Abstract
The development of acquired resistance to ErbB2 tyrosine kinase inhibitors limits the clinical efficacy of this class of cancer therapeutics. Little is known about the mechanism(s) of acquired resistance to these agents. Here we establish a model of acquired resistance to N-{3-chloro-4-[(3-fluorobenzyl) oxy]phenyl}-6-[5-({[2 (methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine (lapatinib), an inhibitor of ErbB2 and ErbB1 tyrosine kinases by chronically exposing lapatinib-sensitive ErbB2-overexpressing breast cancer cells to lapatinib, simulating the clinic where lapatinib is administered on a daily chronic basis. Analysis of baseline gene expression in acquired lapatinib-resistant and parental cells indicates estrogen receptor (ER) signaling involvement in the development of resistance. Using gene interference, we confirm that acquired resistance to lapatinib is mediated by a switch in cell survival dependence and regulation of a key antiapoptotic mediator from ErbB2 alone to codependence upon ER and ErbB2 rather than loss of ErbB2 expression or insensitivity of ErbB2 signaling to lapatinib. Increased ER signaling in response to lapatinib is enhanced by the activation of factors facilitating the transcriptional activity of ER, notably FOXO3a and caveolin-1. Importantly, we confirm that lapatinib induces ER signaling in tumor biopsies from patients with ErbB2-overexpressing breast cancers receiving lapatinib therapy. These findings provided the rationale for preventing the development of acquired resistance by simultaneously inhibiting both ER and ErbB2 signaling pathways. Establishing clinically relevant models of acquired resistance to ErbB2 kinase inhibitors will enhance therapeutic strategies to improve clinical outcomes for patients with ErbB2-overexpressing breast cancers.
Keywords: estrogen receptor, lapatinib, resistance
Aberrant activation of oncogenic tyrosine kinases and steroid receptors plays a key role in breast carcinogenesis. Overexpression or gene amplification of ErbB2, a member of the ErbB receptor tyrosine kinase family, occurs in 25–30% of breast cancers where it predicts for a poor clinical outcome (1). Consequently, therapies targeting ErbB2 represent an attractive strategy in breast cancer (2, 3). Trastuzumab, a humanized anti-ErbB2 monoclonal antibody, is an approved treatment for patients with ErbB2-overexpressing breast cancers (4). ErbB2 signaling can also be blocked using small-molecule tyrosine kinase inhibitors that compete with ATP for binding at the ErbB2 catalytic kinase domain. N-{3-chloro-4-[(3-fluorobenzyl) oxy]phenyl}-6-[5-({[2 (methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine (lapatinib; GW572016), a potent reversible inhibitor of ErbB2 and ErbB1 tyrosine kinases is currently in Phase III clinical trials in breast and other carcinomas (5). Inhibition of ErbB2 tyrosine autophosphorylation by lapatinib abrogates downstream mitogen-activated protein kinase (MAPK)-Erk1/2 and PI3K-Akt growth/survival signaling in ErbB2-overexpressing breast cancer cell lines, xenografts, and in patients with ErbB2-overexpressing breast cancers (6–9). The preponderance of evidence indicates that lapatinib activity in breast cancer cells depends upon ErbB2 overexpression rather than ErbB1 (6–9). The poor prognostic effects of ErbB2 overexpression have been attributed to the concomitant up-regulation of the PI3K-Akt survival pathway (10–13). Although inhibition of ErbB2 autophosphorylation and signaling via MAPK-Erk1/2 and PI3K-Akt pathways may be necessary for clinical response to lapatinib, it is not sufficient (6, 14). In this regard, the antitumor effects of lapatinib on ErbB2-overexpressing breast cancer cells appear to be closely linked to the down-regulation of survivin, a key regulator of mitosis and cell survival (15–17).
Estrogen and its cognate receptors estrogen receptor (ER)α and ERβ also play critical roles in breast carcinogenesis (18). More than 60% of human breast cancers are ERα-positive, which mediates most estrogenic responses, although estrogen can also exert ER-independent biologic effects (19–23). ER negativity may occur as a result of (i) ER promoter hypermethylation (24) or (ii) constitutive inactivation of FOXO3a, a member of the Forkhead family of transcription factors that enhances ER transcriptional activity. FOXO3a is inactive when phosphorylated by Akt (25, 26). Concomitant up-regulation of PI3K-Akt signaling in ErbB2-overexpressing breast cancers inactivates FOXO3a, thereby suppressing ER transcriptional activity (26).
Results from early-phase trials indicate that clinical responses to lapatinib monotherapy in patients with ErbB2-overexpressing breast cancers are generally short-lived (27). Enhancing the clinical efficacy of ErbB2 kinase inhibitors like lapatinib requires the identification of mechanisms responsible for the development of acquired resistance. Here we describe a model of acquired lapatinib resistance that simulates the clinic where patients receive lapatinib on a daily chronic basis. In this model, chronic exposure to lapatinib converts ErbB2-overexpressing breast cancer cells that are initially sensitive to lapatinib-induced apoptosis to resistant cells. Resistance is mediated by enhanced ER signaling, resulting in ER playing a more significant role in regulating cell survival and survivin rather than loss of ErbB2 expression or insensitivity of the ErbB2 pathway to lapatinib. Simultaneous inhibition of ErbB2 and ER signaling prevents the development of acquired resistance to lapatinib in ErbB2−-overexpressing/ER+ breast cancer cells. Importantly, increased ER signaling is confirmed in patients with ErbB2-overexpressing/ER+ breast cancers treated with lapatinib monotherapy. Elucidating mechanisms of acquired resistance to ErbB2 kinase inhibitors provides a rationale for developing therapeutic strategies to enhance their clinical efficacy.
Results
Lapatinib Up-Regulates ER Signaling.
