Abstract
Monoplacophorans are among the rarest members of the phylum Mollusca. Previously only known from fossils since the Cambrian, the first living monoplacophoran was discovered during the famous second Galathea deep-sea expedition. The anatomy of these molluscs shocked the zoological community for presenting serially repeated gills, nephridia, and eight sets of dorsoventral pedal retractor muscles. Seriality of organs in supposedly independent molluscan lineages, i.e., in chitons and the deep-sea living fossil monoplacophorans, was assumed to be a relict of ancestral molluscan segmentation and was commonly accepted to support a direct relationship with annelids. We were able to obtain one specimen of a monoplacophoran Antarctic deep-sea species for molecular study. The first molecular data on monoplacophorans, analyzed together with the largest data set of molluscs ever assembled, clearly illustrate that monoplacophorans and chitons form a clade. This “Serialia” concept may revolutionize molluscan systematics and may have important implications for metazoan evolution as it allows for new interpretations for primitive segmentation in molluscs.
Keywords: Antarctica, deep sea, Mollusca, Monoplacophora, phylogeny
Molluscs (snails, slugs, clams, mussels, squids, octopuses, chitons, etc.) exhibit the largest disparity of all animal phyla and rank second behind arthropods in species diversity. Although the majority of species still remain in the oceans, where they inhabit all types of ecosystems from the upper littoral to the abyss, they are also major components of freshwater and terrestrial habitats. Molluscan diversity can be extraordinary in tropical and temperate regions (1) but can be found at all latitudes.
The phylogenetic position of molluscs within Spiralia is supported by the presence of spiral cleavage and a trochophore larva (2, 3), although their immediate sister group remains uncertain. Although some have proposed a relationship to sipunculans (peanut worms) (4) or entoprocts (5), most researchers still consider molluscs closely related to annelids, in part because of the assumption that they retain traces of segmentation (3). The removal of arthropods and their relatives from the clade Spiralia (6) and the evolutionary importance given to segmentation in annelids have contributed to reengaging the debate about ancestral segmentation in other spiralian clades such as molluscs. This supposed segmentation in molluscs is often justified by the presence of eight sets of pedal retractor muscles and serially repeated gills in both chitons (Polyplacophora) (7) and members of the living fossil class Monoplacophora (8–10), based on the assumption that both groups are basal within their distinct lineages. Certain bivalves also exhibit multiple pedal retractor muscles (11), and caudofoveate larvae show seven transverse rows of calcareous spicules on the dorsal side (3).
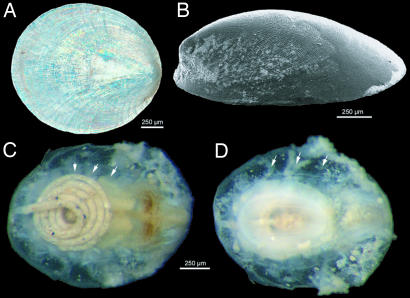
Monoplacophorans are perhaps the least known members of the phylum Mollusca. They have been thought to be “primitive” forms based on their rich fossil record, which dates back to Cambrian–Devonian periods (8). After the recent discovery of the first living monoplacophoran, Neopilina galatheae, during the second Danish Galathea expedition (8), it was suggested that its dorsal uncoiled cap-like shell (Fig. 1) fit the prevalent HAM (hypothetical ancestor mollusc) theories (12). This idea positioned monoplacophorans at the base of “Conchifera,” a clade that includes all molluscs with a true dorsal shell (the classes Monoplacophora, Gastropoda, Cephalopoda, Bivalvia, and Scaphopoda). Neopilina’s newly discovered anatomy [with serially repeated gills and eight sets of dorsoventral pedal retractor muscles, as those found in chitons, and serially repeated nephridia (8, 10)] suggested that serial homology was present at least in two extant molluscan lineages, Aculifera (molluscs with spicules) and Conchifera (molluscs with a true shell).
Fig. 1.
