Abstract
Ciliary neurotrophic factor (Cntf) plays an essential role in postnatal maintenance of spinal motoneurons. Whereas the expression of this neurotrophic factor is low during embryonic development, it is highly up-regulated after birth in myelinating Schwann cells of rodents. To characterize the underlying transcriptional mechanisms, we have analyzed and compared the effects of various glial transcription factors. In contrast to Pit-1, Oct-1, Unc-86 homology region (POU) domain class 3, transcription factor 1 (Oct6/SCIP/Tst-1) and paired box gene 3 (Pax3), SRY-box-containing gene 10 (Sox10) induces Cntf expression in Schwann cells. Subsequent promoter analysis using luciferase reporter gene and EMSA identified the corresponding response elements within the Cntf promoter. Overexpression of Sox10 in primary sciatic nerve Schwann cells leads to a >100-fold up-regulation of Cntf protein, and suppression of Sox10 by RNA interference in the spontaneously immortalized Schwann cell line 32 reduces Cntf expression by >80%. Mice with heterozygous inactivation of the Sox10 gene show significantly reduced Cntf protein levels in sciatic nerves, indicating that Sox10 is necessary and sufficient for regulating Cntf expression in the peripheral nervous system.
Keywords: promoter, Hirschsprung disease, Oct6, neuropathy, Pax3
Schwann cells play an important role for maintenance of motoneurons. Their development is regulated by various transcription factors (1), including paired box gene 3 (Pax3) (2), Pit-1, Oct-1, Unc-86 homology region (POU) domain class 3, transcription factor 1 (Oct-6/SCIP/Tst-1) (3–6), and SRY-box-containing gene 10 (Sox10) (7, 8). Ciliary neurotrophic factor (Cntf) belongs to a family of neuropoietic cytokines that promote neuronal survival (9–13) and peripheral nerve regeneration (14–16). Targeted inactivation of the Cntf gene in mice revealed its role in postnatal maintenance of motoneurons (17, 18). Moreover, endogenous Cntf modulates onset and severity of disease in patients and mouse models for motoneuron disease and other neurological disorders (19–21). In progressive motor neuronopathy and wobbler mice, Cntf treatment protects motoneurons from cell death and improves motor performance (22, 23), indicating that this factor is a major mediator of the protective effects of Schwann cells, both under physiological and pathological conditions.
Myelinating Schwann cells in the peripheral nervous system are the richest source of Cntf in adult mammals (9, 10, 24). Expression of Cntf in the peripheral nervous system rises at the end of the first postnatal week. The control mechanisms that regulate and restrict Cntf expression to myelinating Schwann cells are still not known. To analyze the regulation of Cntf expression, we have cloned a 4.7-kb fragment containing the complete 5′ region of the murine Cntf gene up to the neighboring zinc finger protein (Zfp) gene. Using deletion and mutation analysis, we found that Cntf gene expression is controlled by several glial transcriptional factors. Sox10 was identified as a key regulator of Cntf expression. Sox10 overexpression in cultured primary Schwann cells leads to a >100-fold up-regulation of Cntf protein levels. In addition, Cntf levels are significantly lower in sciatic nerves of Sox10+/− mice, indicating that Sox10 acts as a major physiological regulator of Cntf gene expression in vivo.
Results
Reduced Cntf Expression in Sox10+/− Mice.
