Abstract
About 35% of patients with 22q11 deletion syndrome (22q11DS), which includes DiGeorge and velocardiofacial syndromes, develops psychiatric disorders, mainly schizophrenia and bipolar disorder. We previously reported that mice carrying a multigene deletion (Df1) that models 22q11DS have reduced prepulse inhibition (PPI), a behavioral abnormality and schizophrenia endophenotype. Impaired PPI is associated with several psychiatric disorders, including those that occur in 22q11DS, and recently, reduced PPI was reported in children with 22q11DS. Here, we have mapped PPI deficits in a panel of mouse mutants that carry deletions that partially overlap with Df1 and have defined a PPI critical region encompassing four genes. We then used single-gene mutants to identify the causative genes. We show that PPI deficits in Df1/+ mice are caused by haploinsufficiency of two genes, Tbx1 and Gnb1l. Mutation of either gene is sufficient to cause reduced PPI. Tbx1 is a transcription factor, the mutation of which is sufficient to cause most of the physical features of 22q11DS, but the gene had not been previously associated with the behavioral/psychiatric phenotype. A likely role for Tbx1 haploinsufficiency in psychiatric disease is further suggested by the identification of a family in which the phenotypic features of 22q11DS, including psychiatric disorders, segregate with an inactivating mutation of TBX1. One family member has Asperger syndrome, an autistic spectrum disorder that is associated with reduced PPI. Thus, Tbx1 and Gnb1l are strong candidates for psychiatric disease in 22q11DS patients and candidate susceptibility genes for psychiatric disease in the wider population.
Keywords: mouse model, psychiatric disease, DiGeorge syndrome, sensorimotor gating
Caused by a heterozygous multigene deletion, 22q11 deletion syndrome (22q11DS) is a relatively common genetic disorder (1:4,000 live births). Behavioral and psychiatric disorders are a prominent part of the 22q11DS phenotype. In children, these disorders include cognitive defects, anxiety, attention deficit disorder, and problems of social interaction that are increasingly recognized to meet the criteria of autistic spectrum disorder (1, 2), a neurodevelopmental disorder. In adults, high rates of psychotic disorders, especially schizophrenia, have been reported (2–5).
It is likely that the pathophysiological basis of many psychiatric disorders is heterogeneous involving multiple genes and environmental factors. Therefore, when they occur frequently in association with a defined genetic defect, as in the case of 22q11DS (3, 4, 6, 7), it offers a unique opportunity to identify causative or contributing genes, especially if a good animal model is available. We developed a mouse model of 22q11DS (8), the Df1/+ mouse, which carries a heterozygous deletion encompassing 22 genes. Df1/+ mice recapitulate many of the cardiovascular defects associated with 22q11DS (8), and they also display abnormal behavior, including impaired sensorimotor gating, as measured by prepulse inhibition (PPI) of the startle response (9), a behavioral abnormality that is associated with several psychiatric and behavioral disorders including schizophrenia and schizotypal personality (reviewed in ref. 10), 22q11DS (11), and Asperger syndrome (12). In the present study, we set out to determine whether reduced PPI in Df1/+ mice results from haploinsufficiency of a particular gene or genes. Results unexpectedly revealed the presence of two adjacent, dosage-sensitive genes that significantly affect sensorimotor gating. We also report that psychiatric disorders, in particular Asperger syndrome, can occur in association with inactivating mutations of TBX1, rather than with a 22q11 chromosomal deletion, consistent with our mouse studies.
Results
PPI Analysis of Mouse Mutants.
The reflexive response to the acoustic startle stimulus that is the basis of the PPI assay requires that mice can hear, and hearing loss could confound the results of this test. We therefore evaluated auditory function in Df1/+ mice by measuring distortion product otoacoustic emissions (for methodology, see Supporting Text, which is published as supporting information on the PNAS web site). No difference was found between mutants and wild-type littermates (see Fig. 5, which is published as supporting information on the PNAS web site), indicating that reduced PPI in Df1/+ mutants is not secondary to hearing loss.