Details of L. antarctica Warén & Hain, 1992. (A) Shell, dorsal view. Note the limpet-like shape with anterior apex and light reflection caused by prismatic and inner nacreous layers. (B) Scanning electron micrograph of the shell (dorsolateral view from left side). (C) Soft body (shell removed) (dorsal view). Note the characteristic spiral intestine (left) filled with mineral particles, brown-dotted esophageal pouches (right), and serial shell muscles (arrows). (D) Soft body, ventral view. Note the round sucker-like foot (central), serial gills (arrows), and mouth area with tentacles (right).
Although a generalized mollusc is portrayed as a limpet-like form with a creeping foot and a dorsal shell made of calcium carbonate (as in the class Monoplacophora), other body plans such as those of the worm-like, shell-less fossorial chaetodermomorphs (class Caudofoveata) and neomeniomorphs (class Solenogastres), or the bentho-pelagic cephalopods (class Cephalopoda) differ radically from this prototype. Mussels, clams and their kind (class Bivalvia) are also quite divergent from this model. Furthermore, modern chitons (class Polyplacophora) have a distinct dorsal “shell” formed by eight interlocking plates. In fact, the disparity of mollusc body plans is so great that it is quite difficult to find a single trait shared by all seven classes of molluscs (13).
Our understanding of relationships among the major molluscan lineages is still in its infancy. Recent attempts to resolve their relationships by using morphological data found limitations in character homology definitions and polarization because of uncertainty regarding the molluscan sister group (4, 5, 14). Molecular attempts have not been conclusive, but they have aided to refute the “Diasoma” hypothesis (a clade uniting bivalves and scaphopods). Most recent molecular analyses suggest a relationship of scaphopods to cephalopods and gastropods (15–17), further corroborated through morphological and developmental studies (5, 18). To date, the phylogenetic position of monoplacophorans remained untested using molecular data because of difficulties in collecting live samples of these enigmatic animals.
Results and Discussion
An Antarctic Benthic Deep-Sea Biodiversity oceanographic campaign (ANDEEP III) with the RV Polarstern to the Weddell Sea (Antarctica), 3 km southwest of Wegener Canyon at ≈3,100-m depth, yielded one small specimen (1.7-mm shell length) of the monoplacophoran Laevipilina antarctica Warén & Hain, 1992 (19), one of the 26 known species of this group of molluscs (9, 20). The single specimen was obtained from an epibenthic sledge sample that had been fixed with precooled 96% EtOH for molecular studies and stored at −20°C for 48 h. The shell (ZSM Moll 20050866; Fig. 1 A and B) was removed for gross anatomy and SEM examination, the soft body was photographed (Fig. 1 C and D), and half of the specimen was used for molecular work.
Although monoplacophoran DNA was highly degraded, perhaps because of bulk fixation of the sediment performed in the vessel, we were able to amplify and sequence a 1.2-kb fragment of the large nuclear ribosomal subunit (28S rRNA). This gene has proven to be highly informative in recent studies on metazoan and molluscan evolution (17, 21).