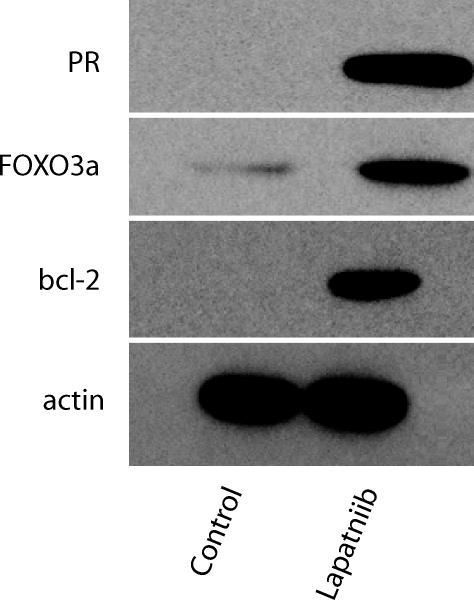
BT474, an ErbB2-overexpressing/ER+ breast cancer cell line, is one of the few breast cancer lines that undergo apoptosis in response to lapatinib (8). Although BT474 constitutively express ERα, baseline expression of ER-regulated gene products, e.g., progesterone receptor (PR) and bcl-2, is not increased (Fig. 1). Treating BT474 cells with 1 μM lapatinib for 24 h, a concentration that is readily achieved in patients (27) and shown to inhibit p-ErbB2 and downstream p-Erk1/2 and p-Akt (8, 9), increased PR and bcl-2 protein levels (Fig. 1), despite little change in ER protein expression (data not shown). Induction of ER signaling by lapatinib was not unique to BT474, because ER was up-regulated in MCF-7/Her-2, an ErbB2 overexpressing/ER+ cell line (28), compared with non-ErbB2-expressing parental MCF-7 cells (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
FOXO3a and ER-regulated gene products are induced by lapatinib. Steady-state protein levels of FOXO3a, PR, and bcl-2 were assessed in BT474 cells treated with lapatinib (1 μM) for 24 h. Cells treated with vehicle (DMSO) served as controls. Steady-state actin protein levels served as controls for equal loading of protein. Results are representative of three independent experiments.
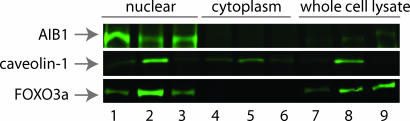
Because PR and bcl-2 are ER-regulated gene products, we examined the effects of lapatinib on cofactors affecting ER transcriptional activity. Protein expression of NCoR, an ER corepressor (data not shown), and the steroid receptor coactivator-3 (SRC-3/NCoA-3/AIB1) remained unchanged in lapatinib-treated cells (Fig. 2). In contrast, caveolin-1, a protein that enhances ER transcriptional activity and regulates cell growth by inhibiting ErbB2 and mitogen-activated protein kinase signaling (29–31), markedly increased in lapatinib-treated cells (Fig. 2). Normally a component of cell-surface membrane structures called caveolae, caveolin-1 was highly expressed in the nucleus of lapatinib-treated cells (Fig. 2). Steady-state protein levels of Oct-1 and IκΒα were used to verify the integrity of the nuclear and cytoplasmic fractions, respectively (data not shown).
Fig. 2.
Lapatinib modulates the expression and cell localization of molecules that promote transcription of ER-regulated genes. Western blot was performed on equal amounts of protein from nuclear, cytoplasmic, and whole-cell extracts. Steady-state protein levels of AIB1, caveolin-1, and FOXO3a were assessed in vehicle-treated BT474 cells (lanes 1, 4, and 7); BT474 cells treated with lapatinib (1 μM) for 24 h (lanes 2, 5, and 8); and B5 cells cultured in the continuous presence of lapatinib (5 μM; lanes 3, 6, and 9). Oct-1 and IκΒα were used to verify the integrity of the nuclear and cytoplasmic extracts, respectively (data not shown). Results were confirmed in three independent experiments.
Derepression of FOXO3a in Response to Lapatinib Contributes to Increased ER Signaling.
Because FOXO3a regulates transcription of caveolin-1 and ER (25, 30), it was tempting to speculate that inactivation of Akt by lapatinib derepresses FOXO3a, which in turn increases caveolin-1 protein expression and ER transcriptional activity. Total FOXO3a protein levels increased in lapatinib-treated BT474 cells (Fig. 2). A faster-migrating form of FOXO3a, its hypophosphorylated transcriptionally active form, was detected by SDS/PAGE in both lapatinib-treated BT474 and Au565 cells, an ErbB2-overexpressing/ER-negative breast cancer line (Fig. 8, which is published as supporting information on the PNAS web site).
Establishing a Model of Acquired Resistance to Lapatinib.
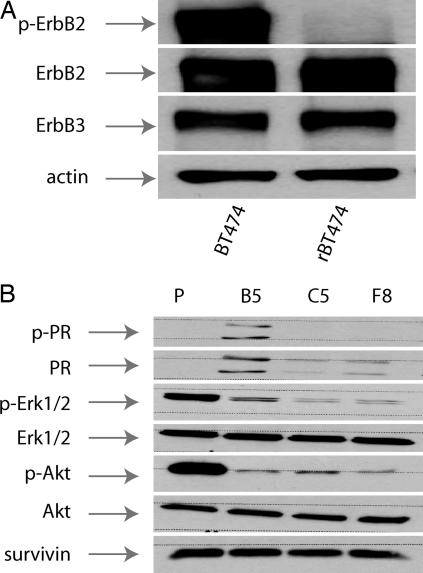
Because lapatinib activated FOXO3a, caveolin-1, and ER signaling, we next addressed whether these changes might be involved in acquired lapatinib resistance. BT474 cells were continuously exposed to lapatinib. As reported (8), marked apoptosis occurred within the first 5 days after lapatinib treatment (data not shown). However, after 21 days, small colonies of viable cells appeared. After 12 weeks, lapatinib-resistant BT474 cells (rBT474) were growing in the continuous presence of 5 μM lapatinib without significant effect on cell viability, e.g., ≈5% spontaneous apoptosis. Three additional lapatinib-resistant cell lines (B5, F8, and C5) were established from single-cell cloning of pooled rBT474 cells. Resistance to lapatinib was not related to loss of ErbB expression, because steady-state ErbB2 and ErbB3 protein levels in rBT474 were comparable to parental cells (Fig. 3A). ErbB1 was not detectable in either rBT474 or parental cells (data not shown), nor was resistance related to refractoriness of the ErbB2 pathway to lapatinib, because p-ErbB2, p-Akt, and p-Erk1/2 inhibition was similar in rBT474 (Fig. 3 A and B) compared with that previously shown in lapatinib-treated parental cells (8).
