Abstract
Osteoporosis and low bone mass are currently estimated to be a major public health risk affecting >50% of the female population over the age of 50. Because of their bone-selective pharmacokinetics, nitrogen-containing bisphosphonates (N-BPs), currently used as clinical inhibitors of bone-resorption diseases, target osteoclast farnesyl pyrophosphate synthase (FPPS) and inhibit protein prenylation. FPPS, a key branchpoint of the mevalonate pathway, catalyzes the successive condensation of isopentenyl pyrophosphate with dimethylallyl pyrophosphate and geranyl pyrophosphate. To understand the molecular events involved in inhibition of FPPS by N-BPs, we used protein crystallography, enzyme kinetics, and isothermal titration calorimetry. We report here high-resolution x-ray structures of the human enzyme in complexes with risedronate and zoledronate, two of the leading N-BPs in clinical use. These agents bind to the dimethylallyl/geranyl pyrophosphate ligand pocket and induce a conformational change. The interactions of the N-BP cyclic nitrogen with Thr-201 and Lys-200 suggest that these inhibitors achieve potency by positioning their nitrogen in the proposed carbocation-binding site. Kinetic analyses reveal that inhibition is competitive with geranyl pyrophosphate and is of a slow, tight binding character, indicating that isomerization of an initial enzyme–inhibitor complex occurs with inhibitor binding. Isothermal titration calorimetry indicates that binding of N-BPs to the apoenzyme is entropy-driven, presumably through desolvation entropy effects. These experiments reveal the molecular binding characteristics of an important pharmacological target and provide a route for further optimization of these important drugs.
Keywords: farnesyl pyrophosphate synthase; osteoclast; slow, tight inhibition; farnesyl diphosphate synthase; trans-prenyltransferase
Farnesyl pyrophosphate synthase (FPPS) is a key regulatory enzyme in the mevalonate pathway. This pathway, ubiquitous in mammalian cells, provides essential lipid molecules, such as cholesterol and isoprenoids, with the latter necessary for posttranslational prenylation of small GTPases (1). The blockade of this pathway is a concept that has found widespread clinical use, with statins as drugs that inhibit hydroxymethylglutaryl-CoA reductase and reduce cholesterol biosynthesis, and nitrogen-containing bisphosphonates (N-BPs) as drugs for osteoporosis therapy that target FPPS and inhibit protein prenylation. In the case of N-BPs, the unique bone-targeting pharmacokinetic properties of these compounds cause selective inhibition of FPPS and loss of prenylated proteins in osteoclasts, thereby inhibiting the bone-destroying function of these cells (2).
FPPS catalyzes the sequential condensation of isopentenyl pyrophosphate (IPP), first with dimethylallyl pyrophosphate (DMAPP) and then with the resultant geranyl pyrophosphate (GPP) to produce the C15 farnesyl pyrophosphate (FPP). FPP is a substrate for geranylgeranyl pyrophosphate synthase, which produces the C20 isoprenoid geranylgeranyl pyrophosphate (GGPP). Posttranslational prenylation of small GTPases with FPP or GGPP is crucial for their correct subcellular localization and function (2). It is now clear that FPPS is the major enzyme target of N-BPs, such as risedronate (RIS) and zoledronate (ZOL) (3). These agents are currently used to treat postmenopausal and steroid-induced osteoporosis, Paget’s disease, hypercalcemia, and osteolysis associated with multiple myeloma and metastatic cancers (4, 5). Because of their ability to bind calcium ions in a multidentate manner, bisphosphonates (BPs) accumulate rapidly in bone tissue, where they inhibit the activity of bone-resorbing osteoclasts. The antiresorptive activity of BPs was described more than 30 years ago, but their molecular mode of action has only become apparent recently. Whereas some of the earlier BPs such as etidronate and clodronate appear to act by reversing pyrophosphorylytic reactions catalyzed by aminoacyl-tRNA synthetases, thereby producing the corresponding bisphosphonate analogs of ATP (6), the action of the more potent N-BPs involves a different mechanism (7–9). Their higher potency results from two key properties: their affinity for bone mineral and their ability to inhibit osteoclast function (10). Several years ago we and others demonstrated that N-BPs inhibit bone resorption by inhibiting FPPS, thereby preventing the prenylation of small GTPases, such as Rho, Rac, and Rabs, which are essential for osteoclast function (3, 11, 12).
There is a highly significant correlation between the order of potency of N-BPs for inhibiting human FPPS in vitro and their antiresorptive potency in vivo, with ZOL being an extremely potent inhibitor of FPPS. Importantly, minor modifications to the N-BP side chain that were known to affect antiresorptive potency have also now been shown to affect the ability to inhibit FPPS (3). Furthermore, the high degree of evolutionary conservation of FPPS explains why N-BPs also inhibit the growth of Dictyostelium and other eukaryotic microorganisms such as Leishmania and Trypanosoma parasites (13–15).
The exact mechanism by which N-BPs inhibit FPPS remains unclear, however. Recent structural studies of prokaryotic FPPS demonstrated that N-BPs can bind to the GPP/DMAPP substrate pocket (16). Studies of RIS in complex with Escherichia coli FPPS revealed that the side chain is positioned in the hydrophobic cleft that normally accommodates an isoprenoid lipid, and the phosphonate groups are bound to a cluster of three Mg2+ ions, chelated by two aspartate-rich motifs that are conserved in FPPS sequences (17).
