverse 5.8S primers designed for this study were JK5.8F (GATACTTGGTGTGAATTGCAGA) and JK5.8R (ATGGTTCACGGGATTCTGCAA). For sequencing of members of the genus Podocarpus, where ITS1 alone exceeds 1.5 kb in length, an additional ITS1 internal forward primer, PodITSf (CTTCCCTTGCACCGTCGTCC), was designed.
The nearest possible confamilial neighbor (typically the same subfamily, tribe, or subtribe) was applied as an outgroup to the relevant ingroup pairing. Appropriate outgroup taxa were identified from DNA-based phylogenetic literature and, when they were available from GenBank, they were applied. When such closely related outgroup species were unavailable from GenBank or the existing ingroup data set, additional collections were made of appropriate species. In a number of instances additional outgroups were secured from the field despite the availability of confamilial ITS lineages in GenBank. Field collections for outgroups were made when it was possible to apply a closer sister to the relevant ingroup pair and thereby improve the outgroup relationship. Potential outgroup sequences from GenBank were avoided when they were not substantially complete for ITS; only one missing nucleotide was permitted at start/stop. Two outgroup lineages were used for both Pennantia and Corynocarpus because each belongs to a monotypic family that appears to be isolated from other angiosperms. On the basis of our own phylogenetic investigations with ITS, the previously applied outgroup Coriaria (34) was found to be only distantly related to Corynocarpus, with Griselinia and Pennantia providing the more proximate sister taxa that we subsequently applied. A second outgroup was applied for Grevillea because the closest sister genus, Buckinghamia, was found to have two large deletions in ITS2. This finding prompted us to sequence and add the next nearest sister, Opisthiolepis, to the analysis to compensate for those regions of phylogenetic uncertainty. The use of Betula (Betulaceae) as outgroup for Nothofagus (Nothofagaceae) follows Manos (35).
Phylogenies were based on maximum likelihood under the general time-reversible model with substitution rate matrix, proportion of invariant sites, and gamma shape parameter all estimated (36). It has been found that variations of the general time-reversible model are able to construct more accurate phylogenies than other comparable models (37). Alignment using the program clustalx (38) followed the hierarchy of classification, so that each congeneric ingroup pair was aligned first and the confamilial outgroups were subsequently aligned to that pair. The resultant branch length estimates from the common node of divergence for each ingroup pair were converted to ratios (1 − shorter branch length/longer branch length). The comparative ratios for branch length estimates provided the means to perform statistical analyses of ranked order (Wilcoxon signed rank sum tests). Ratio values were differentiated into two states (+ or −) depending on whether the tropical or temperate taxon in each paired comparison showed the longer branch length. Analyses were done on the whole data set and four subsets of the data set. These subsets were: first, of genera showing higher (or equivalent) species diversity in the cooler landmass for a given paired comparison; second, of pairings with temperate genera from New Zealand; third, of pairings with temperate genera not from New Zealand; and last, of pairings with tropical genera not from New Guinea.
Acknowledgments
We thank Robert Kiapranis, Lyn Craven, Peter Wilson, Stewart Ware, Michael Lovave, Aaron Liston, Spencer Sealy, and Mark Harrington for help with tissue collection, Professors Richard Gardner and Richard Bellamy, and Russell Gray, Howard Ross, and Ana Tutone. This study was funded by Nga Pae O Te Maramatanga, Maori Centre of Research Excellence under the direction of Professor Michael Walker and Associate Professor Linda Smith. S.W. holds the Michael Horton Lectureship in Biogeography at the University of Auckland.
Abbreviations
- ITS
internal transcribed spacer
- W
Wilcoxon statistic.