Fig. 3.
Acquired resistance is not mediated by loss of target expression or insensitivity of the ErbB2-mitogen-activated protein kinase–PI3K pathways to lapatinib. (A) ErbB2, p-ErbB2, and ErbB3 steady-state protein levels in parental and rBT474. Actin steady-state protein levels served as a control to ensure equal loading of protein. (B) Steady-state levels of the indicated proteins in untreated parental (P) BT474 cells and in B5, C5, and F8 cells (see Materials and Methods).
To gain a broader perspective on potential mechanisms of acquired resistance, we used the Human Affymetrix oligonucleotide array platform (U133 plus 2 chips) comparing gene expression in rBT474 and parental cells. Pathway analysis of genes using proprietary analytical tools (see Supporting Text, which is published as supporting information on the PNAS web site) was used to assist in the biologic interpretation of genes differentially expressed in rBT474 cells. Fifty-seven genes were significantly up-regulation (>3-fold) in rBT474 compared to parental cells (data not shown). Pathway analysis indicated the induction of ER and PR signaling pathways in rBT474 compared with parental cells (Table 1, which is published as supporting information on the PNAS web site). Expression of PR transcript (Table 1) and protein increased in rBT474 compared with parental cells, although the degree of PR protein expression varied among the B5, F8, and C5 cell lines (Fig. 3B). Similarly, bcl-2 transcript and protein levels increased in rBT474 cells (Table 1 and Fig. 9, which is published as supporting information on the PNAS web site). Expression of SRC-3/NCoA-3/AIB1 transcript increased 2.5-fold in rBT474 cells, although protein levels were essentially unchanged (Fig. 2). FOXO3a protein levels, which had increased after 24 h in lapatinib-treated parental cells, also increased in rBT474 cells, particularly in whole-cell extracts, compared with untreated parental BT474 cells (Fig. 2). Gene expression analysis in conjunction with protein expression validation is consistent with activation of ER signaling in rBT474 cells.
Activation of FOXO3a and ER Signaling Occurs in Patients Administered Lapatinib.
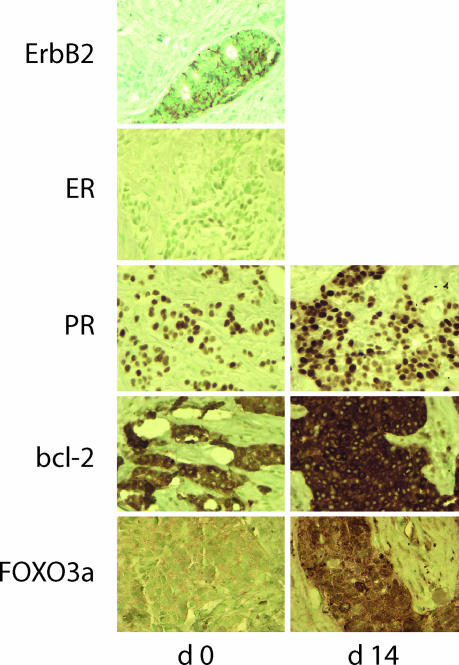
To elucidate the clinical relevance of these findings, we examined sequential tumor biopsies obtained before (d 0) and after 14 days of (d 14) lapatinib therapy as part of a neoadjuvant trial in patients with early-stage breast cancer. Increased expression of FOXO3a, PR, and bcl-2 was observed after 14 days of lapatinib in ErbB2-overexpressing breast cancers that were constitutively ER-positive (Fig. 4 and Fig. 10, which is published as supporting information on the PNAS web site). In response to lapatinib therapy, ER exclusively localized to tumor cell nuclei in ErbB2-overexpressing/ER+ breast cancers where it regulates gene transcription (Fig. 11, which is published as supporting information on the PNAS web site).
Fig. 4.
Lapatinib therapy enhances the expression of ER-regulated gene products in patients with ErbB2-overexpressing/ER+ breast cancers. Sequential tumor biopsies obtained before (day 0) and after 14 days of lapatinib therapy (1,500 mg per day, day 14) were analyzed by IHC for expression of ErbB2, ER, PR, bcl-2, and FOXO3a proteins.
Survival of rBT474 Cells No Longer Depends upon ErbB2.
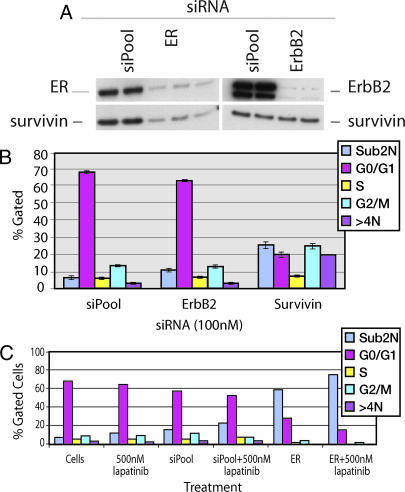
We recently showed that survivin down-regulation by lapatinib or gene interference induced apoptosis in parental BT474 cells (15). siRNA-targeted knockdown of ErbB2 did not affect survivin protein expression or rBT474 cell survival (Fig. 5A and B), a finding consistent with the inability of lapatinib to regulate survivin in rBT474 despite effectively inhibiting p-ErbB2, p-Erk1/2, and p-Akt (Fig. 3).
