Abstract
To investigate multitissue engraftment of human primitive hematopoietic cells and their differentiation in goats, human CD34+Lin− cord blood cells transduced with a GFP vector were transplanted into fetal goats at 45–55 days of gestation. GFP+ cells were detected in hematopoietic and nonhematopoietic organs including blood, bone marrow, spleen, liver, kidney, muscle, lung, and heart of the recipient goats (1.2–36% of all cells examined). We identified human β2 microglobulin-positive cells in multiple tissues. GFP+ cells sorted from the perfused liver of a transplant goat showed human insulin-like growth factor 1 gene sequences, indicating that the engrafted GFP+ cells were of human origin. A substantial fraction of cells engrafted in goat livers expressed the human hepatocyte-specific antigen, proliferating cell nuclear antigen, albumin, hepatocyte nuclear factor, and GFP. DNA content analysis showed no evidence for cellular fusion. Long-term engraftment of GFP+ cells could be detected in the blood of goats for up to 2 yr. Microarray analysis indicated that human genes from a variety of functional categories were expressed in chimeric livers and blood. The human/goat xenotransplant model provides a unique system to study the kinetics of hematopoietic stem cell engraftment, gene expression, and possible stem cell plasticity under noninjured conditions.
Keywords: hematopoietic stem cell, transplantation, plasticity, microarray
Hematopoietic stem cell (HSC) transplantation can compensate for tissue damage elicited by a wide variety of disorders, including malignant and/or inherited diseases (1–5). However, broader clinical application is still limited because of a number of biological and technical problems. For example, allogeneic HSC transplantation may require immunosuppressive treatment to prevent engraftment failure, increasing the risk of life-threatening infection. One new approach is to generate chimeras via in utero transplantation using allogeneic or xenogeneic HSCs (6, 7). The fetus is incapable of rejecting transplanted allogeneic cells because of its immunological incompetence or tolerance of non-self antigens. Thus, the need for immunosuppression and myeloablation used for postnatal transplantation can be avoided. Allogeneic and xenogeneic chimerism has been generated by in utero transplantation procedures, and fetal engraftment of allogeneic and xenogeneic HSC has been tested in mouse, sheep, and monkey and more recently in pigs and goats (8–12). Questions remain regarding the engraftment, homing, and differentiation of allogeneic or xenogeneic HSC, as well as the gene expression of the engrafted cells in different tissues of the recipients. There is also the concern that transplant and host cells may fuse, producing a significant population of undesirably altered or pathogenic hybrid cells. Mouse models were recently developed by using retroviral transduction with an MSCV-IRES-GFP (MIG) vector to study the expansion of adult HSCs and engraftment of lymphoid-myeloid cells from ES cells (13, 14). The resulting GFP+ cells are easily detected and can be directly identified in various organs as cells of human origin in the transplant animals. In the present study we generated human/goat xenogeneic chimeras transplanted with human CD34+Lin−GFP+ cord blood (CB) cells.
Results
Engraftment of GFP+ Cells in Multiple Organs of the Recipient Goats.
Five of 14 recipient goats transplanted with MIG-transduced human CB CD34+Lin− cells were live-born. Engraftment of GFP+ human cells ranged from 1.5% to 4% in blood of all MIG-transplant goats up to 2 yr after birth (Table 1). The engrafted cells specifically expressed surface markers of human myeloid, B- and T-lymphoid, and erythroid lineage cells at 3, 6, 12, and 24 mo of age, indicating the long-term engraftment and slow expansion of primitive human CB cells in this xenograft model.
Table 1.
Percentage of human marker-positive cells detected in blood from individual MIG-transplant goats
| Marker | MIG-1, 3 mo | MIG-2, 3 mo | MIG-3 |
MIG-4 |
MIG-5 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 mo | 6 mo | 1 yr | 2 yr | 3 mo | 6 mo | 1 yr | 2 yr | 3 mo | 6 mo | 1 yr | 2 yr | |||
| GFP | 2.1 | 3.7 | 4.1 | 6.3 | 1.9 | 1.1 | 3.7 | 3.6 | 2.1 | 1.7 | 2.5 | 1.7 | 1.7 | 1.4 |
| CD34 | 0.5 | 0.5 | 0.6 | 1.6 | 0.7 | 0.5 | 1.4 | 1.5 | 1.3 | 1.4 | 2.1 | 1.5 | 1.3 | 1.3 |
| GPA | 3.3 | 7.4 | 6.7 | 9.7 | 3.8 | 3.8 | 6.6 | 3.4 | 2.1 | 1.4 | 9.9 | 4.7 | 4.1 | 6.6 |
| CD14 | 0.4 | 0.8 | 1.7 | 4.8 | 0.6 | 0.9 | 5 | 7.9 | 7 | 7.2 | 1.9 | 1.5 | 1.1 | 1.1 |
| CD20 | 0.1 | 1.2 | 1 | 1.1 | 0.4 | 0.4 | 8.5 | 1.1 | 0.8 | 0.8 | 1.9 | 1 | 0.5 | 0.4 |
| CD15 | 0.3 | 0.2 | 0.5 | 0.6 | 0.4 | 0.4 | 0.3 | 0.3 | 0.2 | 0.2 | 0.6 | 0.7 | 0.6 | 0.4 |
| CD7 | 0.4 | 0.6 | 1 | 1.4 | 0.9 | 1.4 | 1.6 | 1.9 | 1.1 | 1.2 | 0.6 | 2 | 0.5 | 0.5 |
| CD45 | 0.3 | 0.2 | 0.4 | 0.6 | 0.4 | 0.3 | 0.7 | 0.8 | 0.8 | 0.6 | 0.6 | 0.6 | 0.5 | 0.3 |
MIG-1 and MIG-2 goats were dissected at the age of 3 mo. No human marker-positive cells were detected in normal goats of the same age.
