Abstract
We report here that human T cells give much stronger proliferative responses to specific activation via the T cell receptor (TCR) than those from chimpanzees, our closest evolutionary relatives. Nonspecific activation using phytohemagglutinin was robust in chimpanzee T cells, indicating that the much lower response to TCR simulation is not due to any intrinsic inability to respond to an activating stimulus. CD33-related Siglecs are inhibitory signaling molecules expressed on most immune cells and are thought to down-regulate cellular activation pathways via cytosolic immunoreceptor tyrosine-based inhibitory motifs. Among human immune cells, T lymphocytes are a striking exception, expressing little to none of these molecules. In stark contrast, we find that T lymphocytes from chimpanzees as well as the other closely related “great apes” (bonobos, gorillas, and orangutans) express several CD33-related Siglecs on their surfaces. Thus, human-specific loss of T cell Siglec expression occurred after our last common ancestor with great apes, potentially resulting in an evolutionary difference with regard to inhibitory signaling. We confirmed this by studying Siglec-5, which is prominently expressed on chimpanzee lymphocytes, including CD4 T cells. Ab-mediated clearance of Siglec-5 from chimpanzee T cells enhanced TCR-mediated activation. Conversely, primary human T cells and Jurkat cells transfected with Siglec-5 become less responsive; i.e., they behave more like chimpanzee T cells. This human-specific loss of T cell Siglec expression associated with T cell hyperactivity may help explain the strikingly disparate prevalence and severity of T cell-mediated diseases such as AIDS and chronic active hepatitis between humans and chimpanzees.
Keywords: chimpanzees, HIV/AIDS, T cells
Siglecs are sialic acid (Sia)-recognizing Ig-superfamily lectins prominently expressed in immune cells. CD33-related Siglecs (CD33rSiglecs, Siglec-3 and Siglec5–11) are a subset thought to down-regulate innate immune cell activation via cytosolic immunoreceptor tyrosine-based inhibitory motifs (1–4). These immunoreceptor tyrosine-based inhibitory motifs (5) recruit protein phosphatases, Src homology region 2 domain-containing phosphatases (SHPs) SHP-1 and SHP-2, which limit activation pathways stimulated by tyrosine kinases (6, 7).
Chimpanzees and most other mammals express two major Sias at terminal ends of cell surface and secreted glycans: N-acetylneuraminic acid and N-glycolylneuraminic acid (Neu5Gc). Human cells cannot produce Neu5Gc because of an inactivating exon deletion in the CMAH gene encoding the enzyme that converts CMP-N-acetylneuraminic acid to CMP-Neu5Gc (8). The human-specific loss of Neu5Gc occurred ≈3 million years ago (8) and was apparently followed by rapid evolution of multiple human CD33rSiglecs involving gene deletion, gene conversion, or changes in binding specificity (3, 4, 9–11).
It is striking that T cells are the only human immune cell type that expresses little or no Siglecs. Whereas all other human leukocyte types express one or more of the CD33rSiglecs at easily detectable levels, T cells show only very low-level expression of Siglec-7 and Siglec-9 (2, 3, 12). Transfection of Siglec-7 and Siglec-9 into the Jurkat T cell line gave inhibition of T cell receptor (TCR)-mediated signaling, indicating that CD33rSiglecs can potentially regulate T cell activation (12). However, human T cell expression of Siglec-7 and Siglec-9 is present only on a very small subset (12) and not in all individuals (our current observations).
As our closest evolutionary relatives, the common chimpanzee (Pan troglodytes) shares >99% identity in protein sequences with humans (13, 14). Thus, it has long been assumed that the chimpanzee is an effective animal model for human diseases. In fact, chimpanzee diseases may be more disparate than previously envisioned (14–17). Among the obvious differences are the lack of progression to AIDS with maintenance of CD4 T cell counts in the great majority of chimpanzees infected with the CD4 T cell-tropic HIV (18–20) and the rarity of T cell-mediated chronic active hepatitis and cirrhosis after hepatitis B or C infection (21, 22). Moreover, several other common human T cell-mediated diseases, such as bronchial asthma, rheumatoid arthritis, and type 1 diabetes (6, 23), have not been reported in chimpanzees or other “great apes”¶ (14, 16).
We therefore asked whether T cell differences between humans and great apes could relate to differences in Siglec function and/or expression. Here we report a disparity between humans and chimpanzees in T cell activation via the TCR and the correlative expression of the inhibitory CD33rSiglec molecules only in great ape T cells. We demonstrate a potential inhibitory role for Siglec-5 on chimpanzee T cells and show that induced expression of human Siglec-5 in human T cells mimics the chimpanzee phenotype.
Results
Chimpanzee T Cells Are Much Less Responsive to TCR Stimulation than Human T Cells.