Previous analyses based on molecular modeling and binding studies examined the potential mode of inhibition of vertebrate FPPS (13, 18). Several studies suggested that N-BPs bind to the GPP substrate-binding site because N-BPs might mimic the structure of the enzyme’s natural substrates GPP/DMAPP and act as carbocation transition state analogs (18). However, kinetic studies with recombinant human FPPS indicated that both the GPP and IPP substrate-binding sites might be occupied by N-BPs (19). A two-site binding model was further considered in in silico studies because docking analysis of N-BPs into the GPP pocket of a homology model of human FPPS based on the avian structure did not offer a full qualitative explanation for the binding differences of compounds with dramatic differences in potency (19, 20).
To clarify the mode of N-BP drug binding to its human target, we determined high-resolution structures of human FPPS in complexes with the clinically used N-BPs ZOL and RIS and its substrate IPP, and we studied in detail the mode of inhibition and binding by using isothermal titration calorimetry (ITC) and kinetic analysis.
Results
Structure Determination of Human FPPS.
Human FPPS was crystallized, and its structure was determined in complex with Mg2+ and RIS and in complex with Mg2+, ZOL, and IPP at resolutions of 2.0 and 2.3 Å, respectively (Table 3, which is published as supporting information on the PNAS web site).
Human FPPS exhibits the all α-helical prenyltransferase fold described earlier for the avian form of FPPS (17). FPPS is a dimer with 13 α-helices and connecting loops per subunit (Fig. 1A). Extended loop regions on the top of the protein include those connecting α4–α5 (residues 107–127) and α8–α9 (residues 249–268). A large central cavity located within the helical bundle forms an elongated partly hydrophobic ligand-binding site. The bottom of this cavity is delimited by the side chain of Phe-113. Conserved aspartate-rich motifs found in helices α4 (103DDIMD107) and α8 (243DDYLD247) face the central cavity, and a pronounced kink in helix α7 directs the carbonyl of Lys-200 toward the cavity as well.
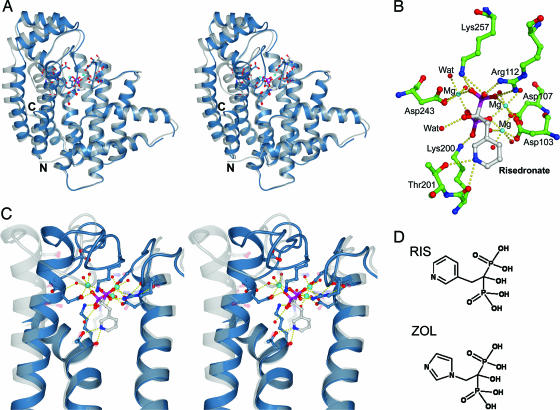
Fig. 1.
Structure of human FPPS. (A) Stereoview of the superimposition of FPPS in complex with RIS and Mg2+ (blue) with the avian apo structure 1FPS (gray), made by superimposing the α-carbons in helices α1–α7. (B) Specific polar interactions of RIS and Mg2+. Amino acid numbering in this paper is offset by −14 residues compared with the Protein Data Bank depositions. (C) Stereoview detail of RIS binding to FPPS (blue). Three Mg2+ ions mediate the interaction between phosphates and conserved aspartate residues. A semitransparent avian apo structure is superimposed in gray to highlight the structural differences. (D) Chemical structures of RIS and ZOL.
The BP moieties of RIS and ZOL are ligated through a cluster of three Mg2+ ions with phosphonate oxygens, aspartate residues, and water molecules contributing to the octahedral Mg2+ coordination (Fig. 1B). Compared with the avian apo structure (Protein Data Bank ID code 1FPS) and the Staphylococcus aureus structure (Protein Data Bank ID code 1RTR), the ligand-bound human structures have a more closed conformation (16, 17). This observation suggests that N-BP binding causes a structural rearrangement accompanied by a decrease in the size of the internal cavity (Fig. 1C). This movement is likely mediated by the Asp-rich motifs on α4 and α8 being drawn together to participate in ligand binding. The region connecting α8–α9 (residues 249–268) creates a lid over the N-BP and is held in place by several polar interactions: loop residue Lys-257 contacts Asp-243 and a phosphonate oxygen; Asp-247 at the end of α8 forms bidentate hydrogen bonds with main-chain amides of Thr-260 and Asp-261; and the main-chain amide of Ile-258 is within hydrogen-bonding distance of Thr-111 Oγ. The heterocyclic ring structures of RIS and ZOL are surrounded mainly by hydrophobic side chains of residues Phe-99, Leu-100, Thr-167, Lys-200, and Tyr-204, and the nitrogen atom of the ring system is found within hydrogen-bonding distance of Oγ of Thr-201 (Fig. 2A: RIS, 3.0 Å; ZOL, 2.8 Å) and the carbonyl oxygen of Lys-200 (≈3.0 Å).