To determine the tissue distribution of engrafted cells, goats MIG-1 and MIG-2 were examined 3 mo after birth. Kidney, muscle, liver, spleen, heart, and lung sections were examined by fluorescence microscopy. As shown in Fig. 1A, a large number of GFP+ cells were observed in various tissues. This and all subsequent tissue sections represent regions containing the highest GFP+ densities observed; GFP+ cells were unevenly distributed in all tissue types tested (see Fig. 2D for an example of similar uneven distribution at lower magnification). There was no fluorescence signal in tissues of the normal goats. Distributions of engrafted human GFP+ cells were further measured by FACS analysis. Recipient livers contained the highest number of grafted cells (>27%) among all tissues. GFP+ cells could also be found in kidney, muscle, lung, and heart and comprised 1.2–36% of total cells examined (Fig. 1B). In perfused liver from goat MIG-3, the number of GFP+ cells remained high (37%) after 2 yr, indicating that the long-term engrafted cells detected in the liver were not due to contamination by peripheral blood or other circulating cells (Fig. 1C; and see supporting information, which is published on the PNAS web site).
Fig. 1.
Detection of human GFP+ cells in various tissues of the MIG goats. (A Upper) Fluorescence emission and hematoxylin/eosin (HE) staining in tissue sections of a representative goat transplanted with MIG-GFP-transduced CD34+Lin− CB cells. (Magnification: ×400.) (A Lower) Tissue sections were prepared from a normal (negative control) goat. (Magnification: ×400.) (B) GFP+ human cells were detected by FACS analysis in hematopoietic and nonhematopoietic organs of the recipient goats (MIG-1 and MIG-2). The GFP+ cells comprised a wide range (1.2–36%) of the examined cell populations. (C) FACS analysis of GFP+ cells from the perfused liver of goat MIG-3 2 yr after birth. The histogram shows number of cells vs. GFP fluorescence units.
Fig. 2.
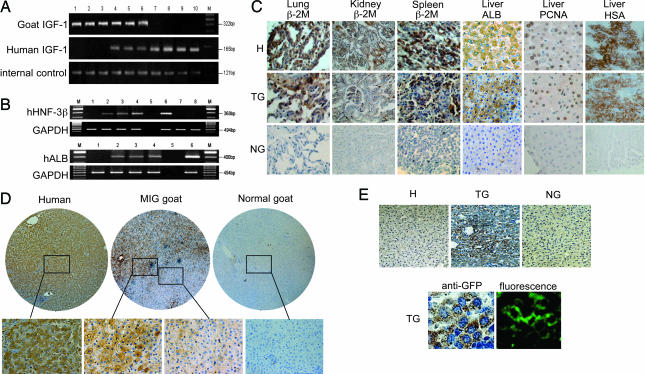
Identification of specific human genes in MIG-transplant goats. (A) IGF-1 genes were detected in MIG-transplant goats and sorted GFP+ cells by PCR analysis using primer sets specific for unique human or goat sequences, or a shared sequence (internal control). M, molecular weight DNA marker. Lanes 1–3, amplicons from liver DNA samples from three different normal goats. Lanes 4–6, liver DNA samples from three different MIG goats. Lane 7, the liver DNA sample from sorted GFP+ cells. Lanes 8–10, liver DNA samples from three humans. (B) RT-PCR analysis of human gene transcripts for hepatocyte nuclear factor 3β and serum albumin (hALB) expressed in the transplant liver tissue but not in the control goats. GAPDH is used as internal control. Lane 1, normal goat RNA. Lanes 2–4, RNA from three MIG goats. Lane 5, blank. Lane 6, positive control from human liver. Lanes 7 and 8, RNA from human CB cells. (C) Immunohistochemistry analysis for human β2 microglobulin antigen, hALB, proliferating cell nuclear antigen, and hepatocyte-specific antigen; brown staining shows positive cells in various tissues of transplant goat (TG) and human (H) samples. No positive cells are found in control goats (NG). (Magnification: ×400.) (D) Staining for hALB was performed on sections of human, MIG-transplant goat, and normal goat livers and is shown at ×50 and ×200 magnification. Although the human tissue is uniformly positive and normal goat is entirely negative, the chimeric liver contains regions of high staining density surrounded by nonstaining cells. The margin of one such region is shown with further magnification of adjacent positive and negative areas. (E Upper) Anti-GFP staining (brown) was present in the liver cells (cytoplasm) of MIG-transplant goat (TG) but not in human or normal goat (NG). (Magnification: ×100.) (E Lower) GFP+ cells in the liver of TG with immunohistochemistry staining, and corresponding images with fluorescence emission. (Magnification: ×400.)
