Abstract
In the adult central nervous system, two distinct populations of glial cells expressing the chondroitin sulfate proteoglycan NG2 have been described: bipolar progenitor cells and more differentiated “synantocytes.” These cells have diverse neurological functions, including critical roles in synaptic transmission, repair, and regeneration. Despite their potential importance, the genetic factors that regulate NG2 cell development are poorly understood, and the relationship of synantocytes to the oligodendroglial lineage, in particular, remains controversial. Here, we show that >90% of embryonic and adult NG2 cells express Olig2, a basic helix–loop–helix transcription factor required for oligodendrocyte lineage specification. Analysis of mice lacking Olig function demonstrates a failure of NG2 cell development at embryonic and perinatal stages that can be rescued by addition of a transgene containing the human OLIG2 locus. These findings show a general requirement for Olig function in NG2 cell development and highlight further roles for Olig transcription factors in neural progenitor cells.
Keywords: Cspg4, oligodendrocyte, synantocyte, glia, regeneration
NG2 cells are central nervous system (CNS) glial cells defined by their expression of the chondroitin sulfate proteoglycan, NG2, which is also known as Cspg4 or AN2 (1–4). A significant proportion of NG2 cells actively proliferate and indeed have been characterized as the most prevalent cycling progenitor cell population in the adult CNS (5, 6). NG2 cells have diverse functions and participate in oligodendrogenesis (7) and neurogenesis (8), as well as the physiologic support of neurons and synaptic signaling (4, 9). They have also been proposed to play critical roles in brain repair and regeneration and are the primary responding neural cell type to CNS injury (10, 11).
Despite their abundance as a progenitor population and potential importance in maintenance and repair of neurological function, the developmental ontogeny of NG2 cells remains controversial (4, 7, 12). Cytologically, they are reportedly a heterogeneous population, and two distinctive cellular morphologies have been described: bipolar progenitor cells that resemble oligodendrocyte progenitors (OLP) (1, 13) and stellate “synantocytes.” Most postnatal NG2 cells are synantocytes, which are defined by a complex “differentiated” morphologic appearance. Even so, they lack expression of most differentiated glial cell-type specific markers (12). Indeed, based on these observations, several studies have suggested that the majority of postnatal NG2+ synantocytes comprise an independent neuroepithelial lineage distinct from neurons, astrocytes, and oligodendrocytes (4, 12, 14), a proposal that has provoked ongoing debate (15, 16).
The genetic pathways, which regulate NG2 cell development, have yet to be defined, and the question of whether bipolar NG2 cells and synantocytes have fundamentally similar or different origins is unresolved. Given their early developmental expression of OLP markers, NG2 cell formation might be regulated by the same genetic pathways that control oligodendrocyte development. During embryogenesis, Olig1 and Olig2 encode basic helix–loop–helix transcription factor proteins that are required for development of motor neuron and OLPs (17–19). Although Olig1 can participate in OLP specification in the developing brain (19), it is more generally required for maturation of OLPs to oligodendrocytes in neonates (20) and in repair of demyelinating lesions (21). In the adult brain, Olig2 expression has been reported in type C cells of the subventricular zone (SVZ) (22). However, outside of the SVZ in the normal CNS, Olig2 is expressed in oligodendroglial progenitor cells and mature oligodendrocytes (23, 24).
If aspects of NG2 cell development occur independently of oligodendroglial development, then a testable prediction is that NG2 cells would be generated in the absence of Olig function. To evaluate this prediction, we focused on expression and function of Olig2 in the rodent brain. Our data indicate that Olig2 expression marks parenchymal NG2 cells, and that Olig gene function is necessary for NG2 cell development throughout the CNS at antenatal stages, including both bipolar progenitor cells (i.e., OLPs) and synantocytes. Together, our findings provide genetic support for a unified NG2/OLP developmental connection.
Results
More Than 90% of NG2 Cells Express Olig2 in the Brain.
We first determined whether Olig2 expression was limited to a particular stage of NG2 cell development. To complement our analysis using existing antibody reagents to Olig2 and NG2, we developed a monoclonal antibody to Olig2, which specifically recognizes mouse Olig2 protein by immunohistochemistry (Fig. 1C). Double immunohistochemical (IHC) analysis of Olig2 expression in NG2 cells of 18.5 days postcoitum (dpc) mouse brain showed colocalization of Olig2 in NG2 cells with both stellate synantocyte and bipolar progenitor morphologies (Fig. 1A). Quantitative analysis of histological sections showed that >90% of NG2 cells were Olig2+ in all subregions of the brain examined (dorsal neocortex, corpus callosum, SVZ/VZ, and ventral forebrain regions). In contrast, only 18% of the overall Olig2+ cell population on average expressed NG2. The number of Olig2+ cells that expressed NG2 varied with the subregion of the brain examined from a low of 8% in the SVZ to a high of 53% within white matter tracts. We found that NG2 glial cells easily could be distinguished from NG2+/Olig2− pericytes associated with blood vessels (25) on the basis of morphology; however, we could not entirely rule out that a minor population of NG2+/Olig2− cells in our counts are pericytes.