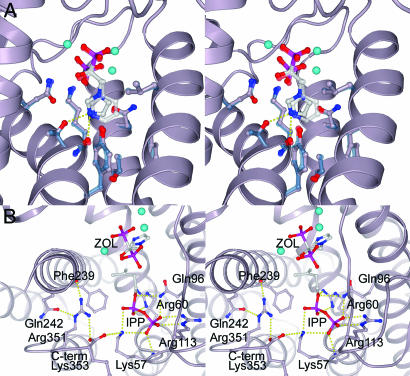
Fig. 2.
Detail of the FPPS·ZOL·IPP ternary complex. (A) Close-up stereoview of the heterocyclic ring binding pocket. ZOL and residues within 4 Å of the BP side chain are shown in ball-and-stick format (lilac). The equivalent residues from the RIS complex are overlaid in blue, and the RIS is shown semitransparently. (B) Close-up stereoview of the IPP-binding site showing the network of electrostatic interactions involving the C terminus.
The production of GPP/FPP is proposed to proceed by a three-step ionization–condensation–elimination reaction in which the double bond of IPP attacks the C1 atom of a dimethylallyl/geranyl carbocation. The carbocation intermediate is predicted to be stabilized through electrostatic interactions with a magnesium-bound pyrophosphate moiety and through interactions with the carbonyl of Lys-200 and the side-chain oxygen of Thr-201. The Lys-200 and Thr-201 interactions are analogous to the N-BP nitrogen binding described above. The catalytic base necessary to abstract the IPP 2R proton is likely to be a pyrophosphate oxygen bound through interactions with conserved Arg-112 and Lys-257 side chains, located on the flexible loops shielding the active site from bulk solvent during catalysis (16).
The N-BPs bind in a nearly identical manner, and root-mean-square deviations for α-carbon positions between the RIS and ZOL structures are 0.44 Å. The largest conformational change involves the basic residues at the C terminus, 350KRRK353, which become ordered at IPP binding. Although they were predicted to bind the IPP phosphates, a direct interaction between these residues and the IPP is not observed. Alternatively, the IPP phosphates are coordinated by six water molecules, Gln-96, Arg-113, Arg-60, and Lys-57. Additionally, there is a network of salt links involving Lys-57, the terminal carboxylate of Lys-353, and Arg-351. This conformation of the C terminus sequesters the active site from bulk solvent, locks the Lys-57 side chain into position to make two polar interactions with the IPP pyrophosphate moiety, and directs the hydrophobic tail of IPP toward ZOL (Fig. 2B).
Inhibition of Human FPPS by N-BPs.
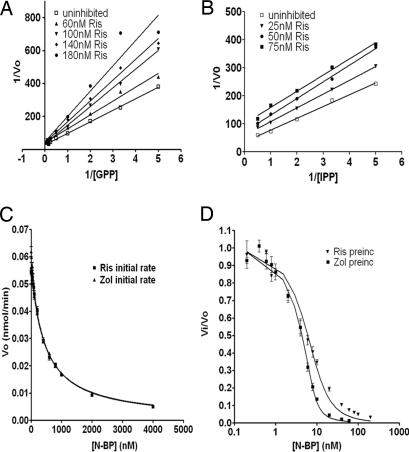
Recombinant human FPPS catalyzes synthesis of FPP with Km values of 1.8 ± 0.33 μM (IPP) and 2.07 ± 0.2 μM (GPP) and a kcat of 0.42 s−1 (Table 1). RIS inhibits the reaction competitively with respect to GPP and in an uncompetitive/mixed-type manner with respect to IPP (Fig. 3A and B). It was found that preincubation of the enzyme with the inhibitor increased the enzyme inhibition, with times >5 min giving the highest inhibition, compatible with observations in ITC experiments (see below). Thereafter, all enzyme assays that required preincubation were done after 10 min of preincubation, usually followed by an initial rate measurement of 3 min that was started by the addition of both substrates. The increased inhibition caused by preincubation could be completely overcome by preincubation in the presence of high [GPP] alone and partially overcome in the presence of high [IPP] alone (data not shown). Because IPP inhibits FPPS at high concentrations by binding competitively to the DMAPP/GPP site (21), this finding implies that N-BPs compete with GPP and IPP for a binding site, but the IPP binding site where this competition occurs is most likely the IPP substrate inhibition site, namely the GPP site. In the absence of GPP, inhibition by 20 nM RIS was maximal after 5 min of preincubation; however, preincubation in the presence of higher [GPP] showed little decrease in initial rate. GPP concentrations <0.5 μM showed significant inhibition at the same concentration of RIS, which would account for our previous observations showing an apparent increase in the inhibition of FPPS by RIS at low substrate concentrations or at high RIS concentrations. This behavior is manifested in the apparent nonlinearity of Lineweaver–Burk and Dixon plots and, when interpreted as classical linear inhibition, suggested that two molecules of inhibitor bind cooperatively. For the above reasons, it is now apparent that our previous interpretation of the kinetic data (19) was incorrect. Given that the equilibrium among enzyme, inhibitor, and substrate takes many minutes to be achieved and that the inhibition of FPPS by RIS occurs at such a low inhibitor concentration (≈50% inhibition of 10 nM FPPS by 10 nM RIS), the inhibition mechanism can be considered to be “slow, tight binding” rather than classical competitive. Because the N-BP can act as a classic competitive inhibitor in the early stages of the enzyme reaction and increases in potency after preincubation, a two-step reaction scheme involving a rate-limiting isomerization step fits the observed enzyme behavior best. The vast majority of known examples of slow, tight binding adhere to this mechanism (22).