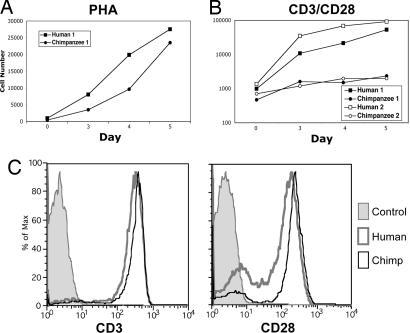
The general responsiveness of freshly isolated human and chimpanzee T cells was evaluated by activation with the lectin phytohemagglutinin-L (PHA), which nonspecifically stimulates T cells by random crosslinking of surface proteins. Both cell types responded robustly, with the proliferation of chimpanzee cells being somewhat lower (Fig. 1A). This finding fits with prior data of others, in which responses of chimpanzee T cells to some superantigens were as robust as responses by human T cells (24). We next examined human and chimpanzee T cell activation by TCR activation using immobilized anti-CD3 along with costimulation by soluble anti-CD28. Under these more physiological conditions, chimpanzee T cells proliferated much less than human T cells (Fig. 1B). After 5 days of activation, chimpanzee T cell numbers were approximately two orders of magnitude lower than in humans. No major differences in CD3 or CD28 levels on human and chimpanzee T cells could account for this finding (Fig. 1C). Thus, although chimpanzee T cells can proliferate upon nonspecific lectin-mediated activation, there is a striking disparity with human T cells after physiologically relevant activation via the TCR.
Fig. 1.
Differences in human and chimpanzee T cell activation by PHA and anti-CD3/anti-CD28. Human and chimpanzee T lymphocytes were stimulated with 10 μg/ml soluble PHA (A) or with immobilized anti-CD3 (2.5 μg/ml coating concentration) plus 0.1 μg/ml soluble anti-CD28 (B). Cells were collected and counted on a FACSCalibur on the indicated days at 60 μl/min for 30 s. A log scale is used on the y axis to accommodate the range of values seen. Human and chimpanzee lymphocytes were labeled with anti-CD3 or anti-CD28 and detected with PE goat anti-mouse IgG (C). The control histogram indicates the human lymphocyte population in the presence of secondary Ab only. One representative pair of three human and chimpanzee comparisons is presented in A. The same donor pair is presented in B as solid symbols, along with another pair of human and chimpanzee samples (filled symbols).
Great Apes Express Higher Levels and Wider Varieties of CD33rSiglecs on Lymphocytes in Comparison to Humans.
Siglec expression on immune cells of great apes and other nonhuman primates has not been previously studied. We compared CD33rSiglec expression on lymphocytes from humans and all four great ape species (chimpanzees, bonobos, gorillas, and orangutans) using previously characterized mAbs against human Siglec-3 and Siglec-5–11. Given the very close genetic similarity of humans and great apes, most or all of the mAbs were expected to cross-react. Indeed, we found that all bound recombinant Siglec human and chimpanzee CD33rSiglecs equally well in ELISAs (data not shown).
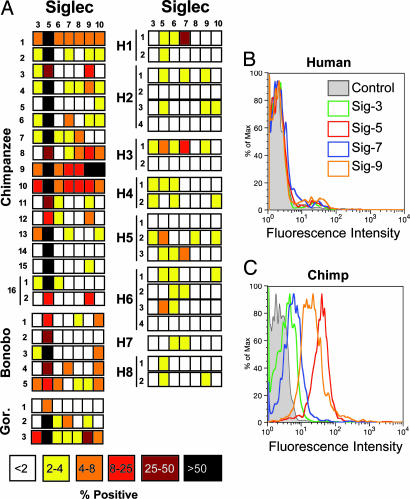
We found striking differences in CD33rSiglec expression between human and great ape lymphocytes. Anti-Siglec-5 staining was consistently found in all chimpanzees studied, and several other CD33rSiglecs were variably expressed (Fig. 2). Positive staining for anti-Siglec-5 ranged from 11% to 98% of total chimpanzee lymphocytes. In contrast, humans representing a range of geographic and ethnic origins demonstrated very weak and transient expression of CD33rSiglecs on lymphocytes (8 humans are shown, representative of 16 tested). One human subject showed Siglec-7 expression in 34% of lymphocytes, but upon retesting no significant expression was observed (<2% positive). This finding could reflect changes in the percentage of natural killer cells, which are known to be Siglec-7-positive (25) and can vary in number. In most humans, expression of any CD33rSiglec rarely exceeded 4% of lymphocytes. Interestingly, chimpanzee 16 was analyzed on two separate occasions and also demonstrated some variability in Siglec expression (Fig. 2A). Overall, although 19 chimpanzee samples showed an average of ≈60% anti-Siglec-5-positive lymphocytes, the average for 28 human samples was 3.4%. Outgroup comparisons revealed a similar expression of multiple CD33rSiglecs (particularly Siglec-5) on lymphocytes of bonobos and gorillas (Fig. 2A). Several other CD33rSiglecs also showed significant and variable expression on great ape, but not human, lymphocytes (Fig. 2A). One orangutan sample demonstrated relatively high expression of Siglec-6 (41% positive) but lower expression of Siglec-3, Siglec-5, Siglec-7, and Siglec-10 (13%, 7%, 5%, and 18% positive, respectively; data not shown). The results for orangutan expression of Siglec-5 are inconclusive because of low detection on monocytes and granulocytes (data not shown), which normally express high levels of Siglec-5 in humans and great apes. This result may be because of poor mAb recognition of Siglec-5 in this great ape species, which is most distantly related to humans.
Fig. 2.
Expression of CD33rSiglecs on human and great ape lymphocytes. (A) Percentage of positive lymphocytes for each Siglec Ab (staining above negative controls) for 16 chimpanzees, 5 bonobos, and 3 gorillas are shown, as well as data for 8 humans (the latter were tested on one or more occasions). Examples of flow cytometry histograms of human (B) and chimpanzee (C) lymphocytes using Abs recognizing Siglec-3, Siglec-5, Siglec-7, and Siglec-9 (y axis: normalized cell numbers expressed as percent of maximum cell number detected). In later samples examined, low levels of Siglec-11 staining (<5% positive) were occasionally detected on lymphocytes in both great apes and humans (data not shown).