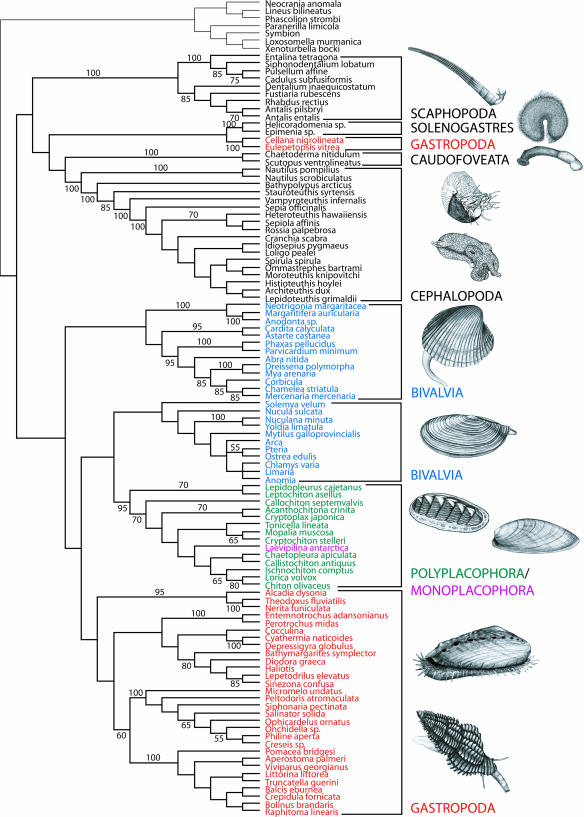
Analysis of the data using a single-step phylogenetic approach with direct optimization (Fig. 2) and a two-step approach using Bayesian phylogenetics (Fig. 4, which is published as supporting information on the PNAS web site) exhibited congruent results suggesting monophyly of molluscs as well as that of the molluscan classes Caudofoveata, Solenogastres, Scaphopoda, and Cephalopoda. Resolution with high jackknife support is found mostly within the main clades of Scaphopoda (Dentaliida and Gadilida), Cephalopoda (Nautiloidea, Coleoidea, and the sister group relationship of the vampire squid to decabrachians, which include the giant squid Architeuthis dux and the pygmy squid Idiosepius pygmaeus), as well as within Bivalvia (Palaeoheterodonta and Euheterodonta) and Gastropoda (Patellogastropoda, Neritopsina, Caenogastropoda, and Heterobranchia). However, the available sequence data do not recover monophyly of Gastropoda or Bivalvia, which are both diphyletic, with patellogastropods separated from the other gastropods and heteroconchs separated from the remainder of the bivalves (protobranchs and pteriomorphians).
Fig. 2.
Phylogenetic tree depicting the relationships of Monoplacophora to other molluscs based on the combined analysis of all molecular loci. Shown is strict consensus of two most parsimonious trees at 64,679 weighted steps (gap opening cost of 3, gap extension cost of 1, all base transformations cost 2) for the analysis of all data under direct optimization with tree fusing. Numbers on branches indicate jackknife support values. Gastropods (in red) and bivalves (in blue) appear diphyletic. Polyplacophora and Monoplacophora form a well supported clade (95% jackknife support). The monoplacophoran species (purple) appears nested within chitons (dark green), but nodal support for its exact position is low. The tree shows monophyly of molluscs, as well as that of Scaphopoda, Cephalopoda, Caudofoveata, and Solenogastres.
Nodal support for interclass relationships or for the relationships of the two clades of bivalves and gastropods is low in general, but a clade containing Monoplacophora and Polyplacophora received strong nodal support (90–100% jackknife support value depending on the analysis, as well as 1.0 posterior probability). Interestingly, this clade, which we name “Serialia,” contains the two classes whose members present a variable number of serially repeated gills and eight sets of dorsoventral pedal retractor muscles. This result clearly contrasts with previous cladistic hypotheses suggesting that Monoplacophora constitute the sister group to the remainder of the conchiferans (4, 5, 14), those molluscs with a true shell unlike that of chitons or the vermiform aplacophorans, although it finds no clear support for the exact position of Serialia. To our knowledge, this is also the first published analysis that demonstrates monophyly of the phylum Mollusca using a range of appropriate outgroups, but we caution the reader to consider that jackknife support for molluscan monophyly is low. The results further support a previous study (22) that indicates that Xenoturbella is not a bivalve mollusc.
All analyses (including different optimality criteria and alternative models of indel and base substitutions) support a Polyplacophora plus Monoplacophora clade. However, L. antarctica appears nested within the chiton tree in some analyses, a result that may look suspicious at first. Evidence for including Monoplacophora within Polyplacophora is restricted to one node, which groups nonlepidopleurid chitons with the monoplacophoran species (70% jackknife support; Fig. 1), but this is not the case when considering only the 1.2-kb region of 28S rRNA amplified for Laevipilina (tree not shown). Furthermore, detailed examination of the DNA sequences clearly illustrates that chitons share unambiguous positions in the alignment not found in L. antarctica (Fig. 3). This fact eliminates the possibility of contaminant DNA in our analysis.
Fig. 3.