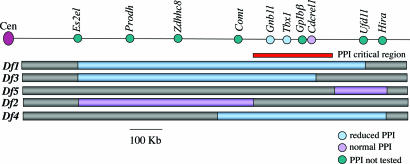
To identify the gene(s) responsible for impaired sensorimotor gating in Df1/+ mice, we performed the PPI assay on five mutant mouse lines (13, 14) that carry heterozygous multigene deletions that are partially overlapping with Df1 (Fig. 1). All mice analyzed were male and female littermates on a N5–6 C57BL/6c−/c− genetic background that was generated by backcrossing mutant C57BL/6c−/c−;129S5/SvEvBrd mice with C57BL/6c−/c− mice for 5–6 generations. Replicating our original observations (9), we found PPI to be significantly impaired in Df1/+ mice (P = 0.044; Fig. 2). In addition, Df3/+ and Df4/+ mice also had reduced PPI (P = 0.004 and 0.001, respectively), whereas the other two deletion mutants tested, Df2/+ and Df5/+, had normal PPI (P = 0.871 and 0.92, respectively; Fig. 2). Collectively, these results identified a PPI critical region that was defined by the distal Df2 deletion breakpoint and the proximal Df5 deletion breakpoint (Fig. 1). The generation of these two chromosomal deletions has been described (13, 14). Briefly, we used retroviral insertion of an Hprt3′ chromosome engineering cassette (15), using as a substrate for the insertion embryonic stem cell lines in which an Hprt5′ cassette (15) had been previously inserted into the genes Es2el (for Df2) and Hira (for Df4 and Df5). The proximal Df5 breakpoint has been localized to the genomic region between Cdcrel1 and Cldn1 (14), whereas the distal Df2 breakpoint had previously only been mapped by in situ hybridization by using a large genomic clone. To localize more precisely the Df2 breakpoint, we used long-range PCR to amplify the breakpoint region (15). Cloning and sequencing of the PCR products identified the breakpoint to be in intron 2 of the Txnrd2 gene. The published mouse sequence (http://www.ensembl.org) indicated that the refined PPI critical region spanned 300 kb of DNA and encompassed four genes: Gnb1l, Tbx1, Gp1bβ, and Cdcrel1 (Fig. 1). Interestingly, this critical region excluded two genes that have been shown to affect PPI in mice, Prodh (16) and Zdhh8c (17), as being responsible for PPI impairments in the affected deletion mutants. A third candidate behavioral gene that maps within the Df2 deletion is Comt. Several population-based studies have reported genetic association between COMT and schizophrenia, although results have been inconsistent. Recently, a COMT low-activity allele, COMT158met (18), was reported to correlate with increased severity of psychosis and reduced prefrontal cortex gray matter volume in a small longitudinal study of 22q11DS patients (19). Interestingly, in mice, a genetic interaction between Comt and Prodh has been recently demonstrated (20). Our finding of normal PPI in Df2/+ mice, which are heterozygous for Comt, is consistent with a previous study that reported normal PPI in Comt+/− and Comt−/− mice (21) and excludes a role for the gene in the Df1/+ PPI phenotype.
Fig. 1.
Deletion mutants and single-gene mutants. The black line at top is a scaled diagram of the mouse chromosome 16 region that is syntenic to human chromosome 22q11.2 showing selected genes. Blue and violet bars below indicate the deletion alleles established in mice. The original nomenclature of these alleles, Df (16)1–Df (16)5 (13, 14), has been abbreviated to Df1–Df5. The red bar indicates the PPI critical region as defined by the deletion mutants. The presence or absence of PPI impairments in mouse mutants is indicated by color, where blue indicates reduced PPI, violet indicates normal PPI, and green indicates PPI not tested.
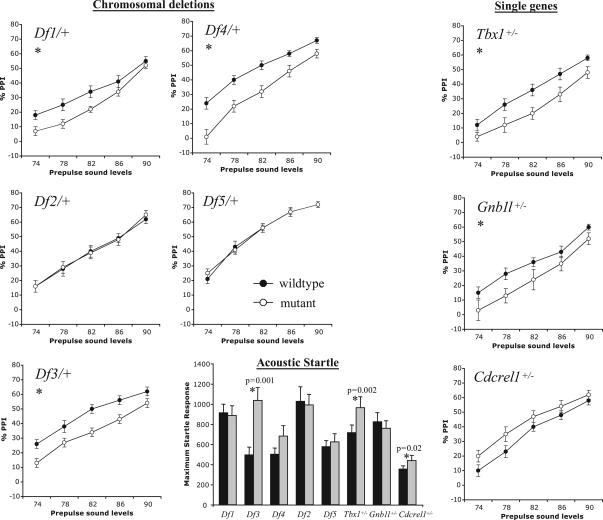
Fig. 2.
PPI of the acoustic startle response (ASR). Reduced PPI was seen in deletion mutants Df1/+, Df3/+, and Df4/+ and in Tbx1+/− and Gnb1l+/− mice. Impairment was most apparent at the lower prepulse sound levels, as previously noted (9). The increased PPI seen in Cdcrel1+/− mice did not reach statistical significance. The magnitude of the ASR was significantly greater (∗) in Df3/+, Tbx1+/−, and Cdcrel1+/− mice than in their respective wild-type littermates, but overall there was no relationship between ASR and PPI.
Genes That Modulate Sensorimotor Gating.