Detection of Human Genomic DNA in GFP+ Cells from Transplant Goats.
GFP+ cells were sorted from the perfused liver of the MIG-transplant goat; enrichment was ≈98% (Fig. 3A). Standard FACS cell-cycle analyses were performed to measure the DNA content of cells from perfused human, normal goat, and MIG-transplant goat livers. Diploid human cells containing 46 chromosomes can be easily distinguished from normal goat cells with 2n = 60 (Fig. 3B a and b). As shown in Fig. 3Bc, chimeric liver produced two peaks representing both human and goat cell populations. Sorted GFP+ cells were highly enriched for human diploid DNA content, with a small shoulder to the right of the main peak, which likely results from the <2% contaminating normal goat cells (Fig. 3Bd).
Fig. 3.
Detection of human DNA in MIG-transplant goat. (A Top) FACS of samples from perfused liver to enrich for the GFP population. The resulting cell pool was reanalyzed by FACS to assess enrichment, producing the cytogram of cell counts vs. fluorescence. (A Middle and Bottom) Individual cells from the sorted pool were observed under light (Middle) and fluorescence (Bottom) microscopy and compared to normal goat liver cells. (B) FACS to measure DNA content discriminated goat and human cells by total chromosome number. DNA content is shown for perfused human liver (a), normal goat liver (b), MIG goat liver (c), and sorted GFP+ cells from perfused MIG goat liver (d). H, human, 2n = 46 chromosomes; G, goat, 2n = 60 chromosomes.
To complement the total DNA content analysis, individual loci were assayed to confirm the presence of human sequences in GFP+ cells. PCR primer sets were designed to amplify the insulin-like growth factor 1 (IGF-1) gene from human, goat, or both species. Human and goat IGF-1 sequences were concurrently amplified from liver DNA of three MIG goats (Fig. 2A), and the FACS-sorted GFP+ cell population (see lane 7) produced predominantly human IGF-1 amplicons with a minor amount of goat PCR product (again likely because of contaminating cells). Sequence analysis of the amplified products confirmed these results.
Human RNA and Protein Expression in Multiple Hematopoietic and Nonhematopoietic Organs.
RT-PCR analysis revealed the presence of human hepatocyte nuclear factor 3β and human serum albumin (hALB) mRNA in livers of MIG-transplant goats, and no such transcripts were detected in the liver of normal goats or human blood cells (Fig. 2B).
Tissue-specific protein expression was detected by immunohistochemistry in lung, kidney, spleen, and liver (Fig. 2C). Cells expressing human β2 microglobulin can be found in the kidney, lung, and spleen of the transplant goats as well as in humans, but not in control goats. hALB, human hepatocyte-specific antigen, and human proliferating cell nuclear antigen-positive cells were found in the chimeric liver, but not in normal goat. As stated previously, human-like cells were distributed unevenly in the various chimeric organs, with patches of high-density engraftment surrounded by normal goat cells (Fig. 2D). GFP+ cells were also detected in the MIG-transplant goats but not in control goats using anti-GFP, which provides better cell resolution than GFP fluorescence detection (Fig. 2E).
Gene Expression Profile from Blood and Liver RNA.
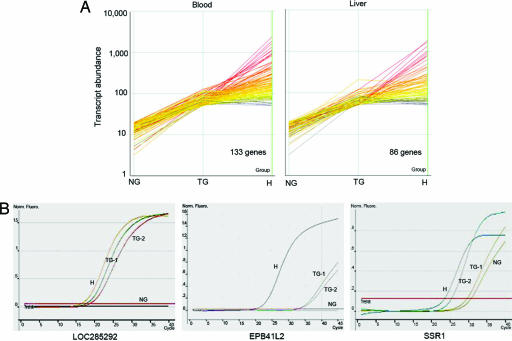
To provide systematic evidence of human gene expression, a microarray analysis was performed. The RNA expression data were filtered to identify human gene probes that hybridized to very little or no RNA from normal goat but detected significantly more hybridization in transplant goat samples. A total of 133 human transcripts were specifically detected in RNA from blood samples, and 86 were detected in liver samples of the transplant goats (Fig. 4A). Interestingly, only five of the detected mRNAs were coexpressed in both liver and blood, indicating that the engrafted human cells express distinct patterns of genes in different tissues. A subset of these human genes showed particularly consistent signals markedly higher than the nontransplant background (Table 2). Each goat is considered a separate transplantation event, so variance between transplant individuals is expected. For example, the KIAA0494 gene was detected with a 7-fold greater signal in TG-1 livers and a 3-fold greater signal in TG-2 livers compared with background signal in normal goat liver; in blood, however, it was not present in TG-1 and TG-2 yet was highly detected in TG-3. Eighteen such genes displayed tissue specificity; the five human transcripts detected in both transplant liver and blood included signal recognition particle (9 kDa), transmembrane protein 14B, histamine N-methyltransferase, KIAA0494, and SSR1. Three candidates were tested by conventional and quantitative RT-PCR (Fig. 4B) for verification. Specific detection of human RNAs, without amplification of goat RNA, was confirmed by sequencing of the PCR products. The results were in agreement with the microarray analysis, confirming expression in both blood and liver of the transplant goats.