Siglec-5 Is Expressed on Chimpanzee, but Not Human, B Cells and CD4+ T Cells.
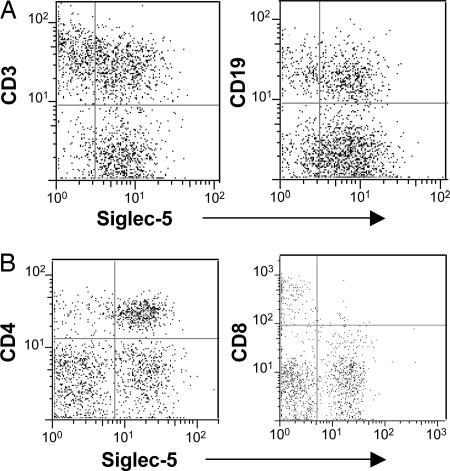
Further characterization of Siglec-5-positive lymphocytes from seven chimpanzees revealed expression on CD3+ T cells as well as CD19+ B cells (see Fig. 3A for representative results). Because B cells already express other inhibitory Siglecs, like CD22 (2, 3), we focused on T cells. Double-staining flow cytometric analysis of chimpanzee lymphocytes revealed that the majority of CD4+ T cells expressed Siglec-5 (Fig. 3B, 83%). In contrast, only 5% of CD8+ T cells were positive for Siglec-5. Corroborating the earlier findings, both CD4 and CD8 T cells in humans were negative for Siglec-5 (<2% positive).
Fig. 3.
Anti-Siglec-5 Abs stain chimpanzee T and B cells. Chimpanzee lymphocytes were double-labeled with anti-Siglec-5 and PE-goat anti-mouse IgG and with APC-anti-CD3 or APC-anti-CD19 (A) or with FITC-anti-CD4 and PE-Cy5-anti-CD8 (B). Results for CD3 and CD19 are representative of seven individuals, and results for CD4 and CD8 are representative of two individuals.
Ab-Induced Siglec-5 Internalization Partially Releases Inhibition of Chimpanzee T Cell Stimulation.
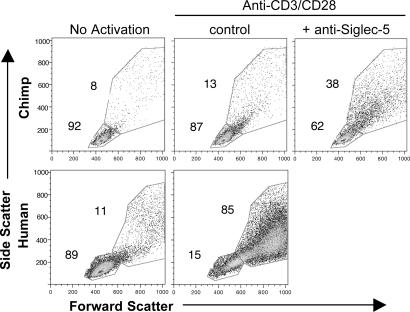
Chimpanzee Siglec-5 contains a mutation that renders it potentially unable to bind Sias compared with the human Siglec-5 orthologue (4). Although this mutation could alter Siglec-5 signaling, similar mutations in Siglec-2 and Siglec-9 do not completely abolish inhibitory function (12, 26). Thus, the prominent expression of Siglec-5 on chimpanzee lymphocytes is predicted to inhibit TCR/CD3-mediated activation signals. To address this possibility, we studied chimpanzee cells in the presence or absence of soluble anti-Siglec-5 mAbs during stimulation with immobilized anti-CD3. Anti-Siglec-5 mAbs induced 40–70% internalization of cell-surface Siglec-5 after 1 h at 37°C while not affecting CD3 levels (data not shown). After 3 days of incubation we saw a significant increase in expanded cells, evident by increases in flow cytometric side and forward scatter (Fig. 4). Although this approach did not increase chimpanzee T cell proliferation to the level seen with humans, the results indicate that Siglec-5 can contribute to regulating the TCR-initiated response in chimpanzee cells.
Fig. 4.
Enhanced chimpanzee T cell response to anti-CD3 after anti-Siglec-5 Ab treatment. Human and chimpanzee T lymphocytes were stimulated with immobilized anti-CD3 plus soluble anti-CD28. For the indicated samples, anti-Siglec-5 was added at 5 μg/ml to chimpanzee lymphocytes in solution. Cells were analyzed by flow cytometry after 3 days of stimulation. Increases in forward scatter and side scatter indicate increases in cell size and internal complexity/granularity, respectively. Dead cells were excluded from analysis. Results are representative of four different samples from one chimpanzee.
Induced Expression of Siglec-5 in Primary Human T Cells Inhibits TCR Responses.