Alignment of one of the regions of 28S rRNA illustrating that L. antarctica does not share unique chiton synapomorphies (asterisks).
Evidence for a clade of serialian molluscs is important for our current understanding of molluscan relationships and may have implications for deeper metazoan evolution. This new evidence may imply that serially repeated structures (e.g., gills and pedal retractor muscles in both monoplacophorans and chitons) are not primitive for molluscs, as was previously thought (9). However, it is fair to mention that additional types of serial repetition of dorsoventral musculature have been reported in other molluscan groups (23), including the eight sets of pedal retractors of the Ordovician lucinoid bivalve Babinka (11), the serially repeated rows of spicules in caudofoveate larvae (3), or the two pairs of gills and nephridia in cephalopods (3). Whether these represent true seriality or not may have profound implications in reconstructing the molluscan common ancestor, but it does not contradict the evidence of our Serialia clade.
The classical hypothesis for the position of monoplacophorans as basal conchiferans relies heavily on the presence of a true dorsal shell with similar mineralogical composition to that of many basal members of each conchiferan class. However, the mode of shell deposition by the mantle edge and the microstructure and composition of the chitinous organic layer in monoplacophorans differ from those of higher conchiferans or polyplacophorans (9, 24, 25), which makes monoplacophorans apomorphic (derived) in the form of shell deposition. The rejection of conchiferan monophyly based on shell deposition would be consistent with our findings, which suggest that serial repetition of anatomical structures such as gills and muscles may have evolved once in the common ancestor of chitons and monoplacophorans. Therefore, serial repetition of these structures could constitute a derived feature that would not support the hypothesis of a segmented ancestral mollusc. Again, other interpretations may exist if the pedal scars of Bibankia were the result of muscles homologous to the serialian dorsoventral pedal muscles.
Molluscs are undoubtedly one of the animal phyla with the largest disparity. Numerous Cambrian forms such as Wiwaxia and Halkieria or the Silurian Acaenoplax have been more or less ambiguously assigned to this animal phylum (26–28). Kimberella is another putative mollusc extending the age of the group back to the Neoproterozoic (29). Although chitons were once thought to have changed little since their first appearance in the Late Cambrian period (30), recent discoveries of articulated polyplacophorans and multiplacophorans from the Ordovician to the Carboniferous (31, 32) suggest that a much larger disparity evolved during the Paleozoic. Perhaps such an episode of diversification is responsible for the two modern anatomies of molluscs with conspicuous serial repetition of organs, but no explanation for their divergent evolution of shell morphologies can be provided at this point. Recognition of a serialian clade comprised of chitons and monoplacophorans broadens our perspective toward new interpretations of molluscan anatomy and once more questions preconceived ideas on molluscan relationships that rely almost entirely on shell morphology.
Here we provide the first molecular test for the phylogenetic position of Monoplacophora by using sequence data from a deep-sea monoplacophoran species from Antarctica. Contrary to all previously published accounts, which placed monoplacophorans as a sister group to higher, i.e., shelled, molluscs, our data strongly support a clade including Monoplacophora and Polyplacophora. This rather surprising result from a conchological perspective is congruent with soft anatomy data. It furthermore reopens the debate about the putative ancestral segmentation of molluscs (3), because serial repetition of gills and pedal retractor muscles may be derived and not primitive features within molluscs. If this were the case, little evidence would remain for the case of homology of segmentation in annelids and serial repetition in molluscs (33), as confirmed in part by recent reevaluation of their early development (34, 35).
Materials and Methods
Species Sampling.
Taxon sampling was carefully designed following original and published work on the internal phylogeny of chitons, bivalves, cephalopods, gastropods, and scaphopods (15, 16, 36–38). Outgroups were selected among other spiralian protostomes (lophotrochozoans) (39). The enigmatic Xenoturbella was also included because it was once postulated to be a derived mollusc, although more recent data consider it to be an ancestral deuterostome (22). In total, we analyzed 101 molluscs including 2 Caudofoveata, 2 Solenogastres, 13 Polyplacophora, 1 Monoplacophora, 9 Scaphopoda, 32 Gastropoda, 24 Bivalvia, and 18 Cephalopoda (see Table 1).