To identify the causative gene(s), we performed the PPI assay in Tbx1+/−, Cdcrel1+/−, and Gnb1l+/− mice. We excluded GpIbβ from our analysis because the gene is only expressed in platelets, and in humans GPIBΒ loss of function causes Bernard–Soulier disease, a bleeding disorder that has no known association with psychiatric disease (22). The generation of Tbx1 and Cdcrel1 mutants has been reported (13). Gnb1l mutant mice were obtained from Lexicon Genetics Inc. (The Woodlands, TX). The Gnb1l gene was inactivated by insertion of a gene-trapping cassette into intron 2,476 base pairs upstream of exon 3, which is the first coding exon. The insertion results in aberrant splicing to β-gal and loss of downstream mRNA (data not shown). Gnb1l+/− mice, which were provided on a mixed C57BL/6c−/c−;129S5/SvEvBrd background, were healthy and fertile. Loss of Gnb1l function is lethal in early embryogenesis, and no Gnb1l−/− embryos were recovered after embryonic day (E)6.5. To test the single-gene mutations on the same background as the deletion mutants, Tbx1+/−, Cdcrel1+/−, and Gnb1l+/− mice were first backcrossed to C57BL/6c−/c− mice for 5–6 generations. Unexpectedly, the PPI assay showed that both Tbx1+/− and Gnb1l+/− mice had reduced PPI (P = 0.013 and 0.046, respectively; Fig. 2), similar to that identified in the affected deletion mutants, whereas Cdcrel1+/− mice were normal (P = 0.341). Thus, normal gene dosage of both Tbx1 and Gnb1l is required for normal sensorimotor gating in mice. We conclude that the deletion of these two genes causes impaired sensorimotor gating in Df1/+, Df3/+, and Df4 mice. Fig. 2 shows that there were significant increases in the acoustic startle response (ASR) in one of the deletion mutants (Df3/+) and in two single-gene mutants, Tbx1+/− and Cdcrel1+/− (P = 0.001, 0.002, and 0.02, respectively). Differences in startle responses have been reported between male and female mice, and estrous can affect PPI. However, in the two-way ANOVA for ASR and the three-way ANOVA for PPI, we did not detect a main effect of gender, and more importantly, no gender-specific effect in any of the genotypes. Thus, it is unlikely that the increased ASR in the abovementioned mutants is caused by gender effects. Overall, there was no direct relationship between levels of PPI and acoustic startle amongst the various mutant mouse lines. Such dissociation between PPI and acoustic startle has been documented by others (reviewed in ref. 23).
Brain Expression of Tbx1 and Gnb1l.
Tbx1 encodes a member of the T-box family of transcription factors. We and others have shown that Tbx1 haploinsufficiency is responsible for cardiovascular, craniofacial, thymic, and parathyroid defects in mouse models of 22q11DS (13, 24, 25), and TBX1 has been confirmed to be a disease gene through the identification of mutations in patients with a classical 22q11DS phenotype but without the common chromosomal deletion (26). Whether TBX1 haploinsufficiency is responsible for neurodevelopmental or psychiatric disorders in 22q11DS patients is not known because neuropsychiatric assessments on patients with the three TBX1 point mutations identified to date have not been reported.
Tbx1 has been shown by RT-PCR to be expressed in postnatal mouse brain (27), but regional brain expression has not been reported. We analyzed Tbx1 expression at various developmental stages by RT-PCR and real-time quantitative RT-PCR and found it to be very low in preterm embryonic brain, whereas levels increased steadily from birth to 3 months (Fig. 3A and B). To analyze regional brain expression we used a lacZ-knockin allele (28), which showed expression to be limited to the vasculature in term embryos (Fig. 3C) and in adult mice (not shown). A role for the microvasculature in the pathophysiology of schizophrenia has been proposed on theoretical grounds, because microvascular damage could satisfy developmental and degenerative models of schizophrenia (reviewed in ref. 29). Such a proposal is on the basis of numerous clinical studies that have reported cerebral blood flow abnormalities and increased prevalence of minor physical abnormalities in schizophrenia patients and the increased prevalence of schizophrenia in individuals who suffered perinatal problems, especially hypoxia.
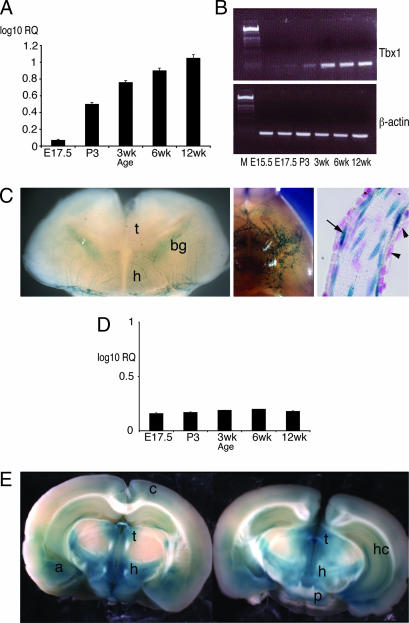
Fig. 3.
Brain expression of candidate behavioral genes. Tbx1 brain expression increases steadily between E17.5 and 12 weeks as measured by real-time quantitative RT-PCR (A) and by semiquantitative RT-PCR (B). β-Gal staining of a Tbx1+/− embryo at E18.5 (C) reveals expression in blood vessels both on the brain surface (Left and Center) and within the brain parenchyma (Left). Tbx1 is expressed in the endothelial cells lining of blood vessels (arrow in Right) but not in the vascular smooth muscle (arrowheads in Right). Gnb1l expression remains steady at the same developmental stages (D). (E) β-Gal-stained thick brain sections (coronal) of an adult Gnb1l+/− mouse showing Gnb1l expression. bg, basal ganglia; t, thalamus; h, hypothalamus; p, pons; a, amygdala; c, cerebral cortex; hc, hippocampus.
Gnb1l encodes an evolutionarily conserved peptide of unknown function, which contains six putative WD40 repeats but no other recognizable functional domains (30, 31). The gene is required for embryonic development and Gnb1l loss of function causes embryonic lethality by E6.5 (P.A. and P.J.S., unpublished data). We analyzed Gnb1l expression by real-time quantitative RT-PCR and found it to be uniformly expressed between E17.5 and 12 weeks (Fig. 3D). The gene is widely expressed in adult mouse brain with striking regional distribution in forebrain, midbrain, and hindbrain structures, including the thalamus, hypothalamus, amygdala, hippocampus, pons (Fig. 3E), medulla, and cerebellum (not shown).