Fig. 5.
Regulation of cell survival and survivin switches from ErbB2 to ER during the development of acquired resistance to lapatinib. (A) siRNA targeted knockdown of ER but not ErbB2 reduces survivin steady-state protein levels in resistant cells. (B) Survivin, but not ErbB2 knockdown, induces apoptosis in rBT474 cells. (C) ER knockdown in the B5 cells induces marked tumor cell apoptosis. The data are representative of three independent experiments.
We sought to determine whether acquired lapatinib resistance was associated with a switch in the regulation of survivin from ErbB2 to ER. ER protein was markedly reduced in rBT474 cells transfected with ER-specific siRNA constructs compared with cells transfected with control siRNA constructs (siPool) (Fig. 5A). PR protein was also reduced as a consequence of ER knockdown (Fig. 12, which is published as supporting information on the PNAS web site). In contrast to ErbB2, knocking down ER in rBT474 cells reduced survivin expression (Fig. 5A). Selective knockdown of survivin led to a 6-fold increase in rBT474 cell apoptosis (sub2N) compared with cells transfected with control siRNA constructs (Fig. 5B). Survivin knockdown also increased the percentage of cells undergoing endoreduplication (>4N), which was reported previously and is consistent with its role in regulating mitosis (15). Importantly, ER knockdown in B5 cells, which appear to particularly depend upon ER signaling based upon their relatively increased expression of PR, induced marked apoptosis (Fig. 5C). Increased apoptosis after ER knockdown was also seen in pooled rBT474, C5, and F8 (data not shown). In contrast to its effect on survivin, ER knockdown in rBT474 cells had relatively little effect on bcl-2 (Fig. 12). Although ER plays a key role in regulating the survival of rBT474 cells, the ErbB2 signaling pathway remains intact. Consequently, inhibiting ER and ErbB2 (500 nM lapatinib) simultaneously in rBT474 cells more effectively induced apoptosis compared with inhibition of either pathway alone (Fig. 5C).
Despite expressing ER, parental BT474 cell survival was unaffected by ER knockdown (Fig. 13, which is published as supporting information on the PNAS web site). The selectivity of the siRNA constructs was demonstrated by its (i) lack of effect on control proteins (e.g., Erk) and (ii) inability to induce IFN-regulated genes, a potential nonspecific consequence of siRNA (data not shown).
Combining Lapatinib with Inhibition of ER Signaling Abrogates the Development of Acquired Lapatinib Resistance.
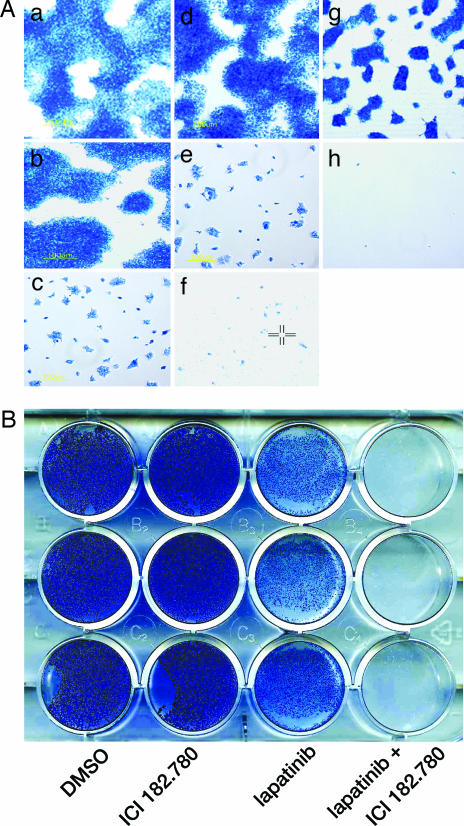
If enhanced ER signaling contributes to the development of acquired lapatinib resistance, then simultaneous blockade of ER and ErbB2 signaling might delay or possibly prevent its onset. BT474 cells were treated according to the following conditions (Fig. 6A): (a) control (DMSO), (b) ICI 182.780 (fulvestrant) simulates siRNA knockdown by inducing ER proteolysis, (c) lapatinib, (d) tamoxifen, (e) tamoxifen plus lapatinib, (f) ICI 182.780 plus lapatinib, (g) estrogen deprivation, and (h) estrogen deprivation plus lapatinib. As shown in Fig. 6A, cell viability was assessed by methylene blue staining after 21 days of treatment. Viability remained essentially unchanged in cells treated with ICI 182.780 or tamoxifen alone compared with vehicle-treated controls. Treatment with lapatinib alone initially resulted in cell death followed by the appearance of viable cells and small colonies by 21 days. The effects of combining tamoxifen plus lapatinib were similar to that observed with lapatinib alone. In contrast, only rare viable cells were seen at day 21 after the combination of ICI 182.780 plus lapatinib. Because it is difficult to study the effects of aromatase inhibitors in cell culture, we simulated their effects by depriving cells of exogenous estrogen using phenol red-free medium supplemented with charcoal-stripped serum. Estrogen deprivation markedly reduced PR protein (data not shown). Although estrogen deprivation alone affected cell growth (Fig. 6A, compare a and g), it did not prevent the outgrowth of viable resistant cells. However, when parental BT474 cells were subjected to estrogen deprivation plus lapatinib, virtually no viable cells were observed at day 21. To determine the durability of these effects, we evaluated cell viability after 6 weeks of treatment. Fig. 6B demonstrates the effects of treating parental BT474 cells with the combination of ICI 182.780 plus lapatinib compared with (i) ICI 182.780 alone or (ii) lapatinib alone where resistance occurred. Although the concentration of ICI 182.780 used in these experiments blocked ER signaling, e.g., marked inhibition of PR expression (data not shown), it did not suppress survivin protein expression (Fig. 14, which is published as supporting information on the PNAS web site). In contrast, the combination of ICI 182.780 plus lapatinib markedly inhibited survivin protein (Fig. 14) and continued to prevent the outgrowth of resistant cells through 6 weeks (Fig. 6B). Similar results were observed when combining estrogen deprivation and lapatinib (data not shown).