Table 1.
Taxon sampling and GenBank accession numbers employed in this study
Molecular Data.
Molecular data were obtained from ethanol-preserved specimens following standard protocols for molluscan samples (15, 37, 38, 40). Monoplacophoran DNA samples were extracted from the half specimen preserved in 96% EtOH. DNA from preserved tissues was extracted by using the Qiagen DNeasy tissue kit. Data include complete sequences of 18S rRNA, a 3-kb fragment of 28S rRNA, the protein-coding nuclear gene histone H3, and two mitochondrial gene fragments for cytochrome c oxidase subunit I and 16S rRNA, totaling ≈6.5 kb per complete taxon (see Table 1). The amplified samples were purified by using the QIAquick PCR purification kit (Qiagen), labeled by using BigDye Terminator 3.0 (Applied Biosystems), and sequenced with an ABI 3730 genetic analyzer (Applied Biosystems) following the manufacturer’s protocols. Chromatograms obtained from the automatic sequencer were read, and “contig sequences” were assembled by using the editing software sequencher 4.0 and further manipulated in gde 2.2 (41).
From the five different molecular loci chosen for this study, only one yielded positive amplification for the monoplacophoran specimen. This fragment corresponds to a 1.2-kb segment of 28S rRNA obtained by amplifying two overlapping fragments using primer pairs 28Sa and 28S rd5b (5′-GACCCGTCTTGAAGCACG-3′ and 5′-CCACAGCGCCAGTTCTGCTTAC-3′) and 28S rd4.8a and 28S rd7b1 (5′-ACCTATTCTCAAACTTTAAATGG-3′ and 5′-GACTTCCCTTACCTACAT-3′).
Data Analyses.
DNA sequence data were analyzed following two approaches. First, a dynamic homology approach (“single-step phylogenetics”) using parsimony as an optimality criterion for direct optimization (42) was undertaken in the computer package poy 3.0.11 (43). Second, a static homology approach (“two-step phylogenetics”) using a model-based approach was executed under Bayesian phylogenetics in mrbayes 3.1.1 (44).
For the direct optimization analysis, tree searches were conducted by a combination of random addition sequences with multiple rounds of tree fusing (45) on a small 50-processor cluster assembled at Harvard University. Support measures were estimated by using jackknifing with a character probability of deletion of e−1 (46). The data were analyzed for all genes in combination as well as restricted to the 28S rRNA fragment sequenced for L. antarctica under different analytical parameter sets (47, 48).
Bayesian posterior probabilities were calculated by using a general time-reversible model with corrections for the proportion of invariant sites and a discrete gamma distribution, as selected in modeltest 3.7 (49) under the Akaike Information Criterion (50). Two runs of 106 generations were performed, storing 1/100th visited trees. Results from mrbayes 3.1.1 were visualized in the program tracer 1.3 (51), which served to determine the burnin, which differed considerably in the two runs. Aligned data were obtained from the implied alignment (52) generated in poy 3.0.11 for the analyses presented in Fig. 2.
Supplementary Material
Acknowledgments
We are indebted to the numerous colleagues who supplied tissue samples, without whom this work would not have been possible. Rebecca Budinoff assisted with laboratory work. Angelika Brandt organized the ANDEEP III expedition. Katrin Linse, Enrico Schwabe, and the onboard sorting team provided the monoplacophoran specimen. Greg Edgecombe, Andy Knoll, Sigurd von Boletzky, Claus Nielsen, Jim Valentine, and an anonymous reviewer provided comments that helped to improve this article. Enrico Schwabe provided pictures for Fig. 1 A and B. This material is based on work supported by the National Science Foundation Assembling the Tree of Life Program (Grant 0334932 to G.G.) and Population Biology Program (Grant 0316516 to M.K.N.). Field activities of M.S. and his team were supported by the GeoBioCenterLMU. This article is ANDEEP contribution no. 58.