Because Gnb1l and Tbx1 lie only 17 kb apart, we considered the possibility that the engineered mutation of either gene may affect expression of its neighbor. We analyzed Gnb1l expression in the brain of Tbx1−/− embryos at term by in situ hybridization and by real-time quantitative RT-PCR and found no difference between mutant and wild-type littermates (data not shown), indicating that the expression of Gnb1l is not regulated by Tbx1 nor is it compromised by the Tbx1 targeting. The complementary experiment cannot be performed because Gnb1l loss of function is lethal before the onset of Tbx1 expression. However, we have shown that development of the fourth pharyngeal arch artery is a sensitive indicator of Tbx1 dosage reduction, and 100% of Tbx1+/− embryos has fourth pharyngeal arch artery hypoplasia at E10.5 (32). This phenotype is also observed in embryos heterozygous for a hypomorphic allele of Tbx1 (33). If the Gnb1l targeting were to inactivate Tbx1, we would expect to see similar defects in Gnb1l+/− mutants. However, this was not the case and intracardiac ink injection revealed normal fourth pharyngeal arch artery development (data not shown). Although this experiment cannot exclude a tissue-specific effect of Gnb1l mutation on Tbx1 expression in brain, these results make it unlikely that the mutation causes a generalized reduction of Tbx1 expression.
Mutation Analysis in Patients.
The few patients identified so far with TBX1 mutations have not undergone neuropsychiatric assessment. We have identified a cohort of patients with a 22q11DS-like phenotype but without the common 22q11.2 microdeletion. From this cohort, we selected for mutational analysis of TBX1 a family with familial velocardiofacial syndrome (VCFS), which is one of the clinical manifestations of 22q11DS (see Supporting Text for detailed clinical information). Briefly, the index patient, V39/02 (Fig. 4A), has the characteristic facial appearance of VCFS and hypernasal speech. She has no known cardiac defect. Both her surviving children have the characteristic facial appearance of VCFS and congenital heart disease: V39/04, a 17-year-old male, has pulmonary stenosis; and V39/03, a 13-year-old male, was diagnosed at birth with tetralogy of Fallot (Fig. 4A). A recent psychiatric assessment of both boys (see Supporting Text for methodology) resulted in the diagnosis of Asperger syndrome in V39/03. Asperger syndrome is an autistic spectrum disorder that is characterized by stereotyped and obsessional behavior and pervasive abnormalities in socioemotional and communicative behavior (34–36). Impaired PPI has been reported in some individuals with Asperger syndrome (12). The presence of Asperger syndrome in a single family member is consistent with the high variability of other features of the 22q11DS, even within families. In addition, psychiatric disorders often appear in adulthood.
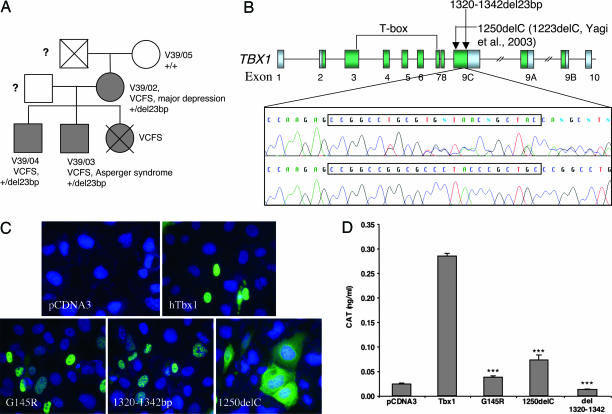
Fig. 4.
Patient data and mutation analysis. (A) Family of index patient V39/02: Individuals with VCFS are shaded; circles symbolize females; squares symbolize males; and crossed symbols represent deceased family members. ?, no information available. (B) Shown at the top is a schematic representation of alternatively spliced TBX1 transcripts (TBX1A, TBX1B, TBX1C); arrows indicate position of known frameshift mutations in TBX1C; green boxes depict TBX1 coding sequences; and gray boxes depict UTRs. Shown at the bottom is a DNA sequence of a patient with the 1320–1342del23bp mutation (upper panel) and the wild-type TBX1 sequence in an unrelated individual (lower panel); the position of the mutation is boxed. (C) Subcellular localization of wild-type and mutant TBX1 constructs expressed in U2-OS cells. hTbx1, wild-type TBX1; 1320–1342del, del23bp mutation described here; G145R, predicted null mutation; 1250delC, point mutation described by Yagi et al. (26). Constructs were detected with anti-TBX1 antibody. Cell nuclei were stained with DAPI. (D) Transcriptional activation of the CAT reporter gene by wild-type and mutant TBX1. Significant differences in transcriptional activation between wild-type TBX1 and a TBX1 construct are indicated by ∗∗∗ (P ≤ 0.001). Data were normalized for transfection efficiency and depicted as average values ± SEM.