Fig. 6.
Therapeutic strategies combining lapatinib with antiestrogens delay or prevent the development of lapatinib resistance in ErbB2-overexpressing/ER+ breast cancer cells. (A) Parental BT474 cells were treated with: (a) vehicle (DMSO); (b) ICI 182.780 (10 nM); (c) lapatinib (500 nM); (d) tamoxifen (1 μM); (e) lapatinib plus tamoxifen (1 μM); (f) ICI 182.780 (10 nM) plus lapatinib; (g) estrogen deprivation (charcoal-stripped serum/phenol red-free medium) alone; (h) estrogen deprivation plus lapatinib. After 21 days, viable cells were assessed by methylene blue staining. Results were confirmed in three independent experiments. (B) Exponentially growing parental BT474 cells were treated according to conditions described in A. After 6 weeks, viable cell colonies were visualized by methylene blue staining. Conditions were repeated in triplicate (rows).
Discussion
The clinical efficacy of small-molecule ErbB2 kinase inhibitors is limited by either primary resistance or the development of secondary acquired resistance. Acquired resistance to lapatinib develops in patients with ErbB2-overexpressing breast cancers who initially responded to therapy (6, 27). Here we describe a model of acquired lapatinib resistance in ErbB2-overexpressing/ER+ breast cancer cells that simulates the clinic, relying on endogenous changes in tumor signaling as a consequence of chronic exposure to lapatinib rather than the forced overexpression of a specific protein(s) using gene transfection. Importantly, we have used this model to identify a therapeutic strategy to prevent its onset.
A number of mechanisms have been proposed for the development of resistance to ErbB targeted therapies. Coexpression of insulin-like growth factor 1 receptor (IGF-IR) in ErbB2-overexpressing breast cancer cells and primary tumors contributes to trastuzumab resistance (32–34) but does not appear to play a role in the development of acquired lapatinib resistance, because IGF-IR expression and activation state remained unchanged in rBT474 compared with parental cells (data not shown). In fact, coexpression of IGF-IR in ErbB2-overexpressing breast cancers appears to predict for a favorable response to lapatinib rather than resistance (6). Similarly, although phosphatase and tensin homologue deleted in chromosome 10 (PTEN) deficiency contributes to trastuzumab resistance (35), it does not appear to play a role in this model of acquired lapatinib resistance, because PI3K-Akt signaling was effectively inhibited in rBT474 cells. In addition, ErbB2 protein levels were comparable in rBT474 and parental cells, making target down-modulation an unlikely explanation for our model of acquired lapatinib resistance. Although it is difficult to exclude other factors, and ER-independent genes were differentially up-regulated in rBT474 compared with parental cells, pathway analysis of the changes in rBT474 cells at the transcriptome and protein levels implicate ER signaling in the development of acquired lapatinib resistance. The ability of genetic knockdown of ER to induce apoptosis in lapatinib-resistant cells coupled with data showing that the combination of lapatinib plus antiestrogen therapy prevents the development of acquired lapatinib resistant is consistent with ER playing a major role in this process.
Activation of ER signaling by lapatinib was not unique to BT474 cells, because it was also observed in ER-positive MCF-7/Her-2 cells and importantly in patients with ErbB2-overexpressing/ER+ breast cancers treated with lapatinib. Whether induction of ER signaling leads to the development of acquired resistance in the clinic cannot be addressed in a neoadjuvant trial and will require further investigation.
Enhanced ER signaling by lapatinib appears to occur as a consequence of FOXO3a activation in response to ErbB2-PI3K-Akt signaling inhibition. It is therefore ironic that lapatinib resistance may occur as a direct consequence of its potent inhibitory effect on the ErbB2 signaling pathway, its intended target. Is the effect of lapatinib on ER signaling ErbB2-dependent as its antitumor activity in breast cancer appears to be (6, 9), or is there a role for ErbB1, which is inhibited by lapatinib with equal potency (5)? We cannot exclude a potential role for ErbB1 in the development of ER-mediated autoresistance to lapatinib, because ErbB1 has also been linked to the development of tamoxifen resistance (36, 37). However, this seems unlikely, because (i) ErbB1 was not detected in either parental BT474 or rBT474 cells, and (ii) enhanced ER signaling by lapatinib occurred in MCF-7/Her-2 but not MCF-7 parental cells despite their equal expression of ErbB1.
Although ER assumes a more prominent role in regulating survivin and cell survival during the development of acquired resistance to lapatinib, the ErbB2 pathway remains intact, making simultaneous inhibition of ER and ErbB2 pathways imperative. Combining lapatinib with therapies that block ER signaling through (i) degradation of ER (e.g., fulvestrant) or (ii) estrogen deprivation (e.g., aromatase inhibitors) prevents the development of lapatinib resistance in cell lines, warranting further testing in clinical trials. Interestingly, tamoxifen was less effective compared with these other antiestrogen approaches, possibly related to the tendency of ErbB2-overexpressing breast cancer cells to express AIB1, which promotes the estrogenic effects of tamoxifen, stimulating rather than inhibiting tumor cell growth (36, 38, 39).