Footnotes
References
- 1.Bouchet P., Lozouet P., Maestrati P., Heros V. Biol. J. Linn. Soc. 2002;75:421–436. [Google Scholar]
- 2.Valentine J. W. Proc. Natl. Acad. Sci. USA. 1997;94:8001–8005. doi: 10.1073/pnas.94.15.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen C. Animal Evolution: Interrelationships of the Living Phyla. 2nd Ed. Oxford: Oxford Univ. Press; 2001. [Google Scholar]
- 4.Scheltema A. H. Biol. Bull. 1993;184:57–78. doi: 10.2307/1542380. [DOI] [PubMed] [Google Scholar]
- 5.Haszprunar G. Am. Malacol. Bull. 2000;15:115–130. [Google Scholar]
- 6.Aguinaldo A. M. A., Turbeville J. M., Lindford L. S., Rivera M. C., Garey J. R., Raff R. A., Lake J. A. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 7.Eernisse D. J., Reynolds P. D. In: Microscopic Anatomy of Invertebrates. Harrison F. W., Kohn A. J., editors. Vol. 5. New York: Wiley-Liss; 1994. pp. 55–110. [Google Scholar]
- 8.Lemche H. Nature. 1957;179:413–416. [Google Scholar]
- 9.Haszprunar G., Schaefer K. In: Microscopic Anatomy of Invertebrates. Harrison F. W., Kohn A. J., editors. Vol. 6B. New York: Wiley-Liss; 1997. pp. 415–157. [Google Scholar]
- 10.Lemche H., Wingstrand K. G. Galathea Rep. 1959;3:9–71. [Google Scholar]
- 11.McAlester A. L. Paleontology. 1965;8:231–246. [Google Scholar]
- 12.Lindberg D. R., Ghiselin M. T. Proc. Calif. Acad. Sci.; 2003. pp. 663–686. [Google Scholar]
- 13.Valentine J. W. On the Origin of Phyla. Chicago: Univ. of Chicago Press; 2004. [Google Scholar]
- 14.Salvini-Plawen L. V., Steiner G. In: Origin and Evolutionary Radiation of the Mollusca. Taylor J. D., editor. Oxford: Oxford Univ. Press; 1996. pp. 29–51. [Google Scholar]
- 15.Giribet G., Wheeler W. C. Invertebr. Biol. 2002;121:271–324. [Google Scholar]
- 16.Steiner G., Dreyer H. Zool. Scripta. 2003;32:343–356. [Google Scholar]
- 17.Passamaneck Y. J., Schander C., Halanych K. M. Mol. Phylogenet. Evol. 2004;32:25–38. doi: 10.1016/j.ympev.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 18.Wanninger A., Haszprunar G. J. Morphol. 2002;254:53–64. doi: 10.1002/jmor.10004. [DOI] [PubMed] [Google Scholar]
- 19.Schrödl M., Linse K., Schwabe E. Polar Biol. 2006 in press. [Google Scholar]
- 20.Schrödl M. Spixiana. 2006 in press. [Google Scholar]
- 21.Mallatt J., Winchell C. J. Mol. Biol. Evol. 2002;19:289–301. doi: 10.1093/oxfordjournals.molbev.a004082. [DOI] [PubMed] [Google Scholar]
- 22.Bourlat S. J., Nielsen C., Lockyer A. E., Littlewood D. T., Telford M. J. Nature. 2003;424:925–928. doi: 10.1038/nature01851. [DOI] [PubMed] [Google Scholar]
- 23.Salvini-Plawen L. V. Z. Wiss. Zool. (Leipzig) 1972;184:205–394. [Google Scholar]
- 24.Schaefer K., Haszprunar G. Zool. Anz. 1997;236:13–23. [Google Scholar]
- 25.Haas W. Biominer. Res. Rep. 1981;6:1–52. [Google Scholar]
- 26.Scheltema A. H., Kerth K., Kuzirian A. M. J. Morphol. 2003;257:219–245. doi: 10.1002/jmor.10121. [DOI] [PubMed] [Google Scholar]
- 27.Vinther J., Nielsen C. Zool. Scripta. 2005;34:81–89. [Google Scholar]