Fig. 4.
Gene expression analysis from human cells in transplant goats. (A) Microarray transcript profiles are shown for selected human genes. The panels plot the expression levels of detected human transcripts in normal goats (NG), transplant goats (TG), and human (H) with low or no detection in normal goats and at least 2.5-fold higher expression in transplant goats. (B) Real-time quantitative RT-PCR confirmed expression levels of three candidate genes from the microarray profiles. LOC285292 was assayed from blood samples, and EPB41L2 and SSR1 were assayed from liver samples. GAPDH cDNA is used as an internal control.
Table 2.
RNA expression levels for selected human genes
| Gene name | GenBank accession no. | Liver |
Blood |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NG | TG1 | TG2 | H | NG | TG1 | TG2 | TG3 | H | ||
| Shared | ||||||||||
| Signal recognition particle 9 kDa | NM_003133 | 19 ± 8 | 44 ± 3 | 64 ± 22 | 867 ± 146 | 6 ± 5 | 52 ± 81 | 37 ± 36 | 574 | 1,120 ± 108 |
| Transmembrane protein 14B | NM_030969 | 6 ± 5 | 137 ± 131 | 55 ± 5 | 260 ± 48 | 28 ± 27 | 53 ± 14 | 57 ± 16 | 257 | 259 ± 76 |
| Histamine N-methyltransferase | BC005907.1 | 14 ± 2 | 94 ± 19 | 43 ± 1 | 239 ± 135 | 17 ± 3 | 71 ± 39 | 85 ± 56 | 51 | 121 ± 17 |
| KIAA0494 gene product | AK001487.1 | 19 ± 13 | 137 ± 1 | 54 ± 11 | 128 ± 6 | 17 ± 3 | 78 ± 76 | 19 ± 1 | 116 | 257 ± 49 |
| SSR1 signal sequence receptor α | AI016620 | 16 ± 8 | 213 ± 183 | 39 ± 11 | 101 ± 13 | 19 ± 15 | 56 ± 31 | 49 ± 39 | 178 | 116 ± 24 |
| Liver-specific | ||||||||||
| Solute carrier family 16 | NM_004731 | 9 ± 0 | 81 ± 78 | 93 ± 33 | 195 ± 32 | |||||
| Microtubule-associated protein 7 | T62571 | 11 ± 8 | 87 ± 38 | 52 ± 53 | 169 ± 21 | |||||
| Erythrocyte membrane protein band 4.1-like 2 | NM_001431 | 8 ± 3 | 122 ± 162 | 89 ± 60 | 168 ± 62 | |||||
| Septin10 | BF981643 | 10 ± 1 | 52 ± 14 | 54 ± 17 | 89 ± 42 | |||||
| Hypothetical protein | NM_024510 | 9 ± 3 | 136 ± 159 | 67 ± 69 | 78 ± 27 | |||||
| Hypothetical protein | BF219234 | 7 ± 4 | 70 ± 42 | 80 ± 63 | 66 ± 12 | |||||
| Glycoprotein VI (platelet) | AB043821.1 | 10 ± 2 | 123 ± 151 | 64 ± 6 | 64 ± 17 | |||||
| Hypothetical protein | NM_017792 | 12 ± 9 | 236 ± 171 | 31 ± 12 | 62 ± 6 | |||||
| cAMP-regulated guanine nucleotide exchange factor | NM_007023 | 3 ± 0 | 78 ± 25 | 59 ± 37 | 53 ± 13 | |||||
| Glioblastoma amplified sequence | NM_001483 | 9 ± 1 | 136 ± 151 | 70 ± 37 | 51 ± 9 | |||||
| Blood-specific | ||||||||||
| Dicer1 homolog (Drosophila) | BG109746 | 7 ± 5 | 86 ± 65 | 56 ± 9 | 24 | 421 ± 34 | ||||
| Similar to heterogeneous nuclear ribonucleoprotein A3 (hnRNPA3) (LOC285292) | BE867771 | 6 ± 4 | 57 ± 72 | 47 ± 52 | 465 | 322 ± 96 | ||||
| Phosphatidylinositol binding clathrin assembly protein | AL135735 | 3 ± 1 | 42 ± 38 | 34 ± 7 | 212 | 225 ± 76 | ||||
| Estrogen-related receptor β-like 1 | NM_018010 | 8 ± 7 | 91 ± 72 | 33 ± 21 | 71 | 82 ± 3 | ||||
| Hypothetical protein | AI809961 | 7 ± 7 | 24 ± 9 | 88 ± 34 | 102 | 89 ± 48 | ||||
| Integrin β5 | AI335208 | 4 ± 0 | 49 ± 39 | 25 ± 11 | 253 | 83 ± 24 | ||||
| 5-Azacytidine-induced gene 2 | NM_022461 | 6 ± 6 | 83 ± 71 | 21 ± 22 | 535 | 79 ± 30 | ||||
| Cofactor required for Sp1 activation subunit 2 | AI971089 | 5 ± 1 | 61 ± 94 | 18 ± 19 | 396 | 51 ± 2 | ||||
Affymetrix signal levels are shown for human genes that were consistently detected in blood or liver from at least one transplant goat. Data columns for normal goats (NG), transplant goats (TG), and normal human (H) are shown with the mean and standard deviation of three replicate GeneChip assays (except for TG3, where only one sample was assayed). Genes that showed specific and consistent expression (low standard deviation for the individual mean) in at least one transplant goat are listed. Additional annotations are available by using the GenBank accession number (www.ncbi.nlm.nih.gov/entrez). Genes confirmed in Fig. 4 are indicated in bold.