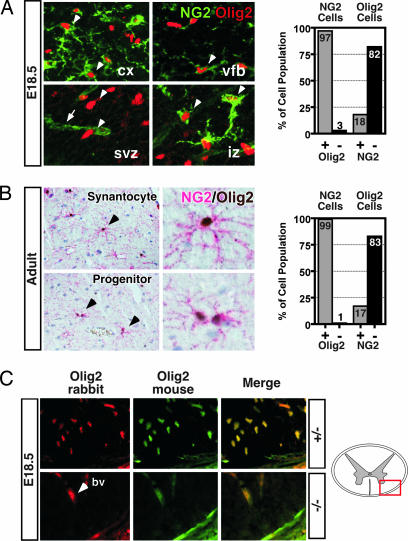
Fig. 1.
NG2 cells express Olig2 in mouse brain. (A) Deconvolution images of immunofluorescent staining showing NG2 (green Cy5) and Olig2 (red Cy2) are coexpressed in neural cells with NG2 synantocyte and progenitor morphology (white arrowheads) in normal 18.5 dpc mouse neocortex (cx), ventral forebrain (vfb), and developing white matter (iz). Olig2+ cells (white arrowheads) in the subventricular zone (svz) rarely expressed NG2. NG2 staining in pericytes around blood vessels (white arrow in svz) served as internal control. Graph shows summed results of manual cell counting at 18.5 dpc (2,949 total cells counted; n = 3 animals). (B) Double IHC staining for NG2 (red, alkaline phosphatase) and Olig2 [brown, diaminobenzidine (DAB)] in adult rat brain. Colocalization was seen in cells with complex synantocyte morphology (Upper) and cells with progenitor morphology (Lower) occurring as pairs of recently divided cells. Graph shows summed results of manual cell counting from adult mouse brain (578 total cells counted; n = 2 animals). The percentage of Olig2+ NG2 cells was >98% in all subregions of the adult brain. Sections are hematoxylin counterstained. (C) Specificity testing of Olig2 mAb TV37–1C10 in spinal cord white matter (red box in diagram) of Olig2+/− mouse at 18.5 dpc shows precise colocalization of signal from established rabbit polyclonal anti-Olig2 antibody DF308 (red) with that from the mouse anti-Olig2 antibody TV37–1C10 (green). Similar findings were obtained at embryonic and adult stages in brain (data not shown). Absence of any specific labeling in Olig2−/− mutant spinal cord (Lower) confirms specificity for Olig2. Arrowhead marks autofluorescence in blood vessels (bv).
Adult NG2+ synantocytes have a highly differentiated morphologic appearance with few similarities to myelinating oligodendrocytes or their perinatal oligodendroglial progenitors. IHC analysis of adult mouse and rat brain showed that at these later stages of development >98% of NG2 cells coexpress Olig2; however, NG2 cells remain a minority of the total Olig2+ cell population (Fig. 1B). As was found at 18.5 dpc, NG2/Olig2 coexpressing cells were seen in all regions of the brain and in NG2 cells with both synantocyte and progenitor morphology. Examination of the Olig2+ cell population showed that the percentage of cells colabeled by NG2 varied again with the subregions of the brain ranging from 6% in white matter tracts to a high of 31% in the ventral forebrain.
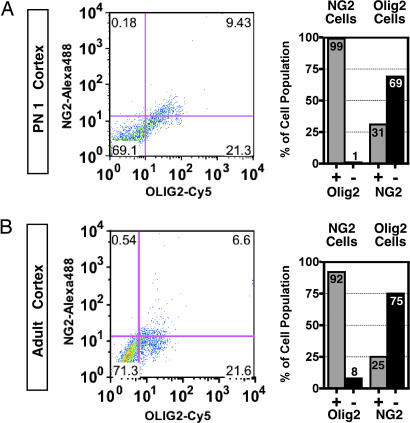
These IHC findings were further validated by flow cytometry. Analysis of freshly dissociated mouse dorsal neocortex at postnatal day 1 (PN1), a period of rapid NG2 cell development, as well as adult stages, confirmed that at least 90% of NG2 cells in the postnatal period coexpress Olig2 (Fig. 2), even without correction for the presence of NG2+Olig2− pericytes within the total NG2 cell population. Although only a minority of Olig2+ cells coexpressed NG2 (31% in PN1 and 25% in adult), more Olig2+ cells were found to coexpress NG2 than in manual counting procedures, possibly because of increased sensitivity of flow cytometry compared with in situ IHC methods. Together, these data show that almost all NG2 cells coexpress Olig2 at prenatal, perinatal, and postnatal stages of development.