The day has arrived when therapeutic decisions for treating patients with breast cancer are based on the molecular profile of tumors rather than histology alone. Patients whose breast cancers overexpress ErbB2 but are ER-negative due to FOXO3a-independent mechanisms appear to develop acquired lapatinib resistance that is ER-independent. For example, SKBR3 cells, an ErbB2-overexpressing/ER-negative breast cancer cell line, are highly sensitive to lapatinib-induced apoptosis (8, 9). However, acquired lapatinib resistance in SKBR3 (rSKBR3) appears to be an ER-independent process (Fig. 15, which is published as supporting information on the PNAS web site). Although p-ErbB2, p-Erk1/2, and p-Akt were inhibited in rSKBR3 cells (data not shown), survivin expression remained essentially unchanged, and the increased expression of activated NF-κB in rSKBR3 cells may provide a clue to their mechanism of resistance (Fig. 16, which is published as supporting information on the PNAS web site). Preliminary data suggest that lapatinib might not induce ER signaling in patients with ErbB2-overexpressing/ER-negative breast cancers (Fig. 17, which is published as supporting information on the PNAS web site), although this is based on a small sample size. It will be important to elucidate ER-independent mechanisms of acquired resistance.
Nonetheless, at least 50% of ErbB2-overexpressing breast cancers are ER-positive at baseline (40), accounting for 10–15% of all breast cancers. The clinical implications of our data are that the efficacy of lapatinib in these patients might be enhanced by combining lapatinib with (i) therapies that degrade ER (e.g., fulvestrant) or (ii) estrogen deprivation (e.g., aromatase inhibitor). This recommendation might also apply to patients whose tumors are technically classified as ER-negative based on established diagnostic criteria yet still express detectable ER. Establishing clinically relevant models of acquired resistance to ErbB2 kinase inhibitors such as lapatinib provides a rationale for designing therapeutic strategies to prevent its development and improve the long-term outcome in patients with ErbB2-overexpressing/ER+ breast cancers.
Materials and Methods
Reagents.
Anti-human survivin antibody was from R & D Systems. Antibodies to phosphotyrosine and actin were from Sigma. The Ab −11 anti-ErbB2 antibody was from NeoMarkers (Fremont, CA). Antibodies to p-Akt (Ser-437), PR, and phospho-PR (Ser-190) were from Cell Signaling Technology (Beverly, MA). Anti-ERα and FKHRL/FOXO3a antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). Anti-AIB1 was from Abcam, Inc. (Cambridge, MA). Antibodies to Akt, p-Erk1/2, Erk1/2, caveolin-1, and bcl-2 were from Santa Cruz Biotechnology. ICI 182.780 was from Tocris Cookson (Ellisville, MO). Tamoxifen citate was purchased from Calbiochem. The guava PCA 96 Nesin kit was purchased from Guava Technologics (Hayward, CA). SuperSignal West Femto Maximum Sensitivity Substrate was purchased from Pierce. Lapatinib was synthesized as described (5). Lapatinib for cell culture work was dissolved in DMSO.
Cell Culture and siRNA Transfection.
BT474, MCF-7, and Au565 cell lines were obtained from the American Type Culture Collection (Manassas, VA) and cultured in RPMI medium 1640 supplemented with 10% FBS (GIBCO/BRL). Cell cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C. For experiments involving estrogen deprivation, cells were cultured in phenol-free medium supplemented with charcoal-stripped serum purchased from Gemini Biological Products (Woodland, CA). Lapatinib-resistant pooled BT474 (rBT474) were established by culturing cells for a minimum of 2 months in RPMI medium 1640 supplemented with increasing concentrations of lapatinib (0.25–5 μM). B5, C5, and F8 cell lines were established by single-cell cloning of pooled rBT474 and then continuously cultured in the presence of 5 μM lapatinib. SMART pools (containing four individual siRNA motifs) against ERα, ErbB2, and control nontargeting pool were purchased from Dharmacon Research (Lafayette, CO). Transfections were performed in a 12-well format using 2 μl of lipofectamine 2000 (Invitrogen Life Technologies) in OPTI-MEM I (Invitrogen) at 2 × 105 cells per well. The concentration of siRNA was 100 nM in a final volume of 1.1 ml according to Invitrogen transfection protocol. After 16–18 h, the transfection media were removed and replaced with complete RPMI medium 1640. For Western blot analysis, cells were washed with PBS at 48 and 72 h posttransfection and lysed in RIPA buffer. Lapatinib was added at the desired concentration 48 h after transfection and FACS analysis performed 72 h after lapatinib.
Cell Fractionation, SDS/PAGE, and Western Blotting.
Whole-cell extracts were prepared by scraping cells off Petri dishes, washing cell pellets twice in PBS, and then resuspending pellets in two-packed cell volumes of RIPA buffer [150 mM NaCl/50 mM Tris·HCl, pH 7.5/0.25% (wt/ol) deoxycholate/1% Nonidet P-40/5 mM sodium orthovanadate/2 mM sodium fluoride/protease inhibitor mixture]. Nuclear and cytoplasmic extracts were isolated by washing cell pellets in buffer A (10 mM Hepes, pH 7.5/10 mM KCl/1.5 mM Mg2Cl/0.5 mM NaF/1 mM glycerol phosphate/protease mixture), then lysis in buffer A + B (buffer A plus 0.5% Nonidet P-40) in a 2:1 mix. After centrifugation (12,000 × g for 10 min), supernatant was collected (cytoplasmic fraction). Pellets were washed in PBS, centrifuged (12,000 × g for 10 min), and then flash-frozen in dry-ice–ethanol in buffer C (20 mM Hepes, pH, 7.5/420 mM NaCl/1.5 mM Mg2Cl/0.5 mM NaF/0.5 mM DTT/1 mM glycerol phosphate/protease mixture), followed by a slow thaw on ice. Pellets were then centrifuged (12,000 × g for 10 min), washed, and membranes lysed in RIPA buffer. The supernatant was centrifuged (12,000 × g) and collected (nuclear fraction). Protein concentrations were determined by using a modification of the Bradford method (Bio-Rad). Equal amounts of proteins (50 μg) were resolved by either 7.5% or 4–15% gradient SDS/PAGE under reducing conditions. Proteins were then transferred to Immobilon-P or nitrocellulose membranes. Efficiency and equal loading of proteins were evaluated by Ponceau S staining. Membranes were blocked for 1 h in TBS (25 mM Tris·HCl, pH 7.4/150 mM NaCl/2.7 mM KCl) containing 4% (wt/vol) low-fat milk or 3% BSA (wt/vol). Membranes were then probed with specific antibodies recognizing target proteins and visualized with enhanced chemiluminescence or the SuperSignal West Femto Maximum sensitivity substrate kit (Pierce). Proteins were visualized with the SuperSignal West Femto maximum sensitivity substrate kit (Pierce) or Odyssey Infrared Imaging System (Li-Cor).