Screening of the TBX1 coding sequence identified a 23-bp frameshift deletion (1320–1342del23bp) in patient V39/02 and in both her sons. The deceased daughter of V39/02 also carried the diagnosis of VCFS, but no DNA was available for analysis. The mutation occurred at the 3′ end of the TBX1 transcript (counting A of the initiation codon as 1; Fig. 4B). This mutation was not detected in 716 controls. The frameshift created by 1320–1342del23bp starts at codon 440 in the C terminus of the TBX1 protein and extends the protein from 504 to 616 amino acids. Although the mutation does not affect the T-box, it disrupts the central domain (amino acids 439–448) of a highly conserved nuclear localization signal (NLS) of the wild-type TBX1 protein (37), where it changes the conserved residues PYP to WPR (see Fig. 6, which is published as supporting information on the PNAS web site).
To identify the functional significance of this mutation, we engineered a human TBX1 cDNA carrying the 1320–1342del23bp mutation and tested the ability of the mutant protein to localize to the nucleus and to transactivate a T-box-binding element construct in a tissue culture system (see Supporting Text for methodology). In parallel, we tested two other engineered constructs: 1250delC, which encodes a previously identified mutant form of TBX1 (26), and G145R, which is the equivalent of a loss-of-function TBX5 T-box mutation G80R that prevents DNA binding (38). Immunocytochemical investigation of transfected cells showed tight nuclear localization of the wild-type TBX1 and G145R proteins (Fig. 4C), whereas the 1250delC mutant protein was mainly found in the cytoplasm, consistent with the findings of Stoller and Epstein (37) (note that 1250delC in our numbering is equivalent to 1223delC in ref. 26). Unexpectedly, the 1320–1342del23bp mutant protein also localized to the nucleus, despite lacking the NLS (Fig. 4C). We therefore examined this peptide sequence for other potential NLS sequences (39) and found that the frameshifted protein contains the sequence RGRRRRCR at amino acids 465–472 (see Fig. 7, which is published as supporting information on the PNAS web site). This sequence corresponds to a known NLS sequence R[GVLIP]RRRRxR that is found in a variety of animal protamine sequences, as well as Epstein–Barr nuclear antigen (http://cubic.bioc.columbia.edu/predictNLS/).
We have shown previously that TBX1 is a transcriptional activator that can induce expression of a CAT-reporter protein under the control of a Brachyury consensus binding site sequence, 1T-CAT (40). Using the same experimental system, we found that wild-type TBX1 activated the CAT reporter, confirming our previous finding (Fig. 4D), whereas the mutant constructs G145R, 1250delC, and 1320–1342del23bp did not. Overall, these data suggest that the mutation identified in V39/02 and her children is a null mutation and that their disease phenotype results from TBX1 haploinsufficiency.
Discussion
The strong association between common psychiatric disorders and the 22q11.2 microdeletion suggests that haploinsufficiency of one or more genes in the region confers susceptibility to these disorders. Three candidate genes from the region, COMT, PRODH, and ZDHH8C, have been shown to cause behavioral abnormalities when the genes are mutated in mice. PPI defects have been reported in Prodh and Zdhhc8 null mutants (very mild in the latter case), but not in heterozygotes, whereas Comt heterozygous and homozygous mice have normal PPI. Our study shows that in a uniform genetic background, combined heterozygosity of all three genes does not affect PPI, although we cannot formally exclude that normal PPI in Df2/+ mice results from a combined effect of two or more genes that positively and negatively modulate PPI. However, our data are consistent with published data that show normal PPI in Zdhh8c and Comt heterozygotes [PPI levels have not been reported for Prodh heterozygotes, but they have normal l-proline levels (20)]. Thus, in the context of the PPI phenotype, there is no evidence of a genetic interaction between any of these genes at heterozygous gene dosage levels. In the future, it will be interesting to see whether a Tbx1 transgene can rescue PPI defects in Df1/+ mutants. Currently, this cannot be tested because the only Tbx1 transgenic lines available carry transgenes that contain multiple genes, including Tbx1.
Our strategy to genetically dissect the Df1 deletion unexpectedly revealed the presence of two adjacent, dosage-sensitive genes that significantly affect sensorimotor gating. Several genes are known to affect sensorimotor gating, and PPI defects are genetically heterogeneous. Importantly from the disease perspective however, Tbx1 and Gnb1l represent previously unrecognized examples of genes that affect PPI in the heterozygous mutant state. Both genes are hemizygous in 22q11DS patients, making them strong candidates for the associated psychiatric and behavioral phenotypes. The two genes are apparently unrelated and have distinct expression patterns in brain, suggesting that they are unlikely to function in the same genetic pathway. Particularly intriguing is our finding of an inactivating mutation in TBX1 in an individual with Asperger syndrome. Because of the small number of patients identified so far with TBX1 mutations, it is difficult to evaluate the relative contribution of TBX1 haploinsufficiency to behavioral disorders and psychiatric disease in 22q11DS. However, our mouse studies show that of 22 genes tested, Tbx1 and Gnb1l are the only ones that cause PPI impairment in the heterozygous state. Therefore, we propose that these two genes are major contributors to the 22q11DS psychiatric phenotype. Furthermore, we propose that in the general population, the 88-kb genomic segment region that harbors TBX1 and GNB1L may represent a susceptibility locus for schizophrenia and other psychiatric disorders characterized by PPI impairment. Future studies into the functions of these two genes in brain should clarify the genetic pathways affected by their mutation and, potentially, may lead to the identification of drug targets aimed at prevention and or treatment of the psychiatric symptoms in 22q11DS patients. Of particular interest is the observation that Tbx1 expression in brain increases postnatally, suggesting that early drug intervention may prevent the onset of Tbx1-related psychiatric disorders.