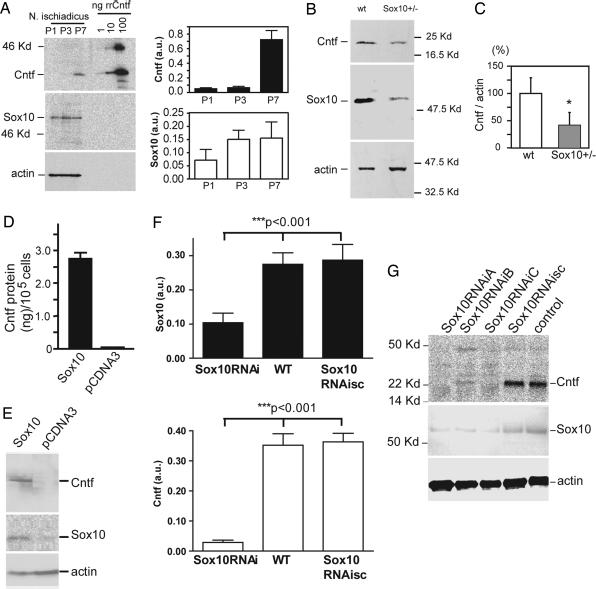
In comparison to other Schwann cell-specific transcription factors, Sox10 expression is most closely linked to Cntf up-regulation during postnatal development (8). We therefore analyzed Cntf expression in Sox10+/− mutant mice (8). Mice with homozygous mutation of the Sox10 gene already start to die around embryonic day 13.5 (25, 26). Sox10 expression increases from postnatal day (P)1 to P3, whereas a strong increase of Cntf expression was first detectable at P7 (Fig. 1A). Cntf protein levels are reduced by >50% (Fig. 1 B and C) in Sox10+/− mice.
Fig. 1.
Expression of Sox10 and Cntf in sciatic nerve of wild-type and Sox10+/− mutant mice. (A Left) Western blot analysis of sciatic nerve (nervus ischiadicus) extracts from P1, P3, and P7 wild-type mice. Western blots were probed for Sox10 and Cntf and reprobed against actin as a control for equal loading. rrCntf, recombinant rat Cntf. (A Right) Semiquantitative analysis of Cntf and Sox10 expression from three independent experiments. Error bars represent SD. (B) Western blot analysis of sciatic nerve extracts from adult wild-type and Sox10+/− mice. Cntf protein was detected as one band at 22 kDa. Sox10 protein content is reduced in sciatic nerve extract of Sox10+/− mice. (C) The density of the Cntf immunoreactive bands was measured in three independent blots. Signals were normalized against the actin signal and are shown as percentage of wild-type levels. Error bars represent SD. ∗, P < 0.05. (D) Cntf expression is elevated in primary Schwann cells after transient transfection of Sox10. Expression levels were estimated by measuring signal density from Western blots. A representative example is shown in C. Expression levels (mean ± SD) were quantified by using the aida software program. (E) Cntf expression in primary rat Schwann cells after transfection of Sox10. (F) Cntf expression is reduced after transfection of Sox10 RNA interference (RNAi) into the IMS32 Schwann cell line. Expression levels for Sox10 and Cntf were estimated by measuring signal density from Western blots. A representative example is shown in G. Expression levels (mean ± SD) were quantified by using the aida software program. (G) Cntf and Sox10 expression in native IMS32 cells and after transient transfection of Sox10RNAiA, Sox10RNAiB, or Sox10RNAiC (see also Table 1). Sox10RNAisc is a scrambled sequence RNA interference corresponding to Sox10RNAiA that was used as a specificity control.
To get more insight into the physiological relevance of this finding, we overexpressed Sox10 in primary Schwann cells from P4 rat sciatic nerves. This treatment resulted in a >100-fold induction of Cntf protein levels (Fig. 1 D and E). Thus, Sox10 appears sufficient for up-regulating Cntf expression in early postnatal Schwann cells. We then investigated the expression of Cntf in the spontaneously immortalized Schwann cell line 32 (IMS32) (27). In contrast to primary Schwann cells, this cell line expresses constitutively relatively high levels of Cntf (Fig. 1G) and therefore was used for this experiment. Suppression of Sox10 by RNA interference leads to significant reduction (P < 0.001) of Cntf expression (Fig. 1F).
Characterization of the Regulatory Elements and Transcription Initiation Sites Within the Cntf Promoter.