Discussion
To investigate whether human HSCs can survive and differentiate in various tissues and organs of in utero transplantation goats, this study used a retroviral vector to deliver the GFP transgene into human CB CD34+Lin− cells. Although the number of GFP+ cells detected in the recipients' blood was relatively low, examination of various organs demonstrated that GFP+ cells were present in liver, spleen, kidney, lung, heart, and skeletal muscle. These GFP+ cells comprised 1.2–36% of the total cells examined by FACS analysis. This finding implies that the transplanted human primitive CB cells can survive and engraft in the recipient goats. A large number of GFP+ cells were detected in a liver perfusion experiment, which avoids GFP signal from contaminating non-liver cells/factors in the circulation system. Immunohistochemical assays and Southern blot analysis (supporting information) further support that human donor cells were seeded and survived in multiple tissues. The GFP+ cells were isolated from perfused chimeric liver by FACS, and DNA content analysis showed that this enriched population is composed mainly of human cells. Human-specific but not goat-specific IGF-1 gene sequences were identified in FACS-enriched GFP+ cells from perfused chimeric liver. This finding further demonstrated that the GFP+ cells engrafted in MIG-transplant goats were of human origin.
Higher levels of engraftment occurred in solid organs compared with blood. Unlike previous transplantation systems that delivered stem cells by i.v. injection into irradiated recipients, in utero transplantation was used to inject CD34+Lin− CB cells into the abdominal cavities of the fetal goats at a preimmune stage. We hypothesize that the transplanted human hematopoietic cells undergo adaptive processes for survival in recipient goats triggered by the local host microenvironment. Despite their CB origin, these stem cells apparently adapt to goat microenvironments through mechanisms that are more efficient for expansion in other tissues, such as liver and kidney, rather than blood. Even within a single organ, engrafted cells were unevenly distributed. It would be interesting to investigate whether this observation reflects the distribution pattern of progenitor cells, the existence of microenvironment niches favorable for engraftment, or other mechanisms. Transgenic tagging and lineage tracking experiments could be performed to determine whether cells within engraftment foci are clonal, indicating they arose from a single xenotransplant progenitor cell. Alternatively, many progenitor cells may populate a fetal organ but only the subset engrafted within a favorable niche survive.
Several lines of evidence have shown that mammalian adult stem cells display “plasticity,” the ability to transdifferentiate into other tissue types (15–18), although the mechanism remains controversial (19). It was hypothesized that stem cell plasticity may result from a cell fusion mechanism (20–22). Recent reports also show that adult stem cells, including HSCs, are able to give rise to nonhematopoietic cells independent of a cell fusion mechanism (23–26). All of these studies transferred adult stem cells into adult recipients, which may lack certain developmental cues that exist only in embryonic/fetal stages. Whether the CD34+Lin− cells derived from CB have the potential to transdifferentiate into other types of nonhematopoietic tissues in the fetal stage remains largely unexplored. Wang et al. (27) reported that human albumin-expressing hepatocyte-like cells can be developed in the livers of immune-deficient and CCl4-injured mice that received transplants of human HSC, suggesting that human “hematopoietic” stem/progenitor cells have the capacity to respond to the injured liver microenvironment by inducing albumin. A more recent study reported that human HSCs can efficiently (20%) generate functional hepatic cells in the human/sheep model (28). In addition, Sato et al. (29) observed that mesenchymal stem cells prepared from human bone marrow could be directly engrafted in allylalcohol-treated rat liver without apparent cell fusion. We show that the engrafted CD34+Lin− cells have a great plasticity in goats because we detected human-like cells in various tissues where they express nonhematopoietic tissue-specific markers, including hALB, hepatocyte-specific antigen, and hepatocyte nuclear factor 3β in goat liver. There was no evidence that this plasticity was a result of cell fusion. FACS analysis was able to discriminate human and goat cells by DNA content, and GFP+ cells clearly showed normal human ploidy levels. Cell fusion typically produces a variety of cells containing portions or full complements of both precursor genomes, and this fusion results in multiple new peaks when analyzed by FACS for DNA content. No peaks other than those for normal goat or human diploid cells were detected. These results support the notion of clinically meaningful transdifferentiation of human hematopoietic cells under appropriate cues and without requiring organ damage. Thus, the present study offers an in vivo model system for exploring the differentiation potential of human adult cells.