Immunohistochemistry.
Sequential tumor biopsies (days 0 and 14) were collected as part of a clinical trial in which patients with early-stage breast cancer were treated with lapatinib therapy alone (1,500 mg per day), conducted in accordance with the 1996 version of the Declaration of Helsinki. The study protocol was approved by Institutional Review Boards at the participating institutions, and all patients provided signed informed consents. Biopsies were stained with hematoxylin/eosin to verify the presence of tumor. ErbB1 immunostaining was performed by using the EGFR (ErbB1) PharmDx kit from DAKO. Anti-ERα (1:200), PR (1:200), and bcl-2 (1:50) antibodies were purchased from DAKO. Anti-ErbB2 (1:80) and FKHRL1/FOXO3a (1:500) were purchased from NovoCastra (Newcastle, U.K.) and Cell Signaling Technology, respectively. ER, PR, and ErbB2 were processed with antigen retrieval by using citrate buffer, pH 6 (DAKO), and bcl-2 and FOXO3a were processed with antigen retrieval by using EDTA buffer, pH 9.0 (DAKO) in the “decloaker” (Biocare, Birmingham, U.K.). All markers were immunostained by using the Autostainer (DAKO). Envision plus dual-link polyper–horseradish peroxidase (DAKO) was used as the detection chemistry, and DAB+ (DAKO) was used as the chromagen. After immunostaining, slides were counterstained manually with methyl green (DAKO).
Cell Cycle Analysis.
Cells were harvested and fixed in 70% methanol in PBS. The pellets were then resuspended in 0.5 ml of PBS containing 50 μg/ml propidium iodide (Molecular Probes) and 100 μg/ml DNase-free RNase (Sigma). Cell cycle analysis was performed by using a FACSCalibur (Becton Dickinson). Data from 20,000 events were collected and analyzed.
Cell Viability Assay.
Cells were treated (see Figs. 6 and 7 for treatment conditions) in six-well plates. After extensive washing with PBS, cells were stained with 1% methylene blue in 50 ml of methanol for 3–5 minutes. After washing, cells with distilled water ×3, and the plates were air-dried and photographed.
Supplementary Material
Acknowledgments
We thank Drs. Allen Oliff, Pearl Huang, and Christine Debouck for helpful suggestions and Janice Spohn for technical expertise.
Abbreviations
- ER
estrogen receptor
- PR
progesterone receptor
- rBT474
lapatinib-resistant BT474 cells.
Footnotes
Conflict of interest statement: W.X., S.B., P.H., I.H., J.S., L. Liu, G.P., J. Harris, and N.L.S. are employees of GlaxoSmithKline.
References
- 1.Slamon D. J., Clark G. M., Wong S. G., Levin W. J., Ullrich A., McGuire W. L. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Yu D., Hung M.-C. Oncogene. 2000;19:6115–6121. doi: 10.1038/sj.onc.1203972. [DOI] [PubMed] [Google Scholar]
- 3.Hortobagyi G. N., Hung M. C., Buzdar A. U. Semin. Oncol. 1999;26:11–30. [PubMed] [Google Scholar]
- 4.Cobleigh M. A., Vogel C. L., Tripathy D., Robert N. J., Scholl S., Fehrenbacker L, Wolter J. M., Paton V., Shak S., Lieberman G., et al. J. Clin. Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 5.Cockerill S., Stubberfield C., Stables J., Carter M., Guntrip S., Smith K., McKeown S., Shaw R., Topley P., Thomsen L., et al. Bioorganic Med. Chem. Lett. 2001;11:1401–1405. doi: 10.1016/s0960-894x(01)00219-0. [DOI] [PubMed] [Google Scholar]
- 6.Spector N. L., Xia W., Burris H., Hurwitz H., Dees C. E., Dowlati A., O'Neil B., Overmoyer B., Marcom P. K., Blackwell K. L., et al. J. Clin. Oncol. 2005;11:2502–2512. doi: 10.1200/JCO.2005.12.157. [DOI] [PubMed] [Google Scholar]
- 7.Xia W., Mullin R. J., Keith B. R., Liu L.-H., Ma H., Rusnak D. W., Owens G., Alligood K. J., Spector N. L. Oncogene. 2002;21:6255–6263. doi: 10.1038/sj.onc.1205794. [DOI] [PubMed] [Google Scholar]
- 8.Xia W., Liu L.-H., Ho P., Spector N. L. Oncogene. 2004;23:646–653. doi: 10.1038/sj.onc.1207166. [DOI] [PubMed] [Google Scholar]
- 9.Konecny G. E., Pegram M. D., Venkatesan N., Finn R., Yang G., Rahmeh M., Untch M., Rusnak D. W., Sephar G., Mullin R. J., et al. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 10.Bacus S. S., Altomare D. A., Lyass L., Chin D. M., Farell M. P., Gurova K., Gudkov A., Testa J. R. Oncogene. 2002;21:3532–3540. doi: 10.1038/sj.onc.1205438. [DOI] [PubMed] [Google Scholar]
- 11.Zhou B. P., Liao Y., Xia W., Zou Y., Spohn B., Hung M.-C. Nat. Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 12.Zhou B. P., Liao Y., Xia W., Spohn B., Lee M. H., Hung M.-C. Nat. Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 13.Zhou B. P., Hu M. C., Miller S. A., Yu Z., Xia W., Lin S. Y., Hung M.-C. J. Biol. Chem. 2001;275:8027–8031. doi: 10.1074/jbc.275.11.8027. [DOI] [PubMed] [Google Scholar]
- 14.Xia W., Gerard C., Lui L., Baudson N., Ory T., Spector N. L. Oncogene. 2005;24:6213–6221. doi: 10.1038/sj.onc.1208774. [DOI] [PubMed] [Google Scholar]