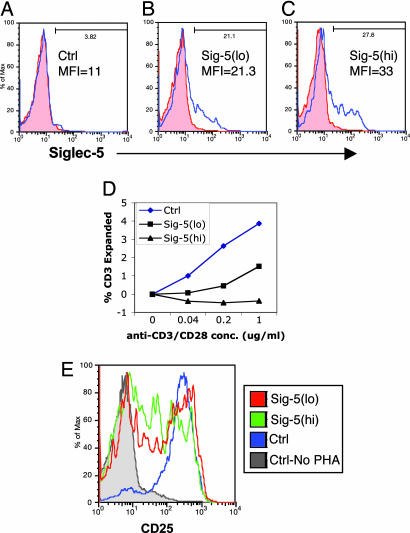
We next asked whether induced expression of human Siglec-5 in human T cells would affect proliferation. Using nucleofection (Nucleofector, Amaxa, Gaithersburg, MD) (27, 28), we induced expression of Siglec-5 in resting human T cells (Figs. 5 and 6). In one experiment, we nucleofected monocyte-depleted peripheral blood mononuclear cells (PBMCs) with 0, 2, or 3 μg of a plasmid construct containing full-length human Siglec-5 (pSig5). The resulting subpopulations were designated as control (Siglec-5−), Sig-5(lo), or Sig-5(hi) based on relative expression of Siglec-5 (Fig. 5 A–C). All three populations demonstrated no significant changes in forward scatter, side scatter, or expression of CD4, suggesting a uniform resting state for each population (data not shown). Twenty-four hours after nucleofection, we added immobilized anti-CD3 and soluble anti-CD28 mAbs at varying concentrations and continued to culture them. After 3 days, we observed significant inhibitory effects on cell proliferation in a Siglec-5 expression-dependent manner (Fig. 5D). With background size-expanded cells normalized to zero in the absence of anti-CD3/anti-CD28, there was no increase in size-expanded cells for high-Siglec-5-expressing cells and a decreased number of size-expanded cells compared with the control at all mAb concentrations (Fig. 5D). The same three cell populations were also stimulated with PHA for 3 days. In contrast to mAb stimulation, a larger percentage of cells in all three populations were activated by PHA, as measured by CD25 expression (Fig. 5E). Sig-5(lo) and Sig-5(hi) cells responded less robustly than control cells, as quantitated by the percentage of cells expanded (50%, 46%, and 86%, respectively) and mean fluorescence intensity of CD25 (50, 24, and 244, respectively). These results correlate well with the differences observed between human and chimpanzee T cells based on Siglec-5 expression. Although we did observe higher cell death with Siglec-5-expressing cells compared with mock-transfected ones, our analyses were gated only on live cell populations (data not shown). The separate but relevant possibility of increased cell death in human cells transfected with Siglec-5 needs to be addressed in future studies.
Fig. 5.
Human T cell Siglec-5 expression inhibits responses to soluble anti-CD3/anti-CD28 and PHA. Unstimulated monocyte-depleted PBMCs were mock-transfected with no DNA (A) or transfected with 2 or 3 μg of pSig5 (B and C) by using the Nucleofector apparatus. After 24 h, cells were labeled with nonspecific mouse IgG (red filled histograms) or anti-Siglec-5 (blue open histograms). MFI, mean fluorescence intensity of Siglec-5 expression. The resulting cell populations were named control (Ctrl), Sig-5(lo), and Sig-5(hi) based on Siglec-5 expression levels. (D) Transfected cells were stimulated with soluble anti-CD3 plus soluble anti-CD28 at the indicated concentrations for 3 days. The percentages of size-expanded cells are plotted with no stimulation background controls subtracted. Similar results were observed with a different PBMC donor. (E) Transfected cells were stimulated with 10 μg/ml PHA for 3 days and then analyzed for CD25 expression. The histograms for CD25 staining are shown.
Fig. 6.
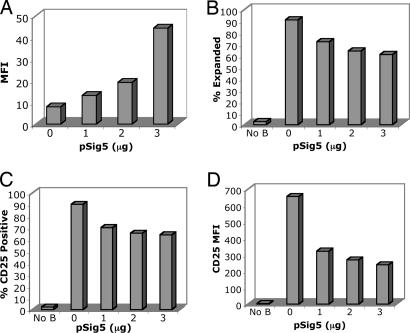
Expression of Siglec-5 in human T cell inhibits responses to anti-CD3/anti-CD28 beads. (A) Unstimulated monocyte-depleted PBMCs were mock-transfected with no DNA or transfected with 1, 2, or 3 μg of pSig5 by using the Nucleofector device. After 24 h, cells were labeled with anti-Siglec-5 and goat anti-mouse IgG Alexa Fluor 488. MFI, mean fluorescence intensity of Siglec-5 expression. Transfected cells were stimulated with anti-CD3/anti-CD28-bearing beads (B–D). After 3 days of stimulation, cells were analyzed for expansion in size and intracellular complexity/granularity (B) and increase in CD25 expression (C and D). Beads and bead-bound cells were excluded from analysis by forward scatter gating and positive autofluorescence of FL3.
We also used a different stimulation method using anti-CD3/anti-CD28-coated beads (Dynabeads CD3/CD28 T Cell Expander, Dynal Biotech, Brown Deer, WI) to stimulate Siglec-5-expressing cells. Nucleofecting primary lymphocytes with 0, 1, 2, and 3 μg of pSig5 produced Siglec-5-expressing cells in a dose-dependent manner (Fig. 6A). After 5 days of incubation at a cell:bead ratio of 1:1, we observed Siglec-5 expression-dependent inhibition of stimulation as measured by the percentage of expanded cells (Fig. 6B), the percentage of cells expressing CD25 (Fig. 6C), and the mean fluorescence intensity of CD25 (Fig. 6D) of each cell population. These results further indicate that Siglec-5 can inhibit anti-CD3/anti-CD28 responsiveness in T cells. Because of varying levels of Siglec-5 expression observed within each cell population after transfection, we wanted to determine whether Siglec-5-expressing cells were truly the “nonresponding” cells. To evaluate this variation, we gated expanded and nonexpanded cells after CD3/CD28 bead stimulation (Fig. 8, which is published as supporting information on the PNAS web site) and stained for Siglec-5 in each subpopulation. Interestingly, nonexpanded cells demonstrated a higher percentage of cells positive for Siglec-5 than expanded cells for all three transfected populations (Fig. 8). Thus, Siglec-5-expressing cells are less responsive, and Siglec-5-negative cells are more likely to respond given the same stimulation.