Fig. 2.
Flow cytometry characterization of NG2 and Olig2 cell populations in postnatal mouse brain. (A) Flow analysis of full thickness dorsal cortical mantle from postnatal day 1 (PN1) mouse for NG2 (rabbit polyclonal; Chemicon) and Olig2 (mouse monoclonal, TV37–1C10) (n = 2 animals). (B) Flow analysis of adult mouse dorsal neocortex for NG2 (rabbit polyclonal; Chemicon) and Olig2 (mouse monoclonal TV37–1C10) (n = 2 animals). Note that, at both stages, >90% of NG2 cells are Olig2+. Conversely, NG2 cells represent <31% of the total Olig2+ cell population.
Olig Function Is Required for Early NG2 Cell Development in the Brain.
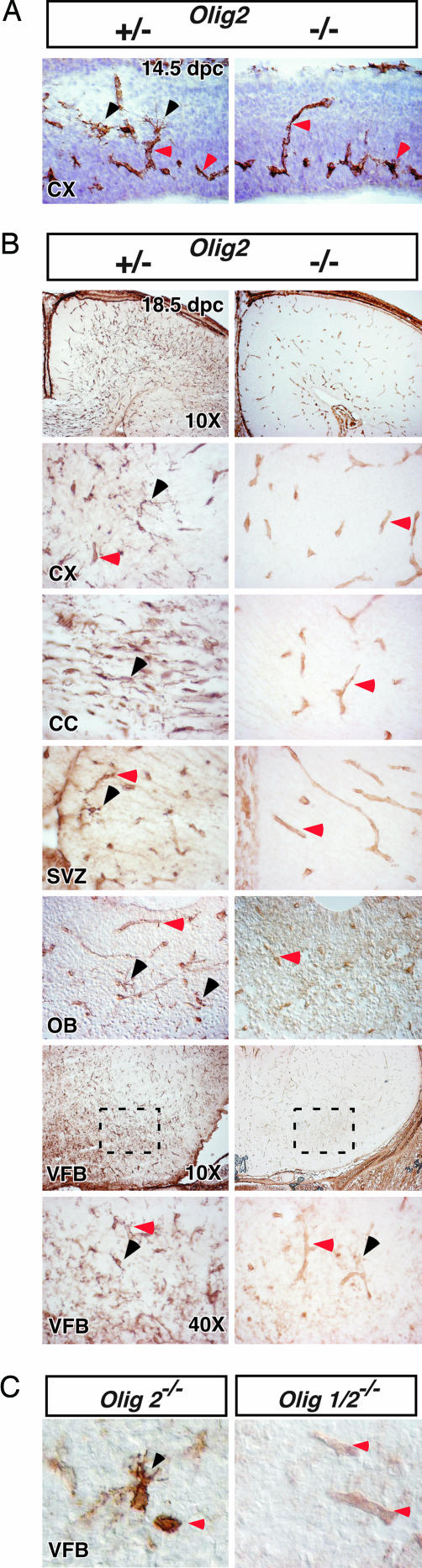
The earliest development of NG2 cells was observed at 14.5 dpc in the mouse CNS (7), so we next tested whether NG2 cell specification could occur in the absence of Olig2 function. Analysis of the brains of Olig2−/− mice at 14.5 dpc showed no NG2+ cells at this stage in mutants, whereas scattered cells were readily noted in the developing cortex of heterozygous littermates (Fig. 3A). To determine whether NG2 cell development might be merely delayed in the absence of Olig2 function, we also examined mutant and heterozygous brains at 18.5 dpc (Fig. 3B), the latest stage of development possible for Olig2−/− mice. Analysis of dorsal neocortex, corpus callosum, SVZ, and olfactory bulb showed no evidence of NG2 cells with normal neural morphology (Fig. 3B). Interestingly, within the ventral forebrain in the region of the anterior commissure, rare NG2+ cells of unusual morphology (average of 6 cells per 16-μm coronal section of brain) were still detected (Fig. 3 B and C), suggesting that NG2 cell development could be partially supported by function of Olig1. Indeed, these cells were in regions where small populations of Olig2-independent OLPs were previously reported to develop in Olig2−/− mice (19). To test this possibility, we thus analyzed Olig1/2−/− compound mutants that lack any evidence of OLP development (17). As indicated in Fig. 3C, no cell bodies with recognizable synantocyte or bipolar morphology were detectable in Olig1/2−/− mutants. Together, these data demonstrate that Olig function is required for NG2 cell specification in the forebrain, largely because of critical roles of Olig2.