Footnotes
References
- 1.Currie D. J., Mittelbach G. G., Cornell H. V., Field R., Guegan J., Hawkins B. A., Kaufman D. M., Kerr J. T., Oberdorff T., O’Brien E. M., Turner J. R. G. Ecol. Lett. 2004;7:1121–1134. [Google Scholar]
- 2.Evans K. L., Warren P. H., Gaston K. J. Biol. Rev. 2005;80:1–25. doi: 10.1017/s1464793104006517. [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson G. E. Am. Nat. 1959;93:145–159. [Google Scholar]
- 4.Palmer M. W. Folia Geobot. Phytotaxon. 1994;29:717–740. [Google Scholar]
- 5.Gillman L. N., Wright S. D. Ecology. 2006 in press. [Google Scholar]
- 6.von Humboldt A. Ansichten der Natur mit Wissenschaftlichen Erlauterungen. Germany: Tubingen; 1808. [Google Scholar]
- 7.Rohde K. Oikos. 1992;65:514–527. [Google Scholar]
- 8.Martin A. P., Palumbi S. R. Proc. Natl. Acad. Sci. USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darwin C. On the Origin of Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- 10.Ohta T. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 11.Stevens G. C. Am. Nat. 1989;133:240–256. [Google Scholar]
- 12.Bromham L., Cardillo M. J. Evol. Biol. 2003;16:200–207. doi: 10.1046/j.1420-9101.2003.00526.x. [DOI] [PubMed] [Google Scholar]
- 13.Gillooly J. F., Allen A. P., West G. B., Brown J. H. Proc. Natl. Acad. Sci. USA. 2005;102:140–145. doi: 10.1073/pnas.0407735101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleiweiss R. Proc. Natl. Acad. Sci. USA. 1998;95:612–616. doi: 10.1073/pnas.95.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooers A. O., Harvey P. H. Mol. Phylogenet. Evol. 1994;3:344–350. doi: 10.1006/mpev.1994.1040. [DOI] [PubMed] [Google Scholar]
- 16.Laird C. D., McConaughy B. L., McCarthy B. J. Nature. 1969;224:149–154. doi: 10.1038/224149a0. [DOI] [PubMed] [Google Scholar]
- 17.Specht A., Specht R. L. Biodiversity Conservation. 1993;2:152–167. [Google Scholar]
- 18.Bruenig E. F. Conservation and Management of Tropical Rainforests: An Integrated Approach to Sustainability. Wallingford, U.K.: CAB International; 1996. [Google Scholar]
- 19.Webster A. J., Payne R. J. H., Pagel M. Science. 2003;301:478. doi: 10.1126/science.1083202. [DOI] [PubMed] [Google Scholar]
- 20.Jobson R. W., Albert V. A. Cladistics. 2002;18:127–136. doi: 10.1111/j.1096-0031.2002.tb00145.x. [DOI] [PubMed] [Google Scholar]
- 21.McKinney M. L. In: The Biology of Rarity. Kunin W. E., Gaston K. J., editors. London: Chapman & Hall; 1997. pp. 110–129. [Google Scholar]
- 22.O’Brien E. M. J. Biogeogr. 1998;25:379–398. [Google Scholar]
- 23.Dobzhansky T. Am. Sci. 1950;38:208–221. [Google Scholar]
- 24.Barraclough T. G., Savolainen V. Evolution. 2001;55:677–683. doi: 10.1554/0014-3820(2001)055[0677:erasdi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Whittaker R. J., Heegaard E. Ecology. 2003;84:3384–3390. [Google Scholar]
- 26.Leigh E. G., Davidar P., Dick C. W., Puyravaud J. P., Terborgh J., ter Steege H., Wright S. J. Biotropica. 2004;36:447–473. [Google Scholar]
- 27.Buzas M. A., Collins L. S., Culver S. J. Proc. Natl. Acad. Sci. USA. 2002;99:7841–7843. doi: 10.1073/pnas.122241499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs J. C. J. Biogeogr. 2004;31:525–530. [Google Scholar]
- 29.Mora C., Chittaro P. M., Sale P. F., Kritzer J. P., Ludsin S. A. Nature. 2003;421:933–936. doi: 10.1038/nature01393. [DOI] [PubMed] [Google Scholar]
- 30.Stehli F. G., Douglas D. G., Newell N. D. Science. 1969;164:947–949. doi: 10.1126/science.164.3882.947. [DOI] [PubMed] [Google Scholar]
- 31.Jablonski D. Nature. 1993;364:142–144. [Google Scholar]
- 32.Laroche J., Bousquet J. Mol. Biol. Evol. 1999;16:441–452. doi: 10.1093/oxfordjournals.molbev.a026126. [DOI] [PubMed] [Google Scholar]
- 33.Laroche J., Li P., Maggia L., Bousquet J. Proc. Natl. Acad. Sci. USA. 1997;94:5722–5727. doi: 10.1073/pnas.94.11.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagstaff S. J., Dawson M. I. Syst. Bot. 2000;25:134–149. [Google Scholar]
- 35.Manos P. S. Am. J. Bot. 1997;84:1137–1155. [PubMed] [Google Scholar]
- 36.Swofford D. L. paup*: Phylogenetic Analysis Using Parsimony (∗ and Other Methods) Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 37.Buckley T. R., Simon C., Chambers G. K. Syst. Biol. 2001;50:67–86. [PubMed] [Google Scholar]
- 38.Thompson T., Gibson T., Plewniak F., Jeanmougin F., Higgins D. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]