Expression of Siglec-5 in Jurkat T Cells Inhibits Anti-CD3-Induced Intracellular Calcium Mobilization.
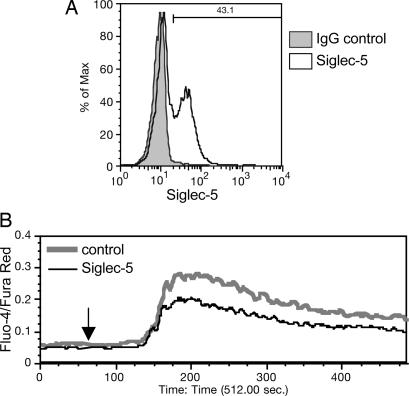
To measure more proximate effects of Siglec-5 expression on CD3 stimulation, we performed intracellular calcium mobilization assays on transfected and mock-transfected control Jurkat T leukemia cells. Using Amaxa nucleofection, we were able to transiently express Siglec-5 in up to 43% of cells 24 h after nucleofection (Fig. 7A). Subsequent intracellular calcium mobilization in response to anti-CD3 mAb was reduced compared with controls, using real-time flow cytometric calcium measurements (Fig. 7B). The inhibitory effects were consistent in three separate experiments. These data further suggest that CD3 activation is regulated by Siglec-5 at the level of signal initiation that leads to calcium flux.
Fig. 7.
Siglec-5 expression on Jurkat T cells inhibits anti-CD3-induced intracellular calcium mobilization. Jurkat cells were mock-transfected with no DNA or transfected with 2 μg of pSig5 per 2 × 106 cells using the Nucleofector device. After 24 h, Siglec-5 expression was analyzed by flow cytometry with anti-Siglec-5 and goat anti-mouse IgG Alexa Fluor 488, by using a nonspecific mouse IgG as background (A). The percentage of Siglec-5-positive cells is indicated. Transfected cells were loaded with calcium-sensing dyes, Fluo-4 and Fura Red, and then analyzed for responses to soluble anti-CD3 by real-time flow cytometric analysis (B). The arrow at 60 s indicates the time of mAb addition. The experiment was repeated twice with different transfectant Jurkat cells and produced similar results.
Mechanism of Down-Regulation of Siglec-5 on Human T Cells.
To explore the mechanism of down-regulation, we studied human T cells for staining by anti-Siglec-5 mAbs, without or with membrane permeabilization, which would allow the mAbs to access intracellular compartments. We found a low level of Siglec-5 staining in human lymphocytes after permeabilization and in only a very minor population of cells (data not shown). Thus, it is likely that the human-specific down-regulation of Siglec-5 expression occurs at a pretranslational level. Given that the human-specific suppression of expression involves multiple CD33rSiglecs, the mechanism is likely to be a general transcriptional repression of the CD33rSiglec cluster. Definitively proving this hypothesis requires FACS sorting of CD4+ human and chimpanzee lymphocytes to a high degree of purity. This approach is rendered difficult by contaminating monocytes, which are known to express high levels of Siglec-5 in both species.
Discussion
Our data indicate that a human-specific suppression of CD33rSiglec expression on T cells occurred at some time before the emergence of modern humans ≈100,000–200,000 years ago (29, 30). In keeping with this finding, the histogram of human T cell responses to increasing TCR stimulation is markedly “shifted to the left” in comparison with chimpanzee T cells. We suggest that this human-specific shift contributes to an intrinsic hyperreactivity of human T cells and may help explain the frequency and severity of T cell-mediated diseases in our species. In this regard, we found that chimpanzee Siglec-5 was particularly abundant on CD4+ T cells. Such T cells are involved in the pathology of many human diseases, including AIDS, chronic active hepatitis, inflammatory bowel disease, rheumatoid arthritis, type 1 diabetes, multiple sclerosis, and psoriasis. (6, 23). The lack of CD33rSiglec expression in humans may contribute to CD4 T cell hyperactivity in these diseases. This finding may also help explain the unexpected interruption of a recent clinical trial in which healthy human volunteers became severely ill upon receiving an anti-CD28 mAb capable of directly stimulating T cell activation (31). The Ab had evidently been previously tested in monkeys at concentrations much higher than those used in the humans, without significant adverse effects. The uniquely human lack of CD33rSiglecs on T cells may have allowed a marked stimulation of these cells in the subjects, perhaps releasing a “cytokine storm.”
CD33rSiglec expression differences on human and great ape B cells also deserve further study. The presence of Siglecs in addition to CD22 may provide more stringent regulation of activation and function in chimpanzee cells compared with humans. In this regard, Ab self-reactivity related to disease (e.g., systemic lupus erythematosis or even a positive lupus Ab test) has not, to our knowledge, been reported in chimpanzees (James G. Else and Elizabeth Strobert, Yerkes National Primate Research Center, personal communication). It remains to be determined whether CD33rSiglec expression down-regulates chimpanzee B cell activation in a manner similar to CD22.