Methods
Mouse Strains, Breeding, and Genotyping.
Behavioral testing was performed on a total of 502 mice (n = 25–35 mutant and wild-type male and female littermates for each mutation). For each genotype, similar numbers of males and females were tested. The mice were all on a N5–6 C57BL/6c−/c− genetic background generated by backcrossing C57BL/6c−/c−;129S5/SvEvBrd mixed-background mutant mice with C57BL/6c−/c− mice for 5–6 generations. Gnb1l+/− mice were provided by Lexicon Genetics Inc., and were generated by blastocyst injection of stem cells from OST35527 (OmniBank). Mice were genotyped by PCR by using DNA extracted from tail biopsies (see Supporting Text for primer sequences).
PPI Assay.
PPI was measured by using the SR-Lab system (San Diego Instruments, San Diego) as described (41). Animals were tested at age 8–16 weeks. Before testing, each mouse was acclimatized to the Plexiglas cylinder for 5 min, during which time the background noise level (70 dB) was continually present. Individual mice were then exposed to six blocks of seven trial types that were presented in a pseudorandom order with an average intertrial interval of 15 seconds. The seven trial types comprised the following: trial 1 (startle-only trial), 40 ms, 120 dB sound burst; trials 2–6 (prepulse trials), 120 dB startle stimulus preceded 100 ms by 20 ms prepulse sounds of 74, 78, 82, 86, or 90 dB; and trial 7, 70 dB background noise. The startle response was recorded for 65 ms, measuring every 1 ms from the onset of the startle stimulus. The maximum startle amplitude recorded during the 65-ms sampling window was used as the dependent variable. We calculated % PPI as: 100 – [(startle response on acoustic prepulse plus startle stimulus trials/startle response-alone trials) × 100]. Data for each deletion and mutation were analyzed independently. Acoustic response amplitude data were analyzed by using two-way (genotype × gender) ANOVAs. PPI data were analyzed by using a three-way (genotype × gender × prepulse sound level) ANOVA with repeated measures.
Gene Expression Analyses.
To visualize β-gal activity, 4% paraformaldehyde-fixed brains were stained in X-gal substrate according to standard procedures. Two- to 3-mm-thick brain sections were photographed as whole-mount specimens and then embedded in paraffin. Sections (10 μm) were counterstained with Nuclear Fast Red. RNA in situ hybridization was performed on 10-μm 4% paraformaldehyde-fixed sections (42). Labeled sense and antisense RNA probes were prepared by reverse transcription of DNA clones in the presence of 35S-UTP (MP Biomedicals, Irvine, CA).
Control Subjects.
Unrelated British Caucasian control subjects were recruited from the Blood Transfusion Service in Wales and England (482 males and 234 females; mean age 41.5 years, SD ± 11.5 years). The sample was not specifically screened for psychiatric illness, but subjects were not taking regular prescribed medications.
Mutation Detection and Sequencing.
A transcript map of TBX1 was created by using the Golden Path Genome Browser (May 2004 freeze) (http://genome.ucsc.edu/). The coding sequence of the TBX1 isoform C [National Center for Biotechnology Information (NCBI) accession no. NM_080647], TBX1 isoform B (NCBI accession no. NM_005992), and TBX1 isoform C (NCBI accession no. NM_080646, NCBI) and at least 50 bp of intronic sequence adjacent to each exon were screened for sequence variants in patient V39/02. Primers were constructed by using the primer3 program (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Mutation detection analysis was performed by denaturing high-performance liquid chromatography (43). All samples with heteroduplex traces were subsequently sequenced with BigDye Terminator 3.0 cycle sequencing kit and analyzed on an ABI3100 sequencer (PE Applied Biosystems).
Genotyping of TBX1 deletions.
The identified deletion was typed by using 5′-fluorescently labeled PCR primers (FAM dye), and PCR products were resolved on an ABI3100 sequencer. The data were analyzed with genescan 3.7 and genotyper 2.5 software.
CAT assays.
U2-OS cells were grown in 12-well dishes to 90% confluency and transfected in quadruplicate with Lipofectamine 2000 (Invitrogen). The 1T-CAT reporter construct (44) was cotransfected with TBX1 DNA and the β-gal expression vector, pCH110 (Amersham Pharmacia), which was used for normalization. The concentration of CAT protein in cell lysates was determined by using the CAT-ELISA kit (Roche). Differences in transcriptional activation between wild-type and mutant TBX1 constructs were evaluated by using a Kruskal–Wallis test (SPSS, Chicago) because the assumption of normality for the variable transcriptional activation was not given.
Immunocytochemistry.