Fig. 3.
Olig genes are required for NG2 cell development in the brain. (A) IHC for NG2 in 14.5 dpc mouse brain demonstrates the normal appearance of multiprocessed NG2 cells in Olig2+/− mice (black arrowheads) and their absence in Olig2−/− mice. NG2 staining in vessel pericytes is unaffected (red arrowheads). (B) IHC at 18.5 dpc demonstrates complete loss of parenchymal NG2 cells (black arrowheads) in the dorsal neocortex (cx), corpus callosum (cc), subventricular zone (svz), and olfactory bulb (ob) of Olig2−/− mice. Scattered surviving cells were detected in the ventral forebrain (vfb; boxed regions shown at higher power below). (C) The rare surviving NG2 cells in Olig2−/− ventral forebrain had abnormal morphology (black arrowheads). However, analysis of ventral forebrain in Olig1/2−/− mice showed complete ablation of normal and abnormal appearing NG2 cells even within the ventral forebrain.
Olig2 Function Is Required for Spinal Cord NG2 Cell Development.
In the embryonic spinal cord, Olig activity is essential for patterning of the ventral “pMN” domain (≈8–9.5 dpc), as well as development of motor neurons (9–10.5 dpc) and an early wave of oligodendrocytes (12.5 dpc) (23). At later stages, a dorsal origin of oligodendrocytes has recently been reported in the fetus (26–29). To determine whether dorsal or ventral spinal NG2 cell development requires Olig function during the embryonic and/or fetal phases, we first investigated Olig2−/− mutant spinal cord at 18.5 dpc. As shown in Fig. 4A, no definitive NG2+ cells were observed in spinal cords of Olig2−/− mice, in contrast to heterozygotes, where NG2 cells were readily detected. Thus, NG2+ cells arising from both ventral and dorsal regions require Olig function.
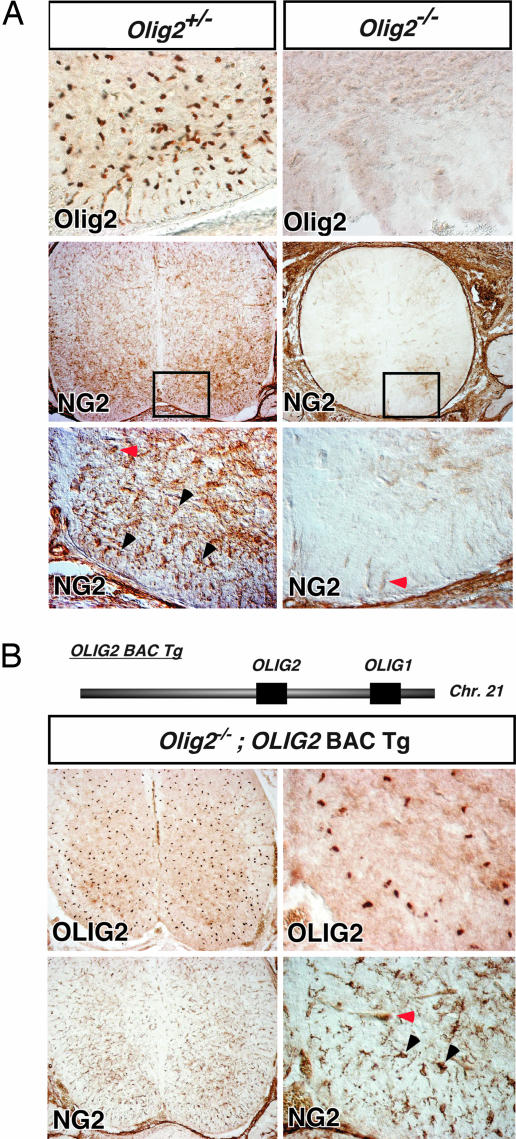
Fig. 4.
Olig2 function is necessary and sufficient for NG2 cell development in the spinal cord. (A) IHC staining for Olig2 and NG2 in 18.5 dpc Olig2+/− and Olig2−/− mouse cervical spinal cord. No cells with NG2 morphology were detected in the absence of Olig2 expression and function. (B) Diagram of 116-kB human BAC clone 2401C4 containing OLIG1 and OLIG2 loci. Crossing of a transgenic mouse line containing this BAC (OLIG2-BAC-Tg) into the Olig2−/− mouse line rescues appropriate expression of OLIG2 and promotes OLP development at 18.5 dpc mouse in the spinal cord (30). Concomitant with OLP rescue, NG2 cells with complex morphology were identified (black arrowhead).