The human gene expression profile in the transplant goats was investigated by microarray analysis. RNA transcript profiling was performed for two purposes: to identify additional human markers indicative of HSC survival and to determine whether the method may have future use for assaying the expression and regulation of human genes against a background of goat RNA. RNA markers are useful because RNAs are generally turned over much more quickly than are proteins and therefore indicate the presence of metabolically active human cells, and RNA expression can report on the activity of genes across a wide range of functions. Even though cross-hybridization between homologous goat and human gene sequences would occur, we expected that many probes to human transcripts would hybridize with more affinity to human than goat targets and would have sufficiently higher detection levels beyond the background of nonspecific or crossreacting goat RNA. The results from comparative analyses of gene expression profiles in blood and liver samples of transplant goats, normal goats, and humans using Affymetrix GeneChips did reveal specific and detectable expression of human genes in the chimeras. Only five transcripts were coexpressed in both liver and blood. This finding suggests that the survival of human donor cells is associated with different expression profiles in different tissues. Transcript profiling on human cell populations enriched by cell sorting or laser capture microdissection from chimeric tissues should dramatically increase signal-to-noise ratios and provide detection of many more genes.
The human transcripts detected represent a variety of functional categories, including signal transduction, membrane proteins and receptors, and transcription factors. These results provide new and specific markers for detecting viable human HSC-derived cells. Short oligonucleotide GeneChip microarrays are capable of discriminating a subset of human transcripts against a background of goat RNA, providing a system for in-depth gene expression profiling and molecular analysis of the responses of human cells in goats.
Using DNA, RNA, and protein assays, the current study shows that a substantial fraction of human cells engraft in goat livers and other tissues and express human proteins. Thus, human/goat chimerism could potentially be used as a bioreactor to produce human proteins for therapeutic or other clinical uses. Xenogeneic chimerism may offer models to evaluate clinical potential for the prenatal treatment of a number of human genetic diseases, cell or tissue repair, and xenogeneic organ transplantation. Human/goat chimerism provides a unique system for studying immune tolerance as well as the kinetics of stem cell engraftment, homing, differentiation, gene expression, and possible plasticity under noninjured conditions.
Materials and Methods
Cell Enrichment, Transduction, and Injection.
CB cells were obtained from consenting mothers undergoing cesarean delivery of normal, full-term male and female infants. Low-density (<1.077 g/ml) cells were isolated by using Ficoll/Hypaque (Amersham Pharmacia Biotech), and a population of enriched cells (82 ± 3% CD34+) was obtained by immunomagnetic removal of lineage marker− (Lin−) cells (StemSep, StemCell Technologies). The monoclonal antibodies for removal of Lin− cells were anti-human CD2, CD3, CD14, CD16, CD19, CD24, CD56, CD66b, and glycophorin A. An MIG vector that contains the internal ribosome entry site and the GFP under the control of the murine stem cell virus LTR was used to introduce GFP into the CD34+Lin− cells. Transduction efficiency was 28 ± 5% (n = 3), which corresponds to the proportion of CD34+Lin− cells after transduction (25 ± 5%). Fourteen recipient goats were obtained from the Experimental Animal Farm (Institute of Medical Genetics, Shanghai Children's Hospital) with approval from the Review Board of Shanghai Children's Hospital. Each fetal goat was injected with 5 × 104 MIG-transduced CD34+Lin− cells into the fetal peritoneal cavity (MIG goat) by using previously described methods (30). The same number of nontransduced CD34+Lin− cells was injected to separately generate “TG” transplant goats for microarray analysis.
GFP+ Cell Distributions.
The liver, kidney, lung, heart, muscle, and spleen were dissected from two goats (MIG-1 and MIG-2) 3 mo after birth, and the left lobe of MIG-3 liver was removed 2 yr after birth for perfusion. The tissue sections were stained with hematoxylin and eosin, and the GFP+ cells were examined under a fluorescence microscope. Suspensions of single cells were prepared as described (31). One hundred thousand cells from each sample were analyzed by flow cytometry (FACSCalibur and FACSVantage SE, Becton Dickinson).
Molecular Detection of Genes.
DNA and RNA were extracted from various tissues of the transplant goats, and GFP, human IGF-1, or goat IGF-1 DNA was detected by PCR and amplicon sequencing. RT-PCR and quantitative RT-PCR were performed on RNA samples to detect human hepatocyte nuclear factor 3β, hALB, and candidates from microarray profiling; these amplicons were also confirmed by sequencing. Immunohistochemistry assays were performed according to the method reported by Tian et al. (32) by using polyclonal antibodies against human β2 microglobulin antigen, hALB, and GFP, as well as monoclonal antibodies specific for proliferating cell nuclear antigen and hepatocyte-specific antigen.