Table 1.
Inhibition and kinetic data for human FPPS and N-BPs
| Constant | IPP | GPP | Inhibitor |
|
|---|---|---|---|---|
| RIS | ZOL | |||
| Km, μM | 1.8 ± 0.33 | 2.07 ± 0.2 | ||
| kcat, s−1 | 0.42 | |||
| IC50, nM | ||||
| Initial | 450 ± 10 | 475 ± 10 | ||
| Preincubated | 5.7 ± 0.54 | 4.1 ± 0.22 | ||
| Ki, nM | 72 ± 4 | 76 ± 3 | ||
| Ki*app, nM | 2.0 ± 0.44 | 0.4 ± 0.1 | ||
| Ki*, nM | 0.34 | 0.07 | ||
| k5/k6 | 210:1 | 1080:1 | ||
Fig. 3.
N-BP inhibition of human FPPS. (A) Competitive inhibition of RIS for the GPP site. Lineweaver–Burk plot of initial rate of FPPS; [IPP] = 10 μM. (B) Uncompetitive inhibition of RIS for the IPP site. Lineweaver–Burk plot of initial rate of FPPS; [GPP] = 5 μM. (C) Initial inhibition of FPPS by RIS and ZOL; [IPP] and [GPP] = 10 μM. Data were fitted to Eq. 1 by nonlinear regression. (D) FPPS preincubated with RIS. Enzyme was assayed after a 10-min preincubation with RIS; [IPP] and [GPP] were 10 μM. Fitting of data to Eq. 2 results in determination of an enzyme concentration of 8 nM.
Based on our observations of the kinetics of inhibition of FPPS we considered the following scheme. First, the inhibitor appears to compete directly with GPP, to form an enzyme–inhibitor complex. This binding step occurs rapidly and causes inhibition. A second step involving the enzyme–inhibitor complex undergoing some change, possibly an isomerization, occurs slowly and increases the inhibition of the enzyme over time (22), which gives rise to the following mechanism:
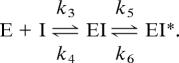 |
The Ki for the first step is the dissociation constant for E·I. The overall dissociation constant is Ki*, and k6 is the rate constant for conversion of FPPS·RIS* back to FPPS·RIS.
The calculated values of Ki suggest that both RIS and ZOL initially bind the enzyme with a similar affinity (Table 1). Values for Ki* indicate that given enough time to allow formation of the E·I* complex, inhibition is increased significantly, with ZOL being somewhat more potent than RIS. The isomerization constant k5/k6 gives an indication of how likely the FPPS·RIS* complex is to return to the FPPS·RIS state. Using the values of Ki and Ki* calculated above, the value of k5/k6 was found to be 210:1 for RIS and 1080:1 for ZOL, which suggests that the equilibrium lies on the side of the FPPS·N-BP* complex, even more so with ZOL than with RIS (accounting for the greater overall potency of ZOL).
Mode of N-BP Inhibitor Binding to Human FPPS.
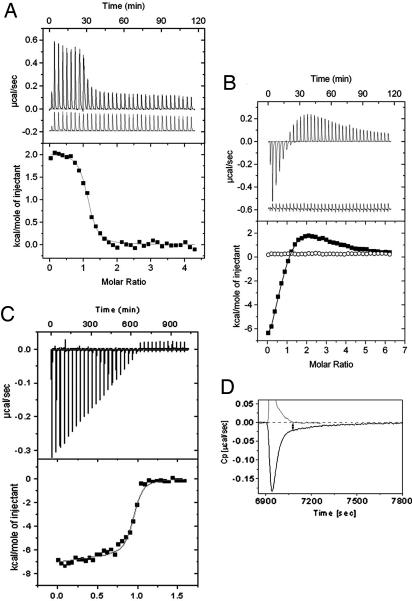
ITC was used to investigate the thermodynamic parameters governing ligand binding to human FPPS. Binding of RIS to apo-FPPS is accompanied by a positive enthalpy change of +1.8 kcal/mol (1 kcal = 4.18 kJ) (Fig. 4A) with an overall stoichiometry of ≈1:1 and an entropy change (TΔS) of 10.5 kcal/mol, resulting in a calculated Kd of 164 nM (Table 2). ZOL titration into FPPS shows a qualitatively similar pattern; however, the overall Kd is significantly lower, with a calculated value of 15 nM, in close agreement with the kinetic data obtained. These data show that N-BPs bind to a single site at FPPS, which almost certainly is the DMAPP/GPP site deduced from these titrations as well as the structural and kinetic data described above. The presence of Mg2+ increases the affinity of RIS for FPPS ≈30-fold, and titration experiments showed that RIS binds one Mg2+ ion with an affinity of 29 μM and an enthalpy change of +1.5 kcal/mol.
Fig. 4.