Activation of T cells can also lead to cell death via apoptosis. Our data may thus explain increased T cell activation and death observed in HIV-infected humans but not chimpanzees. Host proteins such as APOBEC3G and Trim5α are known to differ between Old World monkeys and humans, helping explain species-specific susceptibility to HIV or simian immunodeficiency virus (32–34). However, such differences are not as prominent between humans and chimpanzees. Specifically, the critical amino acid at position 128 of APOBEC3G that confers African green monkey and rhesus macaque resistance to HIV and human resistance to simian immunodeficiency virus is not different between human and chimpanzees (33, 34). Furthermore, the human Trim5α sequence bears much greater similarity to chimpanzees (98% identity) than to rhesus macaques (87% identity) (32). In addition, chimpanzee cells can be effectively infected by HIV (35), and the only major difference is the lack of severe CD4 attrition at later stages of the infectious process in vivo. Thus, the lack of progression to AIDS in chimpanzees may be due to the difference in responsiveness of the CD4 cells in general, the overall inflammatory condition during virus infection (36), and the reduced rate of proliferation and apoptosis of infected CD4 T cells (37). Future studies are needed to fully elucidate whether and how Siglecs contribute to chimpanzee resistance to AIDS.
As with many events during evolution, it is difficult to be certain why humans are the only hominids without prominent expression of CD33rSiglecs on T cells. Our studies suggest that this difference is not because of internal sequestration, but more likely because of promoter-mediated and/or transcription factor-mediated down-regulation of gene expression specific to human T cells. Another possibility could be epigenetic changes affecting the CD33rSiglec cluster in humans. Regardless of the mechanism, we can only speculate on why this occurred in humans. One possibility is that early humans required a higher level of T cell activation to defeat one or more pathogens, which was accomplished by down-regulating CD33rSiglecs. Although this adaptation may have provided a short-term advantage, the long-term consequences may be the various T cell-mediated diseases in humans today. Alternatively, CD33rSiglec loss from human T cells could have occurred in the absence of pathogen pressure and the phenotype propagated in the small early human populations by random chance. In this regard, it is of note that most of the T cell-mediated diseases mentioned occur in adults after the age of reproductive maturity, when selection forces are weak. The trait could have been passed on without deleterious fitness effects on its carriers until recently, when human average lifespan increased. A third possibility arises from the human-specific loss of the Sia Neu5Gc ≈3 million years ago. Possibly because of this dramatic change in human Sia biology multiple human CD33rSiglecs appear to have undergone dramatic changes in other systems involving gene deletion, gene conversion, and/or changes in binding specificity or expression (3, 4, 9–11). Thus, a possible side effect of this human-specific “shakeup” in Sia and CD33rSiglec biology was the almost complete loss of expression of the latter in T cells. Whatever the explanation, the expression of CD33rSiglecs on human T cells has significantly diverged from that of other hominids.
We have presented support for a model to explain differences in human and chimpanzee T cell stimulation, and we postulate that these differences contribute to the involvement of T cells in human diseases, particularly AIDS and chronic active hepatitis. Given that multiple CD33rSiglecs are involved, it is likely that the mechanism of suppression is mediated by changes in transcription and/or epigenetic factors. If so, the hope exists for finding pharmacological treatments that can restore CD33rSiglec expression on human T cells. This approach could potentially result in amelioration of the severity of some human T cell-mediated disorders.
Methods
Cells and Reagents.
Great ape blood samples were collected into EDTA-containing tubes at the San Diego Zoo (San Diego), the Yerkes National Primate Research Center (Atlanta), and the Lincoln Park Zoo (Chicago) and shipped on ice to the University of California at San Diego. Human blood was collected from healthy volunteer donors, with approval from the University of California at San Diego Institutional Review Board. These samples were collected at about the same time at the University of California at San Diego and stored on ice to ensure comparability in handling with the shipped great ape samples. Whole leukocyte preparations were isolated by ACK buffer (0.15 M NH4Cl/10 mM KHCO3/0.1 mM EDTA) lysis of RBCs, or PBMCs were isolated by centrifugation over Ficoll-Paque PLUS (Amersham Pharmacia Bioscience). Jurkat T cell leukemia clone E6.1 cells from the American Type Culture Collection were cultured in RPMI medium 1640 supplemented with 10% FCS (cRPMI). mAbs against Siglec-6 (E20-1232), Siglec-7 (F023-420), and Siglec-9 (clone E10-286) were prepared in collaboration with BD Pharmingen. The following Abs were generously provided by Paul Crocker (University of Dundee, Dundee, Scotland): anti-Siglec-5 (clone 1A5), anti-Siglec-7 (clones 7.5A and 7.7A), anti-Siglec-8 (clone 7C9), and anti-Siglec-10 (clone 5G6). Purified anti-CD33 (clone HIM3-4), anti-CD3 (clone UCHT-1), and anti-CD28 (clone CD28.2) were purchased from BD Pharmingen. R-phycoerythrin (PE) goat anti-mouse IgG (H+L) was purchased from Caltag Laboratories. The plasmid construct pSig5 containing Siglec-5 under control of the CMV promoter was generated by cloning the full-length Siglec-5 cDNA into the multiple cloning site of pcDNA3.1(+) (Invitrogen).
Flow Cytometry.
Cells (1 × 106) were incubated with a 1:100 dilution of Ab supernatant or 1 μg/100 μl purified Ab in 1% BSA in PBS for 30–60 min on ice. Cells were washed with 1% BSA in PBS and resuspended in 100 μl of 1 μg/100 μl PE goat anti-mouse IgG conjugate in 1% BSA in PBS. For some experiments, cells were also labeled with allophycocyanin (APC)-anti-CD3, APC-anti-CD19, FITC-anti-CD4, or APC-anti-CD8 conjugates. Labeled cells were analyzed on a FACSCalibur (BD Biosciences) flow cytometer by using cellquest software. Data are presented with flowjo software (Tree Star).
T Cell Activation.