To further investigate the genetic requirement of Olig2 in NG2 cell development, we introduced a bacterial artificial chromosome (BAC) transgenic allele (OLIG2-BAC-Tg) that carries the human OLIG2 and OLIG1 loci and partial regulatory sequences (30) into the Olig2−/− line by intercross. Expression of human OLIG2 in the Olig2−/−:OLIG2-BAC-Tg line leads to rescue of OLP development (30), in light of the fact that Olig1 is incapable of supporting OLP specification in Olig2 null mutants (19). Concomitant with OLP rescue, we noted effective production of NG2 cells with normal morphology and distribution in the spinal cord of Olig2−/−:OLIG2-BAC-Tg compound animals at all levels examined (Fig. 4B). Together, these findings show that Olig2 activity is critical for NG2 gliogenesis and indicate further developmental parallels between OLPs and NG2 cells.
Discussion
Expanded Roles for Olig Genes in Neural Progenitor Cell Development.
Extensive analysis has shown critical roles for Olig basic helix–loop–helix transcription factors in embryonic oligodendrocyte development (17–19). Recent findings also suggest that Olig2 might have more general roles in other populations of neural stem/progenitor cells, specifically radial glia and adult type C progenitor cells of the SVZ (22, 26). NG2 cells are the most prevalent cycling progenitors in the adult CNS and are the primary responsive CNS cell type to neurological injury and repair (11). Our data clearly link the bulk of the NG2 progenitor cell population to Olig2 expressing cell populations within the CNS by demonstrating that NG2 cells express Olig2 and, more importantly, that Olig2 is genetically required for NG2 cell development in the embryonic and perinatal stages. Given the diverse functions associated with NG2 cells in the literature, these studies expand the potential roles of Olig genes in maintenance of neurologic function and pathological states such as CNS injury (11) and brain tumors (24) where NG2 cells have been suggested to have important roles.
Olig2 Expression in NG2 Synantocytes Supports a Lineage Connection to Oligodendroglia.
Our findings demonstrate >90% of NG2 cells express Olig2, a pan-oligodendroglial cell type marker within the neonatal and adult rodent brain (24). Given that NG2 cells express other oligodendroglial markers including Olig1, PDGFRα, O4, and PLP (7), our findings provide further evidence for the relationship of NG2 cells to OLPs. The finding that only ≈1% of NG2 cells in the adult brain express the oligodendroglial marker tetraspanin (CD9) has been used to argue that NG2 cells comprise a fourth neuroepithelial lineage (31). However, we find that CD9 marks only a minority (≈1%) of Olig2+ cells at 18.5 dpc, whereas more established OLP markers, such as PDGFRα, overlap with Olig2 to a much higher degree (K.L.L. and D.H.R., unpublished observations), consistent with the possibility that CD9 grossly under reports OLP numbers. Thus, our expression analysis suggests that the vast majority of antenatal and postnatal parenchymal NG2 cells express Olig2 and other oligodendroglial markers.
Our analyses were in good agreement with previous studies showing NG2 coexpression with PDGFRα (1), and studies that suggest NG2 cells are a relatively small subpopulation of Olig2+ cells. For example, Buffo et al. (11) recently reported that NG2 cells are <30% of the total Olig2+ cell population in the adult brain. The cell types most likely contributing to the Olig2+NG2− population are newly specified premyelinating OLPs, myelinating oligodendrocyte, radial glia, and type C cells. Another minor population of Olig2+, GFAP+, NG2− cells has also been reported at P7 (32).
Although most NG2 cells definitively coexpress Olig2, rare cells were noted that were NG2+ but were not convincingly associated with nuclear Olig2 staining. These cells were scored as NG2+Olig2− for purposes of our study but often had a morphology that was difficult to reliably distinguish from expression of NG2 within pericytes around blood vessels, suggesting that the percentage of cells that are NG2+Olig2− may in reality be even lower than those we have reported. Further analysis using additional techniques to analyze such potentially small cell populations would be necessary to characterize the nature of these cells and to determine whether they might have unique functions.
Olig Function Is Necessary for Development of NG2-Positive Synantocytes.
We asked whether bipolar NG2 cells and/or synantocytes could develop independently of the oligodendrocyte lineage by analysis of Olig null mutants from 14.5 to 18.5 dpc. Our data demonstrate that Olig gene function is necessary for NG2 cell development throughout the CNS at antenatal stages. Although this result does not prove the existence of a uniform lineage, it establishes a common genetic requirement for Olig function within glial precursors for OLPs and NG2 cells, including both bipolar NG2+ OLPs and synantocytes. In summary, our results, coupled with previous findings, suggest that prenatal OLPs can assume at least two distinct fates: myelinating oligodendrocytes or synantocytes. This idea is supported by two lines of evidence: (i) OLPs and synantocytes are immunophenotypically identical in that there are no markers specific to synantocytes, and (ii) our functional data indicate that both cells derive from an Olig2 precursor.