- 28.Sutton M. D., Briggs D. E. G., Siveter D. J., Siveter D. J. Palaeontology. 2004;47:293–318. [Google Scholar]
- 29.Martin M. W., Grazhdankin D. V., Bowring S. A., Evans D. A., Fedonkin M. A., Kirschvink J. L. Science. 2000;288:841–845. doi: 10.1126/science.288.5467.841. [DOI] [PubMed] [Google Scholar]
- 30.Runnegar B., Pojeta J., Jr, Taylor M. E., Collins D. J. Paleontol. 1979;53:1374–1394. [Google Scholar]
- 31.Pojeta J., Jr, Eernisse D. J., Hoare R. D., Henderson M. D. J. Paleontol. 2003;77:646–654. [Google Scholar]
- 32.Vendrasco M. J., Wood T. E., Runnegar B. N. Nature. 2004;429:288–291. doi: 10.1038/nature02548. [DOI] [PubMed] [Google Scholar]
- 33.Haszprunar G., Schaefer K. Acta Zool. (Stockholm) 1996;77:315–334. [Google Scholar]
- 34.Nielsen C. J. Exp. Zool. B. 2004;302:35–68. doi: 10.1002/jez.b.20001. [DOI] [PubMed] [Google Scholar]
- 35.Wanninger A., Haszprunar G. J. Morphol. 2002;251:103–113. doi: 10.1002/jmor.1077. [DOI] [PubMed] [Google Scholar]
- 36.Giribet G., Distel D. L. In: Molecular Systematics and Phylogeography of Mollusks. Lydeard C., Lindberg D. R., editors. Washington, DC: Smithsonian Books; 2003. pp. 45–90. [Google Scholar]
- 37.Okusu A., Schwabe E., Eernisse D. J., Giribet G. Org. Divers. Evol. 2003;3:281–302. [Google Scholar]
- 38.Lindgren A. R., Giribet G., Nishiguchi M. K. Cladistics. 2004;20:454–486. doi: 10.1111/j.1096-0031.2004.00032.x. [DOI] [PubMed] [Google Scholar]
- 39.Giribet G. Mol. Phylogenet. Evol. 2002;24:345–357. doi: 10.1016/s1055-7903(02)00206-3. [DOI] [PubMed] [Google Scholar]
- 40.Okusu A., Giribet G. J. Moll. Stud. 2003;69:385–387. [Google Scholar]
- 41.Linton E. W. macgde: Genetic Data Environment for Mac OS X. East Lansing: Michigan State Univ; 2005. [Google Scholar]
- 42.Wheeler W. C. Cladistics. 1996;12:1–9. [Google Scholar]
- 43.Wheeler W. C., Gladstein D., De Laet J. poy. New York: Am. Museum of Natural History; 2004. Version 3.0. [Google Scholar]
- 44.Ronquist F., Huelsenbeck J. P. mrbayes: Bayesian Analysis of Phylogeny. Tallahassee: Florida State Univ; 2005. Version 3.1.1. [Google Scholar]
- 45.Goloboff P. A. Cladistics. 1999;15:415–428. doi: 10.1111/j.1096-0031.1999.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 46.Farris J. S., Albert V. A., Källersjö M., Lipscomb D., Kluge A. G. Cladistics. 1996;12:99–124. doi: 10.1111/j.1096-0031.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- 47.Wheeler W. C. Syst. Biol. 1995;44:321–331. [Google Scholar]
- 48.Giribet G. Syst. Biol. 2003;52:554–564. doi: 10.1080/10635150390223730. [DOI] [PubMed] [Google Scholar]
- 49.Posada D. modeltest. Vigo, Spain: Univ. of Vigo; 2005. Version 3.7. [Google Scholar]
- 50.Posada D., Buckley T. Syst. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 51.Rambaut A., Drummond A. tracer: MCMC Trace Analysis Tool. Oxford: University of Oxford; 2003. Version 1.3. [Google Scholar]
- 52.Wheeler W. C. Cladistics. 2003;19:261–268. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.