Thermodynamics of ligand binding to human FPPS measured by ITC. (A) Binding of RIS to FPPS in the presence of 2 mM MgCl2. (Upper) Data obtained for injections of RIS into buffer (lower trace) and into FPPS (upper trace). (Lower) Binding isotherm fit to a one-site binding model after subtraction of blank titration heats. (B) Titration of IPP into FPPS. Biphasic binding is observed, indicating occupation of both GPP and IPP sites. (C) ZOL binding to the FPPS·IPP complex. Displacement of IPP bound in the GPP site is accompanied by negative enthalpies. (D) Binding kinetics of ZOL in the presence of IPP. ZOL showed slow binding kinetics with IPP present (bold trace) but not in the absence of IPP (thin line).
Table 2.
Thermodynamic data determined by isothermal titration calorimetry
| Sample | Kb, ×106 M−1 | Kd, μM | ΔH, kcal/mol | TΔS, kcal/mol | ΔG, kcal/mol | n |
|---|---|---|---|---|---|---|
| RIS/Mg* | 6.1 ± 2 | 0.16 | +1.8 ± 0.4 | 10.5 | −8.8 | 1.14 |
| RIS† | 0.2 ± 0.5 | 5 | +3.9 ± 0.2 | 10.9 | −7 | 1.2 |
| Mg RIS‡ | 0.035 ± 0.004 | 290 | +1.5 ± 0.04 | 6.2 | −4.7 | 0.99 |
| ZOL/Mg§ | 64 ± 2.0 | 0.015 | +2.1 ± 0.03 | 12.1 | −10 | 0.93 |
| IPP 1¶ | 0.33 ± 0.06 | 3 | −13.7 ± 1.3 | −6.6 | −7.1 | 0.85 |
| IPP 2‖ | 0.068 ± 0.008 | 15 | +13.4 ± 1.3 | 19.7 | −6.3 | 1.2 |
| ZOL·IPP** | 55 ± 1.2 | 0.018 | +2.4 ± 2 | ND | ND | 1 |
| RIS·IPP†† | 30 ± 10 | 0.03 | +4.4 ± 2 | ND | ND | 1.1 |
ND, not determined.
*RIS titrated into FPPS with Mg2+ present in both samples.
†RIS titrated into FPPS in the absence of Mg2+.
‡Mg2+ titrated into RIS (no FPPS present).
§ZOL titrated into FPPS with Mg2+ present in both samples.
¶IPP titrated into FPPS with Mg2+ present in both samples, higher-affinity site.
‖IPP titrated into FPPS with Mg2+ present in both samples, lower-affinity site.
**ZOL titrated into an IPP-saturated FPPS, Mg2+ present in both samples.
††RIS titrated into an IPP-saturated FPPS, Mg2+ present in both samples.
Titration of IPP into FPPS resulted in a biphasic binding curve (Fig. 4B). Nonlinear least-squares fits of the measured binding isotherm to a two-site binding model revealed that IPP binding occurs with a 2:1 stoichiometry. Binding to the high-affinity binding site is accompanied by a negative enthalpy change, whereas binding to the second, lower-affinity, site is accompanied by a strongly positive enthalpy change. These data suggest that the primary binding site is the IPP site and that at higher concentrations a second binding site, which is probably the DMAPP/GPP, site gets occupied.
To prove this hypothesis we performed a competition binding experiment titrating ZOL into FPPS saturated with IPP. The binding isotherm showed a large negative enthalpy change for this event, in agreement with the hypothesis that IPP occupies also the DMAPP/GPP site and that the binding event associated with the large positive binding enthalpy change is indeed a result of IPP binding to the DMAPP/GPP-binding site (Fig. 4C). A competition for binding-site model resulted in a satisfying fit and in similar binding constants as determined in experiments in the absence of IPP (Table 2).
Interestingly, N-BP binding to FPPS in the presence of IPP was an extremely slow event, which made it necessary to use exceptionally long spacing times between injections (Fig. 4D). This finding is in agreement with the enzyme kinetic data that characterized the binding of ZOL to FPPS as a slow, tight binding event. The calorimetric data showed that the kinetic effect on binding is coupled to the presence of IPP, whereas binding of ZOL or RIS in the absence of IPP showed fast binding kinetics. A possible scenario explaining this behavior would be that binding of IPP is coupled to a structural change that induces a conformation in the FPPS·IPP complex that is not adequate for ZOL or RIS binding. The extremely large positive enthalpy change associated with the occupation of DMAPP/GPP site by IPP supports such a structural rearrangement on IPP binding.
Discussion
The structural changes occurring in FPPS with N-BP binding can be seen by three methods: (i) preincubation of FPPS with N-BPs increases the degree of inhibition; (ii) long equilibration times are required between ITC injections; and (iii) a more closed conformation is observed in the N-BP-bound crystal structures compared with apo-FPPS structures. Binding of N-BPs is magnesium-dependent and occurs in the GPP/DMAPP site. The cyclic nitrogen interactions linking ZOL and RIS with Thr-201 and Lys-200 support the hypothesis that N-BPs gain potency by positioning their nitrogen in the proposed carbocation-binding site.