- 15.Xia W., Bisi J., Strum J., Liu L., Carrick K., Graham K. L., Treece A. L., Hardwicke M. A., Liao Q., Westlund R. E., et al. Cancer Res. 2006;66:1640–1647. doi: 10.1158/0008-5472.CAN-05-2000. [DOI] [PubMed] [Google Scholar]
- 16.Ambrosini G., Adida C., Altieri D. C. Nat. Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 17.Li F., Ackermann E. J., Bennett F., Rothermel A. L., Plescia J., Tognin S., Villa A., Marchisio P. C., Altieri D. C. Nat. Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- 18.Mangelsdorf D. J., Thummel C., Besto M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborne C. K. Breast Cancer Res. Treat. 1998;51:1344–1359. doi: 10.1023/a:1006132427948. [DOI] [PubMed] [Google Scholar]
- 20.McCarty K. S., Jr., Barton T. K., Fetter B. F., Woodard B. H., Mossler J. A., Reeves W., Daly J., Wilkinson W. E., McCarty K. S., Sr. Cancer. 1980;46:2851–2858. doi: 10.1002/1097-0142(19801215)46:12+<2851::aid-cncr2820461424>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 21.Vollenwieder-Zerargui I., Barrelet I., Wong Y., Lemarchand-Beraud I., Gomez F. Cancer. 1986;57:1171–1180. doi: 10.1002/1097-0142(19860315)57:6<1171::aid-cncr2820570618>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 22.Tsui E. M., Wang S. C., Lee J. N., Hung M. C. Cancer Res. 2001;61:8390–8392. [PubMed] [Google Scholar]
- 23.Yu X., Rajala R. V., McGinnis J. F., Li F., Anderson R. E., Yan X., Li S., Elias R. V., Knapp R. R., Cao W. J. Biol. Chem. 2004;279:13086–13094. doi: 10.1074/jbc.M313283200. [DOI] [PubMed] [Google Scholar]
- 24.Keen J. C., Garrett-Mayer E., Pettit C., Mack K. M., Manning J., Herman J. G., Davidson N. E. Cancer Biol. Ther. 2004;3:1304–1312. doi: 10.4161/cbt.3.12.1458. [DOI] [PubMed] [Google Scholar]
- 25.Guo S., Sonenshein G. Mol. Cell. Biol. 2004;24:8681–8690. doi: 10.1128/MCB.24.19.8681-8690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cahill C. M., Tzivion G., Nusrin N., Ogg S., Dore J., Ruvkun G., Alexander-Bridges M. J. Biol. Chem. 2001;276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- 27.Burris H., Dees C., Hurwitz H., Dowlati A., Blackwell K., Marcom K., Overmoyer B., Smith D., Koch K., Stead A., et al. J. Clin. Oncol. 2005;23:5305–5313. doi: 10.1200/JCO.2005.16.584. [DOI] [PubMed] [Google Scholar]
- 28.Peles E., Ben-Levy R., Tzahar E., Liu N., Wen D., Yarden Y. EMBO J. 1993;12:961–971. doi: 10.1002/j.1460-2075.1993.tb05737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smart E. J., Graf G. A., McNiven M. A., Sessa W. C., Engelman J. A., Scherer P. E., Okamoto T., Lisanti M. P. Mol. Cell. Biol. 1999;19:7289–7304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlegel A., Wang C., Pestell R. G., Lisanti M. Biochem. J. 2001;359:203–210. doi: 10.1042/0264-6021:3590203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Heuvel A. P. J., Schulze A., Burgering M. T. Biochem. J. 2005;385:795–802. doi: 10.1042/BJ20041449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Zi X., Zhao Y., Mascarenhus D., Pollak M. J. Natl. Cancer Inst. 2001;93:1852–1857. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- 33.Smith B. L., Chin D., Maltzman W., Crosby K., Hortobagyi G. N., Bacus S. S. Br. J. Cancer. 2004;91:1190–1194. doi: 10.1038/sj.bjc.6602090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nahta R., Yuan L. X., Zhang B., Kobayashi R., Esteva F. J. Cancer Res. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 35.Nagata Y., Hsueh-Keng L., Zhou X., Tan M., Esteva F. J., Sahin A. A., Klos K. S., Li P., Monia B. P., Nguyen N. T., et al. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Shou J., Massarweh S., Osborne C. K., Wakeling A. E., Ali S., Weiss H., Schiff R. J. Natl. Cancer Inst. 2004;96:926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 37.Britton D. J., Hutcheson I. R., Knowlden J. M., Barrow D., Giles M., McClelland R. A., Gee J. M., Nicholson R. I. Breast Cancer Res. Treat. 2005;27:1–16. doi: 10.1007/s10549-005-9070-2. [DOI] [PubMed] [Google Scholar]
- 38.Osborne C. K., Bardon V., Hopp T. A., Chamness G. C., Hilsenbeck S. G., Fuqua S. A. W., Wong J., Allred D. C., Clark G. M., Schiff R. J. Natl. Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 39.Smith C. I., Nawaz Z., O'Malley B. W. Mol. Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 40.Arpino G., Green S. J., Allred D. C., Lew D., Martino S., Osborne C. K., Elledge R. M. Clin. Cancer Res. 2004;10:5670–5676. doi: 10.1158/1078-0432.CCR-04-0110. [DOI] [PubMed] [Google Scholar]
- 41.Rajagopalan D, Agarwal P. Bioinformatics. 2005;21:788–793. doi: 10.1093/bioinformatics/bti069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.