Isolated human and chimpanzee PBMCs were cultured in RPMI medium 1640 with 5% human AB serum (RPMI-5HS). For plate-bound Ab-mediated stimulation, cells (2 × 106 per milliliter) were added to wells of a 12-well plate coated with 2.5 μg/ml anti-CD3. Anti-CD28 was then added to cells in solution at 0.1 μg/ml. For some experiments, anti-Siglec-5 was added at 1 μg/ml. Cells were cultured for 5 days before being transferred to tubes and counted by flow cytometry at 60 μl/min for 30 s or for a maximum of 1 × 106 cells. PBMCs were also stimulated with equal amounts of anti-CD3 and anti-CD28 in solution (0.04–1.0 μg/ml) or with 10 μg/ml PHA (Sigma-Aldrich) in solution for 3–5 days. Lymphocytes were also stimulated with anti-CD3/anti-CD28-bearing beads (Dynabeads CD3/CD28 T Cell Expander; 4.5 μm).
T Cell Transfection.
PBMCs were monocyte-depleted by incubation in a polystyrene T175 tissue culture flask at 1–2 × 106 per milliliter in RPMI-5HS for 1 h. Nonadherent cells were removed into a separate flask and confirmed to be mostly lymphocytes by flow cytometry. Lymphocytes or Jurkat cells were transfected by using nucleofection technology. Lymphocytes were resuspended with the Human T Cell Nucleofector Kit (Amaxa), and Jurkat cells were resuspended in the Nucleofector Kit V (Amaxa), following the manufacturer’s guidelines for cell line transfection. Briefly, 100 μl of 2–5 × 106 cell suspension mixed with 1–3 μg of plasmid DNA (pSig5) was transferred to the provided cuvette and nucleofected with a Nucleofector apparatus. Lymphocytes were transfected by using the U-14 program settings, and Jurkat cells were transfected with the S-18 program settings. Controls were mock-transfected by using the same conditions with no DNA. Cells were immediately transferred into wells containing 37°C prewarmed culture medium in 12-well plates. After transfection, cells were cultured for 24 h before analysis by flow cytometry.
Intracellular Calcium Mobilization Assay.
Mobilization of intracellular calcium was measured by using a real-time flow cytometric assay. Briefly, Fluo-4, acetoxymethyl ester (1 mM), and Fura Red (1 mM) calcium-sensing dyes (Molecular Probes) were mixed with Pluronic F-127 solution (Molecular Probes) at a volume ratio of 1:2:3. The calcium-sensing dye solution (2.5 μl) was added to Jurkat cells (4 × 106 per 200 μl of PBS) and incubated at 37°C for 45 min. Cells were then washed with PBS, resuspended in 1 ml of PBS, and allowed to rest at room temperature for 30 min before stimulation. For analysis, cells were acquired by using the time parameter on the FACSCalibur and analyzed for FL1 and FL3 fluorescence. The cell flow rate was 60 μl/s (100–200 cells per second). Anti-CD3 (0.5 μg) was added 60 s after beginning cell acquisition. Cells were collected for a total of 512 s. Postcollection analysis was performed by using flowjo software. The ratio of FL1:FL3 was derived and plotted over time. Kinetic plots are expressed as median of the FL1:FL3 ratio, which has been smoothed based on moving average.
Supplementary Material
Acknowledgments
We thank Dr. Paul Crocker for anti-Siglec mAbs and Aaron Carlin for generating the pSig5 plasmid as well as for helping with blood draws, along with Laarni Gapuz. We also thank Vicki Pomidoro and Liz Horton at Amaxa Biosystems for technical assistance and the helpful staff at the Zoological Society of San Diego, the Yerkes National Primate Research Center, and the Lincoln Park Zoo. We especially thank the humans and great apes who provided blood for study. This study was supported by National Institutes of Health Grants P01 HL057345 and R01 GM32373 (to A.V.), American Heart Association Fellowship 0525345Y (to D.H.N.), and the G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations
- Sia
sialic acid
- Neu5Gc
N-glycolylneuraminic acid
- TCR
T cell receptor
- PHA
phytohemagglutinin-L
- PBMC
peripheral blood mononuclear cell
- PE
phycoerythrin
- APC
allophycocyanin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Crocker P. R., Varki A. Trends Immunol. 2001;22:337–342. doi: 10.1016/s1471-4906(01)01930-5. [DOI] [PubMed] [Google Scholar]
- 2.Crocker P. R. Curr. Opin. Pharmacol. 2005;5:431–437. doi: 10.1016/j.coph.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Varki A., Angata T. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 4.Angata T., Margulies E. H., Green E. D., Varki A. Proc. Natl. Acad. Sci. USA. 2004;101:13251–13256. doi: 10.1073/pnas.0404833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravetch J. V., Lanier L. L. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 6.Singer A. L., Koretzky G. A. Science. 2002;296:1639–1640. doi: 10.1126/science.1071551. [DOI] [PubMed] [Google Scholar]