A second conformational change is revealed by comparing the N-BP-bound crystal structures. N-BP binding induces a closed conformation, creates a more ordered IPP-binding site, and provides a surface for IPP to pack against. The second isomerization occurs once IPP is bound and entails the C terminus curling back toward the pyrophosphate moiety, securing the ligands into position and sequestering the site from bulk solvent.
Structure determination, detailed kinetic analysis of N-BP inhibition, and the thermodynamic characterization of ligand binding to human FPPS provide a unique framework for understanding the mode of action of this widely used, important class of antiosteoporosis drugs. From both kinetic and thermodynamic experiments it is evident that RIS and ZOL binding to FPPS is a slow, tight binding process, accompanied by large positive enthalpies when binding to apo-FPPS. These enthalpic components describe largely the ligand–enzyme interactions, and it is likely that these positive enthalpic changes are derived from breaking polar interactions with water molecules both within the enzyme active site and around the ligand. This “counterproductive” event is more than compensated by the gain in entropy. Entropy changes reflect two distinct contributions: changes in solvation entropy and changes in conformational entropy (23, 24). The structural changes occurring with occupation of the DMAPP/GPP site, i.e., closing of the active-site cavity, reduce the large solvation entropy effect occurring at the ligand site, but the overall change is still large enough to drive high-affinity binding, likely by the primarily hydrophobic ring system of the N-BPs. Another example of this unusual drug-binding behavior is given by the first generation of HIV-1 protease inhibitors, displaying enthalpically unfavorable characteristics, with binding driven by large, positive entropy changes (25).
In titration experiments of N-BPs to apo-FPPS and FPPS·IPP complexes, the slow component in binding to FPPS is observed for N-BPs but not for IPP, indicating that specific rearrangements occur around the N-BP site. The exact nature of the slow component remains obscure; however, it is possible that any of the ligand-induced changes (i.e., desolvation effects and structural changes within FPPS) forms the basis for the slow binding kinetics.
On the basis of the data described, it appears feasible to increase binding affinity by designing novel N-BPs targeted to human FPPS. These N-BPs could be conformationally restrained, partly hydrophobic, with functional groups branching further into the DMAPP/GPP site. This site has additional polar residues (Tyr-204 and Thr-167) that could serve as interacting residues for a pharmacophore model. These features would possibly enhance both the entropy and enthalpy effects to drive N-BP ligand binding.
During the course of the evaluation of this paper, an independent report on structure determination of human FPPS was released (26), confirming our conclusions on the structural basis of N-BP binding.
Methods
Expression and Purification of Recombinant Human FPPS.
A clone encoding human FPPS residues 1–353 (P14324) as an N-terminally His6-tagged fusion protein with a tobacco etch virus (TEV) protease cleavage site was expressed in E. coli BL21(DE3). Cells were lysed by using a high-pressure cell disruptor, and the protein was purified to near homogeneity by using nickel nitrilotriacetate resin (Qiagen). Initial crystallization experiments were performed directly with this protein. For further experiments, the histidine tag was removed by incubation with TEV protease, and gel filtration chromatography was performed with a Superdex 200 column (GE/Amersham Pharmacia).
Crystallization and Data Collection of Human FPPS·RIS.
FPPS including the N-terminal tag was concentrated to 13 mg/ml in 10 mM Hepes (pH 7.5)/0.5 M NaCl/5% (vol/vol) glycerol/2 mM RIS/2 mM MgCl2. Crystals were grown at 20°C in sitting drops by mixing 100 nl of protein solution and 50 nl of precipitant consisting of 40% (vol/vol) polyethylene glycol 300 and 0.1 M phosphate/citrate buffer, pH 4.2. Diffraction data were collected from a single crystal at 100 K at the Advanced Light Source Beamline 8.3.1 (Lawrence Berkeley Laboratory, Berkeley, CA).
Crystallization and Data Collection of Human FPPS·ZOL·IPP.
Tobacco etch virus protease-cleaved FPPS was concentrated to 16 mg/ml, and ZOL, IPP, and MgCl2 were added to final concentrations of 2 mM each. Crystals were grown at 20°C in sitting drops by mixing 150 nl of protein solution and 150 nl of precipitant consisting of 14% (vol/vol) polyethylene glycol 6000, 0.7 M LiCl, and 70 mM citrate, (pH 4.0). A single crystal was transferred to a solution composed of 20% (vol/vol) polyethylene glycol 300 and 80% well solution and flash-cooled in liquid nitrogen. Diffraction data were collected at the Swiss Light Source Beamline X10SA (Paul Scherrer Institute, Villigen, Switzerland).
Data Processing and Refinement.
The data sets were processed with mosflm and scala (27). Initial phases for the FPPS·RIS complex were calculated by molecular replacement implemented in phaser (28) using the avian FPPS structure 1UBV as a search model (20). The FPPS·RIS structure minus ligands and solvent molecules was used as a starting model for the FPPS·ZOL·IPP structure. Iterative rounds of model building in coot (29) and refinement using refmac5 (30) resulted in the final models for which statistics appear in Table 3.
Kinetics and Inhibition of Human FPPS by N-BPs.