To characterize the Cntf promoter region, we cloned a 6.5-kb DNA fragment containing the complete Cntf gene including the 4.7-kb 5′ flanking region up to Zfp, the neighboring gene (28). The stop codon of Zfp was located at −4,647 bp from the start codon of Cntf. We performed 3′ RACE to identify the poly(A) signal for Zfp and found a canonical AATAAA site located at −4,549 bp from the Cntf ATG start codon. Because the Cntf promoter sequence does not contain a TATA box (29), we carried out 5′ RACE. For human and rat Cntf, only one initiation site has been identified so far (29). We found three additional initiation sites (−10, −32, −68, and −74 bp from the ATG start codon) for mouse Cntf (see Fig. 5, which is published as supporting information on the PNAS web site). Five, four, six, and one clone of 16 analyzed were identified for these four transcription start sites, respectively. In addition, using sequence analysis, we identified several potential binding sites for specific transcription factors including Pax3, Oct6/SCIP/Tst-1, and Sox proteins (see Figs. 5 and 6A, which are published as supporting information on the PNAS web site). Comparison of the human and mouse Cntf promoter regions revealed that the putative Sox10 binding sites are highly conserved (Fig. 6 B and C).
Analysis of the Cntf Promoter by Luciferase Reporter Assays.
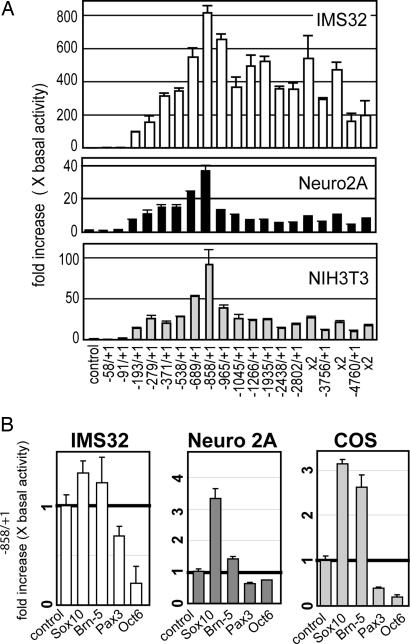
To characterize the role of the identified putative response elements for Pax3, Oct-6/SCIP/Tst-1, and Sox10 (see Figs. 5 and 6) in the Cntf promoter, various reporter gene constructs were established (see Fig. 7A, which is published as supporting information on the PNAS web site) and transfected into the Schwann cell line IMS32, the neuroblastoma cell line Neuro2A, and the green monkey simian virus 40-transfected kidney fibroblast cell line Cos7. We have included these different cell lines in our study to investigate the regulation of Cntf expression in a neural cell line (Neuro2A), in a Schwann cell line (IMS32), and in a cell line not derived from the nervous system (Cos7) as a control. The Schwann cell line IMS32 was used instead of primary Schwann cells because it was not possible to expand the primary Schwann cells at sufficient numbers for the promoter analyses. In all cell lines, the −858/+1 construct showed strongest expression of the reporter (Fig. 2A) and therefore was used for subsequent analyses. Stepwise induction of gene expression was found in three regions: from base pair −91 to base pair −193, from base pair −193 to base pair −371, and from base pair −538 to base pair −858 from the ATG codon (Fig. 2A). In contrast, the region between base pair −371 and base pair −538 did not contribute to increased reporter gene expression. Any construct longer than −858/+1 revealed no additive up-regulation. In Neuro2A and immortalized fibroblasts (NIH3T3), highly reduced reporter gene expression was observed with constructs including the base pair −858 to base pair −965 region. In IMS32 cells, strong reduction was recognized within a region from base pair −965 to base pair −1,045. This observation could indicate that regulatory elements for transcriptional repression exist distal to base pair −858. For further studies, the −4,760/+1 construct was also used because it contained the complete Cntf promoter including these putative repressor elements.
Fig. 2.