U2-OS cells were grown on poly(d-lysine)-coated glass coverslips to 90% confluency and transfected as before with TBX1 constructs in pCDNA3. Cells were fixed in 4% paraformaldehyde after 24 h and permeabilized with 0.05% Nonidet P-40 in PBS. Cells were incubated with rabbit anti-Tbx1 antibody (Zymed) at 1:100 then with donkey anti-rabbit IgG Alexa Fluor 488 (Molecular Probes) at 1:200. Cells were mounted in Vectashield (Vector Laboratories) with DAPI and photographed by using a Zeiss AxioVision microscope.
Supplementary Material
Acknowledgments
We thank M. Reese, P. Terrell, G. Ji, and C. Gerken for technical support and S. Reed for clinical assistance. This work is supported by the National Institutes of Health, National Institutes of Mental Health (E.L. and R.P.), the March of Dimes (E.L.), and the Ministero dell’Instruzione, dell’Università e della Ricerca (E.L.). This work was also supported by the Mental Retardation and Developmental Disabilities Research Center Neurobehavioral Core, Baylor College of Medicine. E.L. is an Associate Telethon Scientist. P.A. and P.J.S. were supported by the British Heart Foundation and the Health Foundation. K.C.M., N.W., M.C.O., and M.J.O. were supported by a grant from the Wellcome Trust. Gnbl1 mutant mice were generated by and purchased from Lexicon Genetics Inc.
Abbreviations
- 22q11DS
22q11 deletion syndrome
- PPI
prepulse inhibition
- En
embryonic day n
- ASR
acoustic startle response
- VCFS
velocardiofacial syndrome
- NLS
nuclear localization signal.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fine S. E., Weissman A., Gerdes M., Pinto-Martin J., Zackai E. H., McDonald-McGinn D. M., Emanuel B. S. J. Autism Dev. Disord. 2005;35:461–470. doi: 10.1007/s10803-005-5036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niklasson L., Rasmussen P., Oskarsdottir S., Gillberg C. Genet. Med. 2001;3:79–84. doi: 10.1097/00125817-200101000-00017. [DOI] [PubMed] [Google Scholar]
- 3.Bassett A. S., Hodgkinson K., Chow E. W., Correia S., Scutt L. E., Weksberg R. Am. J. Med. Genet. 1998;81:328–337. [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy K. C., Jones L. A., Owen M. J. Arch. Gen. Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 5.Bassett A. S., Chow E. W., AbdelMalik P., Gheorghiu M., Husted J., Weksberg R. Am. J. Psychiatry. 2003;160:1580–1586. doi: 10.1176/appi.ajp.160.9.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shprintzen R. J., Goldberg R., Golding-Kushner K. J., Marion R. W. Am. J. Med. Genet. 1992;42:141–142. doi: 10.1002/ajmg.1320420131. [DOI] [PubMed] [Google Scholar]
- 7.Lindsay E. A., Morris M. A., Gos A., Nestadt G., Wolyniec P. S., Lasseter V. K., Shprintzen R., Antonarakis S. E., Baldini A., Pulver A. E. Am. J. Hum. Genet. 1995;56:1502–1503. [PMC free article] [PubMed] [Google Scholar]
- 8.Lindsay E. A., Botta A., Jurecic V., Carattini-Rivera S., Cheah Y.-C., Rosenblatt H. M., Bradley A., Baldini A. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 9.Paylor R., McIlwain K. L., McAninch R., Nellis A., Yuva-Paylor L. A., Baldini A., Lindsay E. A. Hum. Mol. Genet. 2001;10:2645–2650. doi: 10.1093/hmg/10.23.2645. [DOI] [PubMed] [Google Scholar]
- 10.Braff D. L., Geyer M. A., Swerdlow N. R. Psychopharmacology (Berlin) 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- 11.Sobin C., Kiley-Brabeck K., Karayiorgou M. Am. J. Psychiatry. 2005;162:1090–1099. doi: 10.1176/appi.ajp.162.6.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAlonan G. M., Daly E., Kumari V., Critchley H. D., van Amelsvoort T., Suckling J., Simmons A., Sigmundsson T., Greenwood K., Russell A., et al. Brain. 2002;125:1594–1606. doi: 10.1093/brain/awf150. [DOI] [PubMed] [Google Scholar]
- 13.Lindsay E. A., Vitelli F., Su H., Morishima M., Huynh T., Pramparo T., Jurecic V., Ogunrinu G., Sutherland H. S., Scambler P. J., et al. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 14.Vitelli F., Lindsay E. A., Baldini A. Cold Spring Harbor Symp. Quant. Biol. 2002;67:327–332. doi: 10.1101/sqb.2002.67.327. [DOI] [PubMed] [Google Scholar]
- 15.Su H., Wang X., Bradley A. Nat. Genet. 2000;24:92–95. doi: 10.1038/71756. [DOI] [PubMed] [Google Scholar]