An independent function for Olig2 in glial subtype determination has been suggested, based on the ability of Olig2 to suppress the astrocyte fate in cultures of spinal cord progenitors established after motoneuron differentiation is no longer occurring (33). Such in vitro data do not address the issue of secondary effects of the early patterning defects on oligodendrogenesis that occur in Olig mutants in vivo. Although the OLIG2-BAC-Tg transgene lacks regulatory elements for expression of OLIG2 at early stages of neural tube formation, and therefore fails to rescue the normal patterning of the pMN domain (30), we found that expression from this line rescues the OLP/NG2 developmental defect in Olig2−/− mice. These data therefore suggest that Olig2 is required for NG2 fate specification, independently of its role in neural tube patterning.
NG2 cells have been proposed to be the in vivo correlate of O2A glial-restricted progenitors, which, under in vitro culture conditions, can give rise to oligodendrocytes and type II astrocytes. One possibility, therefore, is that Olig function is only required within NG2 cells to suppress their astrocytic potential and promote their later development into oligodendrocytes. If so, developmental analysis of Olig-null mutants would be expected to show little to no effects on NG2+ cell specification or development, but would exhibit compromised potential of NG2 cells to become OLPs. In fact, we found a failure to produce any parenchymal NG2 cells, suggesting that Olig function is required for initial NG2 cell development per se. Although our data do not definitively distinguish whether Olig2 is required to specify or maintain an early NG2 population, given the known functions of Olig2 in specification of the oligodendrocyte lineage, we speculate that Olig2 is also required for specification of NG2 cells. Further, our results do not establish a function for Olig2 in regulation of NG2 expression.
Our findings suggest the possibility that postnatal NG2 progenitor cell development may also depend on Olig function. Although recent work has shown that Olig1 function is dispensable for NG2 cell development in the adult neocortex (20, 21), this finding would still be consistent with our observations of nearly or totally absent NG2 cell development in Olig2 and Olig1/2 null mutants, respectively. Because Olig2 mutants die at birth, we were unable to determine the later roles for Olig2 in adult progenitor populations. However, this important issue could be further addressed by using conditional knockout approaches. Additionally, our work suggests that use of transgenic mouse lines with a targeted Olig2 locus (i.e., Olig2-cre) may allow determination of the in vivo fate of postnatal NG2 cells and may be used as a molecular handle to address the role Olig2/NG2 progenitors play in the setting of brain injury and brain cancer.
Materials and Methods
Transgenic Mice.
Mice that carry targeted mutations of the Olig2 and Olig1/2 loci have been described in refs. 17 and 19. Heterozygous Olig2 mutant mice that were hemizygous for two copies of a human BAC transgene encompassing the OLIG1 and OLIG2 locus on chromosome 21 (line #34) (30) were sibling-mated to generate homozygous Olig2−/−:BAC-Tg/+ and heterozygous Olig2+/−:BAC-Tg/+ offspring. Genotypes were determined by PCR on tail-tip DNA. To evaluate OLP and NG2 cell rescue, two animals of each genotype were analyzed.
Production of Olig Monoclonal Antibody TV371C10.
A GST-fusion protein containing the amino-terminal 108 amino acids of murine Olig2 was injected into mice by the Monoclonal Antibody Core (Dana-Farber Cancer Institute, Boston). At 8 weeks after the initial injection of antigen, the spleen was harvested to produce hybridomas, and these hybridomas were screened for reactivity to Olig2 by immunoblot and immunostaining. This procedure identified the subclone TV37–1C10 as having specific and effective activity.
Tissue Preparation and IHC.
Mouse embryos were collected at different ages according to the assumption that mating occurred at midnight with plug detection at the following noon (0.5 dpc). Embryos were fixed in 4% paraformaldehyde (PFA) in PBS (pH 7.0), whereas adult mice and rats were cardiac perfused with 4% PFA, and the brain was harvested and further fixed in PFA for 24 h. All tissues were washed in 0.5 M sucrose overnight and embedded in optimal cutting temperature (OCT) compound for cryosectioning at 16 μm.
IHC was performed on cryosections using the following primary antibodies: (i) for immunofluorescence, monoclonal guinea pig anti-NG2 (gift of W. Stallcup, Burnham Institute, La Jolla, CA) and polyclonal rabbit anti-Olig2 (DF308; ref. 24); (ii) for diaminobenzidine (DAB), brightfield staining polyclonal rabbit anti-NG2 (Chemicon; AB5320); (iii) for brightfield, double staining polyclonal rabbit anti-NG2 (Chemicon; AB5320) was combined with mouse monoclonal anti-Olig2 (TV73–1C10) for all counts except for one adult brain, which used rabbit polyclonal anti-Olig2 (DF308). Identical results were obtained regardless of the Olig2 antibody used. Rabbit on rabbit combination of antibody was performed sequentially and with inactivation of the first primary antibody before application of the second primary antibody. DAB brightfield IHC was performed by using the Envision+ and Envision Doublestain Systems (DAKO; K4011, K1395) according to the manufacturer's instructions. Double IHC staining was detected by using horseradish peroxidase-DAB reaction and AP-Fast Red reaction. All IHCs were performed by using heat antigen retrieval.