FPPS was assayed by the method of Reed and Rilling with modifications (31). For kinetic analysis, 40 ng of pure FPPS was assayed in a final volume of 100 μl buffer containing 50 mM Tris (pH 7.7), 2 mM MgCl2, 1 mM tris(2-carboxyethyl)phosphine, and 5 μg/ml BSA. The concentrations of GPP and IPP ([14C]IPP, 400 kBq/μmol) were as described in the text but were typically 0.2–20 μM. Reactions were started with the addition of enzyme and allowed to proceed for an appropriate period at 37°C. When a preincubation of the enzyme with the inhibitor was required, the enzyme was incubated with inhibitor in a 60-μl volume. After 10 min, 40 μl of substrate in water was added to start the reaction. Assays were terminated by the addition of 0.2 ml of concentrated HCl/methanol (1:4) and incubated for 10 min at 37°C. The reaction mixtures were then extracted with 0.4 ml of ligroin, and the amount of radioactivity in the upper phase was determined by mixing 0.2 ml of the ligroin with 4 ml of general-purpose scintillant. The radioactivity was measured by using a Packard Tri-Carb 1900CA scintillation counter. At the concentration of FPPS used, half the amount of enzyme gives half the activity. Data were fitted to the following kinetic models by nonlinear regression using prism (GraphPad, San Diego).
To calculate the Ki for the formation of the E·I complex, the enzyme inhibition data from initial rate experiments were fitted to the equation for classical linear competitive inhibition (22):
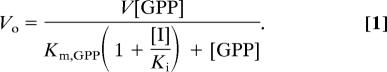 |
Data from experiments that included preincubation of enzyme and inhibitor were fitted to the following equation (32):
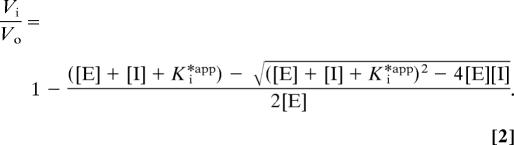 |
The overall dissociation constant, Ki*, was calculated from Ki*app by using the model for competitive inhibition (32):
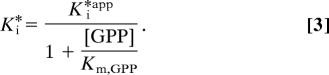 |
The isomerization constant, k5/k6, was calculated by using the following equation (32):
ITC.
Calorimetric measurements were carried out by using a VP-ITC titration calorimeter from Microcal (Amherst, MA). Samples were dialyzed extensively against 10 mM Hepes (pH 7.5)/100 mM NaCl. Each titration experiment consisted of a first (5-μl) injection of the ligand followed by 8-μl injections. Heats of dilution were measured in blank titrations by injecting the ligand into buffer, and the dilution heats were subtracted from the binding heats. Data were analyzed by using single and multiple binding site models implemented in the origin software package (OriginLab, Northampton, MA) provided with the instrument. Competition experiments were analyzed by using a competition for binding site model (33) implemented in the software package.
Supplementary Material
Acknowledgments
We thank Frank von Delft at the Swiss Light Source for data collection. The DNA was amplified from I.M.A.G.E. Consortium Clone ID 4132071. The Structural Genomics Consortium is a registered charity (Number 1097737) funded by the Wellcome Trust, GlaxoSmithKline, Genome Canada, the Canadian Institutes of Health Research, the Ontario Innovation Trust, the Ontario Research and Development Challenge Fund, the Canadian Foundation for Innovation, Vinnova, the Swedish Strategic Research Foundation, the Knut and Alice Wallenberg Foundation, and the Karolinska Institutet.
Abbreviations
- BP
bisphosphonate
- FPPS
farnesyl pyrophosphate synthase
- N-BPs
nitrogen-containing bisphosphonates
- IPP
isopentenyl pyrophosphate
- DMAPP
dimethylallyl pyrophosphate
- FPP
farnesyl pyrophosphate
- GPP
geranyl pyrophosphate
- GGPP
geranylgeranyl pyrophosphate
- ITC
isothermal titration calorimetry
- RIS
risedronate
- ZOL
zoledronate.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1YV5 (FPPS·RIS) and 1ZW5 (FPPS·ZOL·IPP)].