Promoter constructs and analysis of reporter expression in various cell types. (A) Sixteen luciferase reporter constructs with different lengths of the Cntf promoter were generated for transient transfection in various cell lines (Fig. 7A). Transcriptional activity was determined after transfection into the Schwann cell line IMS32, Neuro2A, and NIH 3T3 cells. Relative activity was highest in IMS32 (up to 800× basal activity). The construct −858/+1 revealed highest activity in all cell lines. One microgram of the reporter plasmid was used for transient transfection. As a control, empty vector was transfected. For constructs longer than 2.5 kb (−2,802/+1, −3,856/+1, and −4,760/+1), double amounts of plasmid were transfected to adjust for reduced number of molecules. For each construct, at least three independent transfection experiments with at least three independent plates in each experiment were performed, and luciferase activities are shown as mean ± SD. As a control, empty vector was transfected. (B) Expression plasmids for Sox10, Brn-5, Oct-6, and Pax3 were transfected into Neuro2A, Cos7, and IMS32 cells. Data show promoter activity of the cotransfected −858/+1 reporter construct relative to control (pCDNA3) transfected cells. Similar data were obtained with the −4,760/+1 construct (data not shown).
In the IMS32 Schwann cell line, basal levels for Cntf reporter gene activity were ≈27-fold higher than in Neuro2A cells and 20-fold higher than in Cos7 cells. We then transfected the −856/+1 reporter gene construct with expression plasmids for Pax3, Oct-6/SCIP/Tst-1, Sox10, and Pit-1, Oct-1, Unc-86 conserved region (POU) domain, class 6, transcription factor 1 (Brn5) (Fig. 2B). Cos7 cells do not express endogenous Pax3 (30), Oct-6/SCIP/Tst-1 (31), and Sox10 (32). Neuro2A cells do not express Pax3, Oct-6/SCIP/Tst-1, and Sox10 (2, 33). In both cell lines, transfection of Sox10 induced reporter gene expression ≈3-fold above baseline level (Fig. 2B). In IMS32 cells, Sox10, Pax3, Brn5, and Oct6/SCIP/Tst-1 are expressed endogenously (27, 34). This finding could explain why only a slight induction was observed after Sox10 or Brn5 transfection in IMS32 cells. Results were virtually identical with the −858/+1 construct and the −4,760/+1 promoter construct (data not shown). Brn5 led to enhanced promoter activity in Cos7 cells but not in Neuro2A or IMS32 cells. Highest induction was observed in Sox10-transfected cell lines, whereas Pax3 and Oct6 repressed Cntf reporter activity in all cell lines.
Identification of Sox10 Responsive Elements in the Cntf Promoter.
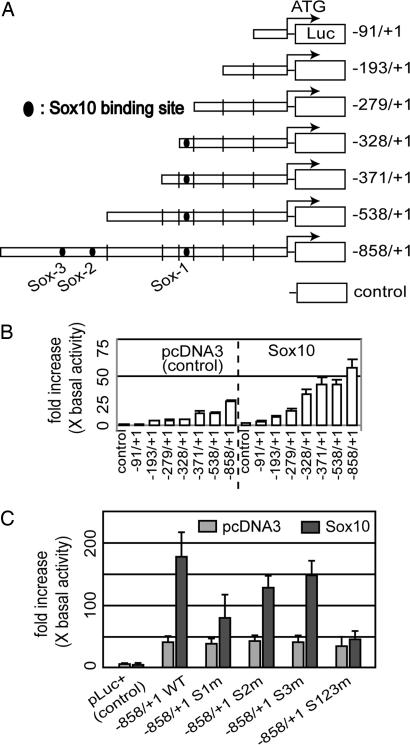
We then performed reporter assays to further characterize Sox10-dependent activation. The Neuro2A cell line was used because it does not express Sox10 endogenously and thus could be used for cotransfection of truncated Cntf promoter-luciferase plasmids and Sox10-expression vectors (Fig. 3A). Sox10-dependent induction was found with constructs including regions between base pair −279 and base pair −328 and between base pair −538 and base pair −858 (Fig. 3B).
Fig. 3.