Cell Counting.
Expression analysis of NG2 and Olig2 was performed by using multiple techniques for cross validation of results. To achieve more precise colocalization information, immunofluorescent analysis was performed by using deconvolution microscopy and three-dimensional reconstruction of image stacks for manual cell counting and accurate distinction of NG2 staining in pericytes from neural cells with processes. Pericytes were identified based on morphology alone, because positive identification by colabeling with antibody against PDGFRβ (Santa Cruz Biotechnology; sc-432) was technically unsuccessful in our hands. Immunofluorescent analysis was performed at the rostral and medial forebrain levels at several subregions (n = 1). To better evaluate the total NG2 population, quantitation was also performed by using the more sensitive technique of double brightfield IHC. Manual cell counting was done at rostral and medial forebrain levels in normal mouse brains from 18.5 dpc and adult (n = 2 animals each). Data from fluorescent and brightfield methods at 18.5 dpc were summed for purposes of reporting.
Mouse Brain Dissociation.
Normal C57BL/6J postnatal day 1 dorsal cortical mantle (full thickness) and adult mouse dorsal neocortex (all layers, but excluding subcortical structures) were dissected by using a dissecting microscope and placed in DMEM solution on ice. Tissue was washed in artificial cerebrospinal fluid (aCSF) and acutely dissociated as described (34), with modification in tissue dissociation medium-collagenase IV (1 mg/ml; Worthington), hyaluronidase (0.67 mg/ml; Sigma), DNase I (0.4 mg/ml; Worthington), and kynurenic acid and N-acetylcysteine (60 μg/ml; Sigma). Cells were washed and stained in the fresh state for FACS analysis. Trypsin was avoided because of higher proteolytic activity and potential cleavage of cell surface proteins (35).
Flow Cytometry.
Fluorescent activated cytometry was performed on a FACSCalibur machine (BD Biosciences). Antibodies included: mouse anti-Olig2 (TV37–1C10) and polyclonal rabbit anti-NG2 (Chemicon; AB5320). Cells were first stained for surface NG2 (antibody dilution 1:100) for 30 min at 4°C and then washed, fixed, and permeabilized to allow access to intracellular antigens (Cytofix/Cytoperm solution; BD Biosciences; no. 554714). Cells were then further incubated with Olig2 antibody (dilution 1:100) for 30 min at 4°C and washed. Cells were incubated with secondary antibodies (anti-rabbit Alexa488 and anti-mouse Cy5; Molecular Probes) for 30 min at 4°C and washed before FACS analysis. Single positives and isotype stains were used as controls for compensation and gating thresholds. Data were analyzed by using flowjo software v6.2.1 (Tree Star, Ashland, OR).
Acknowledgments
We thank D. Yuk and S. Kaing (both of the Dana-Farber Cancer Institute, Boston) for expert technical assistance and W. Stallcup (Burnham Institute, La Jolla, CA) for kindly providing NG2 antibodies. This work was supported by National Institutes of Health Grants K08NS047213 (to K.L.L.) and R01NS40511 (to D.H.R.). D.J.A. is a Howard Hughes Medical Investigator, and D.H.R. is a Harry Weaver Scholar of the National Multiple Sclerosis Society.