References
- 1.Goldstein J. L., Brown M. S. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 2.Rogers M. J. Curr. Pharm. Des. 2003;9:2643–2658. doi: 10.2174/1381612033453640. [DOI] [PubMed] [Google Scholar]
- 3.Dunford J. E., Thompson K., Coxon F. P., Luckman S. P., Hahn F. M., Poulter C. D., Ebetino F. H., Rogers M. J. J. Pharmacol. Exp. Ther. 2001;296:235–242. [PubMed] [Google Scholar]
- 4.Rodan G. A., Reszka A. A. Curr. Mol. Med. 2002;2:571–577. doi: 10.2174/1566524023362104. [DOI] [PubMed] [Google Scholar]
- 5.Russell R. G., Rogers M. J. Bone. 1999;25:97–106. doi: 10.1016/s8756-3282(99)00116-7. [DOI] [PubMed] [Google Scholar]
- 6.Frith J. C., Monkkonen J., Auriola S., Monkkonen H., Rogers M. J. Arthritis Rheum. 2001;44:2201–2210. doi: 10.1002/1529-0131(200109)44:9<2201::aid-art374>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 7.Fisher J. E., Rogers M. J., Halasy J. M., Luckman S. P., Hughes D. E., Masarachia P. J., Wesolowski G., Russell R. G., Rodan G. A., Reszka A. A. Proc. Natl. Acad. Sci. USA. 1999;96:133–138. doi: 10.1073/pnas.96.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luckman S. P., Hughes D. E., Coxon F. P., Graham R., Russell G., Rogers M. J. J. Bone Miner. Res. 1998;13:581–589. doi: 10.1359/jbmr.1998.13.4.581. [DOI] [PubMed] [Google Scholar]
- 9.van Beek E., Lowik C., van der Pluijm G., Papapoulos S. J. Bone Miner. Res. 1999;14:722–729. doi: 10.1359/jbmr.1999.14.5.722. [DOI] [PubMed] [Google Scholar]
- 10.van Beek E. R., Lowik C. W., Ebetino F. H., Papapoulos S. E. Bone. 1998;23:437–442. doi: 10.1016/s8756-3282(98)00120-3. [DOI] [PubMed] [Google Scholar]
- 11.Bergstrom J. D., Bostedor R. G., Masarachia P. J., Reszka A. A., Rodan G. Arch. Biochem. Biophys. 2000;373:231–241. doi: 10.1006/abbi.1999.1502. [DOI] [PubMed] [Google Scholar]
- 12.van Beek E., Pieterman E., Cohen L., Lowik C., Papapoulos S. Biochem. Biophys. Res. Commun. 1999;255:491–494. doi: 10.1006/bbrc.1999.0224. [DOI] [PubMed] [Google Scholar]
- 13.Martin M. B., Grimley J. S., Lewis J. C., Heath H. T., III, Bailey B. N., Kendrick H., Yardley V., Caldera A., Lira R., Urbina J. A., et al. J. Med. Chem. 2001;44:909–916. doi: 10.1021/jm0002578. [DOI] [PubMed] [Google Scholar]
- 14.Sanders J. M., Gomez A. O., Mao J., Meints G. A., Van Brussel E. M., Burzynska A., Kafarski P., Gonzalez-Pacanowska D., Oldfield E. J. Med. Chem. 2003;46:5171–5183. doi: 10.1021/jm0302344. [DOI] [PubMed] [Google Scholar]
- 15.Szabo C. M., Martin M. B., Oldfield E. J. Med. Chem. 2002;45:2894–2903. doi: 10.1021/jm010279+. [DOI] [PubMed] [Google Scholar]
- 16.Hosfield D. J., Zhang Y., Dougan D. R., Broun A., Tari L. W., Swanson R. V., Finn J. J. Biol. Chem. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 17.Tarshis L. C., Yan M., Poulter C. D., Sacchettini J. C. Biochemistry. 1994;33:10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 18.Martin M. B., Arnold W., Heath H. T., III, Urbina J. A., Oldfield E. Biochem. Biophys. Res. Commun. 1999;263:754–758. doi: 10.1006/bbrc.1999.1404. [DOI] [PubMed] [Google Scholar]
- 19.Ebetino F. H., Roze C. N., McKenna C. E., Barnett B. L., Dunford J. E., Russell R. G. G., Mieling G. E., Rogers M. J. J. Organomet. Chem. 2005;690:2679–2687. [Google Scholar]
- 20.Tarshis L. C., Proteau P. J., Kellogg B. A., Sacchettini J. C., Poulter C. D. Proc. Natl. Acad. Sci. USA. 1996;93:15018–15023. doi: 10.1073/pnas.93.26.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnard G. F., Popjak G. Biochim. Biophys. Acta. 1981;661:87–99. doi: 10.1016/0005-2744(81)90086-3. [DOI] [PubMed] [Google Scholar]
- 22.Morrison J. F., Walsh C. T. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 23.Velazquez-Campoy A., Kiso Y., Freire E. Arch Biochem. Biophys. 2001;390:169–175. doi: 10.1006/abbi.2001.2333. [DOI] [PubMed] [Google Scholar]
- 24.Velazquez-Campoy A., Freire E. J. Cell Biochem. Suppl. 2001;37:82–88. doi: 10.1002/jcb.10068. [DOI] [PubMed] [Google Scholar]
- 25.Velazquez-Campoy A., Luque I., Todd M. J., Milutinovich M., Kiso Y., Freire E. Protein Sci. 2000;9:1801–1809. doi: 10.1110/ps.9.9.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rondeau J. M., Bitsch F. M., Boirgier E., Geiser M., Hemmig R., Kroemer M., Lehmann S., Ramage P., Rieffel S., Strauss A., et al. Chem. Med. Chem. 2006;1:267–273. doi: 10.1002/cmdc.200500059. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project 4 Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 28.Storoni L. C., McCoy A. J., Read R. J. Acta Crystallogr. D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov G. N. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Reed B. C., Rilling H. C. Biochemistry. 1976;15:3739–3745. doi: 10.1021/bi00662a015. [DOI] [PubMed] [Google Scholar]
- 32.Copeland R. A. Enzymes: A Practical Guide to Structure, Mechanism, and Data Analysis. New York: Wiley; 2000. pp. 305–349. [Google Scholar]
- 33.Sigurskjold B. W. Anal. Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.