- 16.Gogos J. A., Santha M., Takacs Z., Beck K. D., Luine V., Lucas L. R., Nadler J. V., Karayiorgou M. Nat. Genet. 1999;21:434–439. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- 17.Mukai J., Liu H., Burt R. A., Swor D. E., Lai W. S., Karayiorgou M., Gogos J. A. Nat. Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- 18.Chen J., Lipska B. K., Halim N., Ma Q. D., Matsumoto M., Melhem S., Kolachana B. S., Hyde T. M., Herman M. M., Apud J., et al. Am. J. Hum. Genet. 2004;75:807–821. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gothelf D., Eliez S., Thompson T., Hinard C., Penniman L., Feinstein C., Kwon H., Jin S., Jo B., Antonarakis S. E., et al. Nat. Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- 20.Paterlini M., Zakharenko S. S., Lai W. S., Qin J., Zhang H., Mukai J., Westphal K. G., Olivier B., Sulzer D., Pavlidis P., et al. Nat. Neurosci. 2005;8:1586–1594. doi: 10.1038/nn1562. [DOI] [PubMed] [Google Scholar]
- 21.Gogos J. A., Morgan M., Luine V., Santha M., Ogawa S., Pfaff D., Karayiorgou M. Proc. Natl. Acad. Sci. USA. 1998;95:9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budarf M. L., Konkle B. A., Ludlow L. B., Michaud D., Li M., Yamashiro D. J., McDonald-McGinn D., Zackai E. H., Driscoll D. A. Hum. Mol. Genet. 1995;4:763–766. doi: 10.1093/hmg/4.4.763. [DOI] [PubMed] [Google Scholar]
- 23.Geyer M. A., McIlwain K. L., Paylor R. Mol. Psychiatry. 2002;7:1039–1053. doi: 10.1038/sj.mp.4001159. [DOI] [PubMed] [Google Scholar]
- 24.Merscher S., Funke B., Epstein J. A., Heyer J., Puech A., Min Lu M. M., Xavier R. J., Demay M. B., Russell R. G., Factor S., et al. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 25.Jerome L. A., Papaioannou V. E. Nat. Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 26.Yagi H., Furutani Y., Hamada H., Sasaki T., Asakawa S., Minoshima S., Ichida F., Joo K., Kimura M., Imamura S., et al. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 27.Maynard T. M., Haskell G. T., Peters A. Z., Sikich L., Lieberman J. A., LaMantia A. S. Proc. Natl. Acad. Sci. USA. 2003;100:14433–14438. doi: 10.1073/pnas.2235651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitelli F., Morishima M., Taddei I., Lindsay E. A., Baldini A. Hum. Mol. Genet. 2002;11:915–922. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- 29.Hanson D. R., Gottesman I. I. BMC Med. Genet. 2005;6:7. doi: 10.1186/1471-2350-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong L., Liu M., Jen J., Yeh E. T. Biochim. Biophys. Acta. 2000;1494:185–188. doi: 10.1016/s0167-4781(00)00189-5. [DOI] [PubMed] [Google Scholar]
- 31.Funke B., Pandita R. K., Morrow B. E. Genomics. 2001;73:264–271. doi: 10.1006/geno.2000.6506. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay E. A., Baldini A. Hum. Mol. Genet. 2001;10:997–1002. doi: 10.1093/hmg/10.9.997. [DOI] [PubMed] [Google Scholar]
- 33.Xu H., Morishima M., Wylie J. N., Schwartz R. J., Bruneau B. G., Lindsay E. A., Baldini A. Development (Cambridge, U.K.) 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 34.Gillberg C. Br. J. Psychiatry. 1998;172:200–209. doi: 10.1192/bjp.172.3.200. [DOI] [PubMed] [Google Scholar]
- 35.Gillberg C., Rastam M., Wentz E. Autism. 2001;5:57–66. doi: 10.1177/1362361301005001006. [DOI] [PubMed] [Google Scholar]
- 36.Wing L. J. Autism Dev. Disord. 2005;35:197–203. doi: 10.1007/s10803-004-1998-2. [DOI] [PubMed] [Google Scholar]
- 37.Stoller J. Z., Epstein J. A. Hum. Mol. Genet. 2005;14:885–892. doi: 10.1093/hmg/ddi081. [DOI] [PubMed] [Google Scholar]
- 38.Basson C. T., Huang T., Lin R. C., Bachinsky D. R., Weremowicz S., Vaglio A., Bruzzone R., Quadrelli R., Lerone M., Romeo G., et al. Proc. Natl. Acad. Sci. USA. 1999;96:2919–2924. doi: 10.1073/pnas.96.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cokol M., Nair R., Rost B. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ataliotis P., Ivins S., Mohun T. J., Scambler P. J. Dev. Dyn. 2005;232:979–991. doi: 10.1002/dvdy.20276. [DOI] [PubMed] [Google Scholar]
- 41.Paylor R., Crawley J. N. Psychopharmacology (Berlin) 1997;132:169–180. doi: 10.1007/s002130050333. [DOI] [PubMed] [Google Scholar]
- 42.Albrecht U., Eichele G., Helms J. A., Lu H. C. In: Molecular and Cellular Methods in Developmental Toxicology. Daston G. P., editor. New York: CRC; 1997. pp. 23–48. [Google Scholar]
- 43.Jones A. C., Austin J., Hansen N., Hoogendoorn B., Oefner P. J., Cheadle J. P., O’Donovan M. C. Clin. Chem. 1999;45:1133–1140. [PubMed] [Google Scholar]
- 44.Conlon F. L., Sedgwick S. G., Weston K. M., Smith J. C. Development (Cambridge, U.K.) 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.