DNA Content Measurements.
Samples were derived from the perfused livers of MIG goat, nontransplant goat, human, and sorted GFP+ cells from perfused liver. A total of 700 μl of cold ethanol was added dropwise to 1 × 106 cells in 300 μl of PBS while vortexing, then incubated on ice for 2 h. After washing with PBS, the cells were suspended in 1 ml of dye solution (PBS containing 20 μg of propidium iodide and 200 μg of DNase-free RNase) and incubated at 37°C for 30 min in the dark. The cellular DNA content was determined by flow cytometry cell-cycle analysis with modfit software. In detail, two dot plots, forward scatter vs. side scatter and FL2-W vs. FL2-A, were created. Gate R1 was drawn to enclose the majority of liver cells on forward scatter/side scatter and then reported on FL2-/FL2-A, whereas gate R2 was drawn around singlet cells (to exclude contamination by doublet or triplet cells). We defined gate G1 = R1 and R2 and show data for G1 on a histogram with FL2-A as the x axis. We used propidium iodide to stain DNA and the FL2 channel to detect propidium iodide. The more signal detected in FL2 channel, the greater the DNA content.
Gene Expression Profile Analysis Using Microarrays.
Total RNA was extracted by the TRIzol method from the blood and liver tissues of three transplant goats 6 mo after birth and submitted along with normal goat samples and human liver biopsies to the Penn Microarray Facility for target preparation and hybridization to human U133A GeneChips (Affymetrix) followed by microarray analysis as described previously (33). Triplicate RNA samples from each tissue were assayed. GeneChip tabular data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo), accession number GDS1023.
Additional details for all methods are provided in the supporting information.
Supplementary Material
Acknowledgments
We thank Dr. Yi-Tao Zeng for helpful discussion in conducting this research; Dr. C. P. Pang (Chinese University of Hong Kong) for technical advice in immunohistochemistry; and Dr. Ming-hui Zhang, Dr. Zhen Hong Guo, Dr. Rui Zhang, Mr. Lian Wang, and Ms. Ju Zhang for helpful technical assistance in sorting GFP+ cells. This work was supported by Chinese National “863” Research Program Grant 2002AA216091.
Abbreviations
- HSC
hematopoietic stem cell
- hALB
human serum albumin
- MIG
MSCV-IRES-GFP
- IGF-1
insulin-like growth factor 1
- CB
cord blood.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The data described in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GDS1023).
References
- 1.Elfenbein G. J., Sackstein R. Exp. Hematol. 2004;32:327–339. doi: 10.1016/j.exphem.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Hongeng S., Pakakasama S., Chaisiripoomkere W., Chuansumrit A., Sirachainan N., Ungkanont A., Jootar S. Bone Marrow Transplant. 2004;33:377–379. doi: 10.1038/sj.bmt.1704361. [DOI] [PubMed] [Google Scholar]
- 3.Orofino M. G., Argiolu F., Sanna M. A., Tuveri T., Scalas M. T., Badiali M., Cossu P., Puddu R., Lai M. E., Cao A. Lancet. 2003;362:41–42. doi: 10.1016/S0140-6736(03)13806-8. [DOI] [PubMed] [Google Scholar]
- 4.McDonough C. H., Jacobsohn D. A., Vogelsang G. B., Noga S. J., Chen A. R. Bone Marrow Transplant. 2003;31:1073–1080. doi: 10.1038/sj.bmt.1704071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckner C. D., Epstein R. B., Rudolph R. H., Clift R. A., Storb R., Thomas E. D. J. Hematother. Stem Cell Res. 2001;10:201–208. doi: 10.1089/15258160151134845. [DOI] [PubMed] [Google Scholar]
- 6.Zanjani E. D., Anderson W. F. Science. 1999;285:2084–2088. doi: 10.1126/science.285.5436.2084. [DOI] [PubMed] [Google Scholar]
- 7.Surbek D. V., Holzgreve W., Nicolaides K. H. Hum. Reprod. Update. 2001;7:85–91. doi: 10.1093/humupd/7.1.085. [DOI] [PubMed] [Google Scholar]
- 8.Zanjani E. D., Pallavicini M. G., Ascensao J. L., Flake A. W., Langlois R. G., Reitsma M., MacKintosh F. R., Stutes D., Harrison M. R., Tavassoli M. J. Clin. Invest. 1992;89:1178–1188. doi: 10.1172/JCI115701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison M. R., Slotnick R. N., Crombleholme T. M., Golbus M. S., Tarantal A. F., Zanjani E. D. Lancet. 1989;2:1425–1427. doi: 10.1016/s0140-6736(89)92036-9. [DOI] [PubMed] [Google Scholar]