- 7.Avril T., Floyd H., Lopez F., Vivier E., Crocker P. R. J. Immunol. 2004;173:6841–6849. doi: 10.4049/jimmunol.173.11.6841. [DOI] [PubMed] [Google Scholar]
- 8.Varki A. Am. J. Phys. Anthropol. Suppl. 2001;33:54–69. doi: 10.1002/ajpa.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinkman-Van der Linden E. C. M., Sjoberg E. R., Juneja L. R., Crocker P. R., Varki N., Varki A. J. Biol. Chem. 2000;275:8633–8640. doi: 10.1074/jbc.275.12.8633. [DOI] [PubMed] [Google Scholar]
- 10.Sonnenburg J. L., Altheide T. K., Varki A. Glycobiology. 2004;14:339–346. doi: 10.1093/glycob/cwh039. [DOI] [PubMed] [Google Scholar]
- 11.Hayakawa T., Angata T., Lewis A. L., Mikkelsen T. S., Varki N. M., Varki A. Science. 2005;309:1693. doi: 10.1126/science.1114321. [DOI] [PubMed] [Google Scholar]
- 12.Ikehara Y., Ikehara S. K., Paulson J. C. J. Biol. Chem. 2004;279:43117–43125. doi: 10.1074/jbc.M403538200. [DOI] [PubMed] [Google Scholar]
- 13.Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 14.Varki A., Altheide T. K. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 15.Varki A. Genome Res. 2000;10:1065–1070. doi: 10.1101/gr.10.8.1065. [DOI] [PubMed] [Google Scholar]
- 16.Olson M. V., Varki A. Nat. Rev. Genet. 2003;4:20–28. doi: 10.1038/nrg981. [DOI] [PubMed] [Google Scholar]
- 17.McConkey E. H., Varki A. Science. 2005;309:1499–1501. doi: 10.1126/science.1113863. [DOI] [PubMed] [Google Scholar]
- 18.Alter H. J., Eichberg J. W., Masur H., Saxinger W. C., Gallo R., Macher A. M., Lane H. C., Fauci A. S. Science. 1984;226:549–552. doi: 10.1126/science.6093251. [DOI] [PubMed] [Google Scholar]
- 19.Novembre F. J., Saucier M., Anderson D. C., Klumpp S. A., O’Neil S. P., Brown C. R. I., Hart C. E., Guenthner P. C., Swenson R. B., McClure H. M. J. Virol. 1997;71:4086–4091. doi: 10.1128/jvi.71.5.4086-4091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp P. M., Shaw G. M., Hahn B. H. J. Virol. 2005;79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker C. M. Springer Semin. Immunopathol. 1997;19:85–98. doi: 10.1007/BF00945027. [DOI] [PubMed] [Google Scholar]
- 22.Gagneux P., Muchmore E. A. Methods Mol. Med. 2004;96:289–318. doi: 10.1385/1-59259-670-3:289. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi P. S. Nat. Rev. Immunol. 2002;2:427–438. doi: 10.1038/nri822. [DOI] [PubMed] [Google Scholar]
- 24.Bavari S., Hunt R. E., Ulrich R. G. Clin. Immunol. Immunopathol. 1995;76:248–254. doi: 10.1006/clin.1995.1123. [DOI] [PubMed] [Google Scholar]
- 25.Falco M., Biassoni R., Bottino C., Vitale M., Sivori S., Augugliaro R., Moretta L., Moretta A. J. Exp. Med. 1999;190:793–801. doi: 10.1084/jem.190.6.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poe J. C., Fujimoto Y., Hasegawa M., Haas K. M., Miller A. S., Sanford I. G., Bock C. B., Fujimoto M., Tedder T. F. Nat. Immunol. 2004;5:1078–1087. doi: 10.1038/ni1121. [DOI] [PubMed] [Google Scholar]
- 27.van Baalen C. A., Kwa D., Verschuren E. J., Reedijk M. L., Boon A. C., de Mutsert G., Rimmelzwaan G. F., Osterhaus A. D., Gruters R. A. J. Infect. Dis. 2005;192:1183–1190. doi: 10.1086/444546. [DOI] [PubMed] [Google Scholar]
- 28.Lai W., Chang C. H., Farber D. L. J. Immunol. Methods. 2003;282:93–102. doi: 10.1016/j.jim.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 29.Gagneux P., Wills C., Gerloff U., Tautz D., Morin P. A., Boesch C., Fruth B., Hohmann G., Ryder O. A., Woodruff D. S. Proc. Natl. Acad. Sci. USA. 1999;96:5077–5082. doi: 10.1073/pnas.96.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood B., Collard M. Science. 1999;284:65–66. doi: 10.1126/science.284.5411.65. [DOI] [PubMed] [Google Scholar]
- 31.Wadman M. Nature. 2006;440:388–389. doi: 10.1038/440388a. [DOI] [PubMed] [Google Scholar]
- 32.Song B., Gold B., O’Huigin C., Javanbakht H., Li X., Stremlau M., Winkler C., Dean M., Sodroski J. J. Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogerd H. P., Doehle B. P., Wiegand H. L., Cullen B. R. Proc. Natl. Acad. Sci. USA. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariani R., Chen D., Schrofelbauer B., Navarro F., Konig R., Bollman B., Munk C., Nymark-McMahon H., Landau N. R. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 35.Pischinger K., Zimmermann K., Eibl M. M., Mannhalter J. W. Arch. Virol. 1998;143:2065–2076. doi: 10.1007/s007050050444. [DOI] [PubMed] [Google Scholar]
- 36.Gougeon M. L., Lecoeur H., Boudet F., Ledru E., Marzabal S., Boullier S., Roue R., Nagata S., Heeney J. J. Immunol. 1997;158:2964–2976. [PubMed] [Google Scholar]
- 37.Gougeon M. L., Lecoeur H., Heeney J., Boudet F. Immunol. Lett. 1996;51:75–81. doi: 10.1016/0165-2478(96)02558-8. [DOI] [PubMed] [Google Scholar]
- 38.Goodman M. Am. J. Hum. Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.