Abbreviations
- BAC
bacterial artificial chromosome
- dpc
days postcoitum
- IHC
immunohistochemical
- OLP
oligodendrocyte progenitors
- SVZ
subventricular zone.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nishiyama A., Lin X. H., Giese N., Heldin C. H., Stallcup W. B. J. Neurosci. Res. 1996;43:299–314. doi: 10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 2.Levine J. M., Card J. P. J. Neurosci. 1987;7:2711–2720. doi: 10.1523/JNEUROSCI.07-09-02711.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stegmuller J., Schneider S., Hellwig A., Garwood J., Trotter J. J. Neurocytol. 2002;31:497–505. doi: 10.1023/a:1025743731306. [DOI] [PubMed] [Google Scholar]
- 4.Greenwood K., Butt A. M. Mol. Cell. Neurosci. 2003;23:544–558. doi: 10.1016/s1044-7431(03)00176-3. [DOI] [PubMed] [Google Scholar]
- 5.Dawson M. R., Polito A., Levine J. M., Reynolds R. Mol. Cell. Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 6.Horner P. J., Thallmair M., Gage F. H. J. Neurocytol. 2002;31:469–480. doi: 10.1023/a:1025739630398. [DOI] [PubMed] [Google Scholar]
- 7.Dawson M. R., Levine J. M., Reynolds R. J. Neurosci. Res. 2000;61:471–479. doi: 10.1002/1097-4547(20000901)61:5<471::AID-JNR1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 8.Aguirre A. A., Chittajallu R., Belachew S., Gallo V. J. Cell Biol. 2004;165:575–589. doi: 10.1083/jcb.200311141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S. C., Huck J. H., Roberts J. D., Macklin W. B., Somogyi P., Bergles D. E. Neuron. 2005;46:773–785. doi: 10.1016/j.neuron.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 10.Levine J. M., Reynolds R., Fawcett J. W. Trends Neurosci. 2001;24:39–47. doi: 10.1016/s0166-2236(00)01691-x. [DOI] [PubMed] [Google Scholar]
- 11.Buffo A., Vosko M. R., Erturk D., Hamann G. F., Jucker M., Rowitch D., Gotz M. Proc. Natl. Acad. Sci. USA. 2005;102:18183–18188. doi: 10.1073/pnas.0506535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters A. J. Neurocytol. 2004;33:345–357. doi: 10.1023/B:NEUR.0000044195.64009.27. [DOI] [PubMed] [Google Scholar]
- 13.Ye P., Bagnell R., D'Ercole A. J. J. Neurosci. 2003;23:4401–4405. doi: 10.1523/JNEUROSCI.23-11-04401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiyama A. Hum. Cell. 2001;14:77–82. [PubMed] [Google Scholar]
- 15.Nishiyama A., Yang Z., Butt A. J. Anat. 2005;207:687–693. doi: 10.1111/j.1469-7580.2005.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butt A. M., Hamilton N., Hubbard P., Pugh M., Ibrahim M. J. Anat. 2005;207:695–706. doi: 10.1111/j.1469-7580.2005.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Q., Anderson D. J. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 18.Takebayashi H., Nabeshima Y., Yoshida S., Chisaka O., Ikenaka K. Curr. Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q. R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C. D., Rowitch D. H. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 20.Xin M., Yue T., Ma Z., Wu F. F., Gow A., Lu Q. R. J. Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnett H. A., Fancy S. P., Alberta J. A., Zhao C., Plant S. R., Kaing S., Raine C. S., Rowitch D. H., Franklin R. J. M., Stiles C. D. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- 22.Hack M. A., Saghatelyan A., de Chevigny A., Pfeifer A., Ashery-Padan R., Lledo P. M., Gotz M. Nat. Neurosci. 2005;8:865–872. doi: 10.1038/nn1479. [DOI] [PubMed] [Google Scholar]
- 23.Rowitch D. H. Nat. Rev. Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- 24.Ligon K. L., Alberta J. A., Kho A. T., Weiss J., Kwaan M. R., Nutt C. L., Louis D. N., Stiles C. D., Rowitch D. H. J. Neuropathol. Exp. Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 25.Ozerdem U., Grako K. A., Dahlin-Huppe K., Monosov E., Stallcup W. B. Dev. Dyn. 2001;222:218–227. doi: 10.1002/dvdy.1200. [DOI] [PubMed] [Google Scholar]
- 26.Fogarty M., Richardson W. D., Kessaris N. Development. 2005;132:1951–1959. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- 27.Cai J., Qi Y., Hu X., Tan M., Liu Z., Zhang J., Li Q., Sander M., Qiu M. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 28.Vallstedt A., Klos J. M., Ericson J. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 29.Miller R. H. Neuron. 2005;45:1–3. doi: 10.1016/j.neuron.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 30.Sun T., Hafler B. P., Kaing S., Kitada M., Ligon K. L., Widlund H. R., Yuk D. I., Stiles C. D., Rowitch D. H. Dev. Biol. 2006;292:152–164. doi: 10.1016/j.ydbio.2005.12.047. [DOI] [PubMed] [Google Scholar]
- 31.Berry M., Hubbard P., Butt A. M. J. Neurocytol. 2002;31:457–467. doi: 10.1023/a:1025735513560. [DOI] [PubMed] [Google Scholar]
- 32.Marshall C. A., Novitch B. G., Goldman J. E. J. Neurosci. 2005;25:7289–7298. doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabay L., Lowell S., Rubin L. L., Anderson D. J. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- 34.Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 35.Anderson R. C., Elder J. B., Brown M. D., Mandigo C. E., Parsa A. T., Kim P. D., Senatus P., Anderson D. E., Bruce J. N. Clin. Immunol. 2002;102:84–95. doi: 10.1006/clim.2001.5152. [DOI] [PubMed] [Google Scholar]