- 10.Pixley J. S., Tavassoli M., Zanjani E. D., Shaft D. M., Futamachi K. J., Sauter T., Tavassoli A., MacKintosh F. R. Pathobiology. 1994;62:238–244. doi: 10.1159/000163916. [DOI] [PubMed] [Google Scholar]
- 11.Fujiki Y., Fukawa K., Kameyama K., Kudo O., Onodera M., Nakamura Y., Yagami K., Shiina Y., Hamada H., Shibuya A., et al. Transplantation. 2003;75:916–922. doi: 10.1097/01.TP.0000057243.12110.7C. [DOI] [PubMed] [Google Scholar]
- 12.Zeng F., Chen M. J., Katsumata M., Huang E. Y., Gong Z. J., Hu W., Qian H., Xiao Y. P., Ren Z. R., Huang S. Z. DNA Cell Biol. 2005;24:403–409. doi: 10.1089/dna.2005.24.403. [DOI] [PubMed] [Google Scholar]
- 13.Antonchuk J., Sauvageau G., Humphries P. K. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 14.Kyba M., Perlingeiro R. C., Daley G. O. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 15.Petersen B. E., Bowen W. C., Patrene K. D., Mars W. M., Sullivan A. K., Murase N., Boggs S. S., Greenberger J. S., Goff J. P. Science. 1999;284:1168–1170. doi: 10.1126/science.284.5417.1168. [DOI] [PubMed] [Google Scholar]
- 16.Krause D. S., Theise N. D., Collector M., Henegariu O., Hwang S., Gardner R., Neutzel S., Sharkis S. J. Cell. 2001;105:369–377. doi: 10.1016/s0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
- 17.Jackson K. A., Mi T., Goodell M. A. Proc. Natl. Acad. Sci. USA. 1999;96:14482–14486. doi: 10.1073/pnas.96.25.14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almeida-Porada G., El Shabrawy D., Porada C., Zanjani E. D. Exp. Hematol. 2002;30:1454–1462. doi: 10.1016/s0301-472x(02)00967-0. [DOI] [PubMed] [Google Scholar]
- 19.Dalakas E., Newsome P. N., Harrison D. J. FASEB J. 2005;19:1225–1231. doi: 10.1096/fj.04-2604rev. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Willenbring H., Akkari Y., Torimaru Y., Foster M., Al-Dhalimy M., Lagasse E., Finegold M., Olson S., Grompe M. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- 21.Vassilopoulos G., Wang P. R., Russell D. W. Nature. 2003;422:901–904. doi: 10.1038/nature01539. [DOI] [PubMed] [Google Scholar]
- 22.Pells S., Di Domenico A. I., Gallagher E. J., McWhir J. Cloning Stem Cells. 2002;4:331–338. doi: 10.1089/153623002321025005. [DOI] [PubMed] [Google Scholar]
- 23.Newsome P. N., Johannessen I., Boyle S., Dalakas E., McAulay K. A., Samuel K., Rae F., Forrester L., Turner M. L., Hayes P. C., et al. Gastroenterology. 2003;124:1891–1900. doi: 10.1016/s0016-5085(03)00401-3. [DOI] [PubMed] [Google Scholar]
- 24.Jang Y. Y., Collector M. I., Baylin S. B., Diehl A. M., Sharkis S. J. Nat. Cell Biol. 2004;6:532–539. doi: 10.1038/ncb1132. [DOI] [PubMed] [Google Scholar]
- 25.Harris R. G., Herzog E. L., Bruscia E. M., Grove J. E., Van Arnam J. S., Krause D. S. Science. 2004;305:90–93. doi: 10.1126/science.1098925. [DOI] [PubMed] [Google Scholar]
- 26.Wurmser A. E., Nakashima K., Summers R. G., Toni N., D'Amour K. A., Lie D. C., Gage F. H. Nature. 2004;430:350–356. doi: 10.1038/nature02604. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Ge S., McNamara G., Hao Q. L., Crooks G. M., Nolta J. A. Blood. 2003;101:4201–4208. doi: 10.1182/blood-2002-05-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almeida-Porada G., Porada C. D., Chamberlain J., Torabi A., Zanjani E. D. Blood. 2004;104:2582–2590. doi: 10.1182/blood-2004-01-0259. [DOI] [PubMed] [Google Scholar]
- 29.Sato Y., Araki H., Kato J. Blood. 2005;106:756–763. doi: 10.1182/blood-2005-02-0572. [DOI] [PubMed] [Google Scholar]
- 30.Zeng F., Chen M. J., Huang W. Y., Yan J. B., Xiao Y. P., Gong Z. J., Ren Z. R., Huang S. Z. Eur. J. Obstet. Gynecol. Reprod. Biol. 2005;118:170–173. doi: 10.1016/j.ejogrb.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Almeida-Porada G., Porada C. D., Tran N., Zanjani E. D. Blood. 2000;95:3620–3627. [PubMed] [Google Scholar]
- 32.Tian B., Han L., Kleidon J., Henke C. Am. J. Pathol. 2003;163:789–801. doi: 10.1016/S0002-9440(10)63706-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng F., Baldwin D. A., Schultz R. M. Dev. Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.