Effect of Sox10 on Cntf promoter activity. (A) Cotransfection of Sox10 with Cntf promoter-luciferase constructs of different lengths into Neuro2A cells. The predicted Sox10 responsive regions are shown as filled circles. (B) Sox10-dependent induction was recognized between base pair −279 and base pair −328 and between base pair −538 and base pair −858. (C) Three Sox10 binding sites, S1 (base pair −287 to base pair −293), S2 (base pair −592 to base pair −600), and S3 (base pair −660 to base pair −666) were identified with mutant reporter constructs (Fig. 7B) by cotransfection with Sox10. S1m showed remarkable reduction (44%), S2m showed lower reduction (72%), and S3 showed only modest reduction (83% activity in comparison to nonmutant wild-type construct). Mutation of all three Sox10 binding sites virtually abolished Sox10-dependent promoter activity.
Sox10 is a high mobility group type DNA-binding protein. High mobility group proteins bind to a 7-bp consensus sequence (A/T)(A/T)CAA(A/T)G (33, 35). Such consensus elements are found at base pair positions −287 and −293 of the Cntf promoter, and two additional elements with one mismatch are localized at base pair −592 to base pair −600 and base pair −660 to base pair −666 (Fig. 5). We designated these sequences as “Sox10 high mobility group type DNA-binding motif” 1 (S1) (base pair −287 to base pair −293), S2 (base pair −592 to base pair −600), and S3 (base pair −660 to base pair −666), respectively. To evaluate the influence of Sox10 on these promoter sites, we produced mutant reporter gene constructs by replacing the consensus sequences with GCGC base pairs (see Table 1, which is published as supporting information on the PNAS web site). These mutant reporter constructs (Fig. 7B) were cotransfected with Sox10 expression plasmids into Neuro2A cells, and promoter activity was measured by luciferase assays (Fig. 3C). In comparison to the wild-type Cntf promoter construct, mutated S1 (S1m) showed a 56% reduction, S2m showed a modest reduction (28%), and S3 showed a weak reduction (17%). These results indicate that these putative Sox10 binding sites are important for Cntf expression but that their contributions are not equivalent. The S1 site appears most critical for Sox10-dependent induction. Combined mutation of all three putative Sox10 binding sites (S123m) resulted in a virtually complete loss of luciferase reporter gene induction by Sox10 (Fig. 3C).
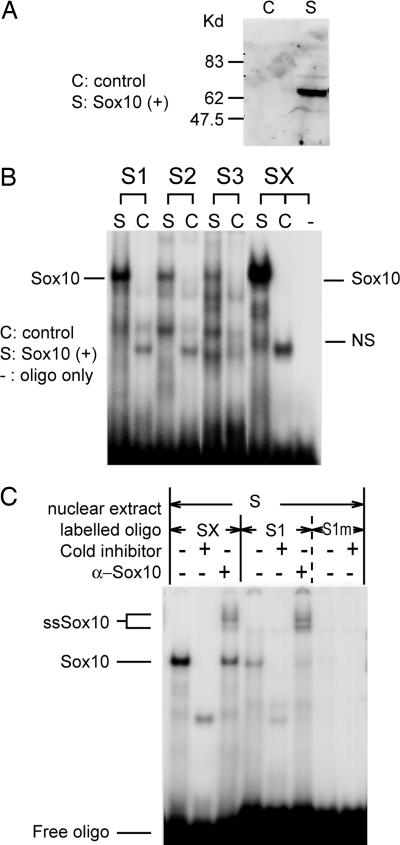
We also performed EMSA to demonstrate direct binding of Sox10 to the S1, S2, and S3 sites. Nuclear extract from COS cells expressing Sox10 protein (Fig. 4A) showed shifted bands with a positive control [Sox10 consensus high mobility group type DNA-binding motif (SX)] and S1-, S2-, and S3-specific oligonucleotides (Fig. 4B). Furthermore, a supershifted band was recognized with a Sox10 antiserum for the S1 site (Fig. 4C). Weak but specific supershifted bands were also recognized for the S2 and S3 sites (results not shown). There was no oligo–protein complex recognized (Fig. 4C) with mutated oligos for the S1 site (see Table 1), indicating specificity of the supershift experiment.
Fig. 4.
Sox10 binding to S1, S2, and S3 sites in the Cntf promoter. (A) Western blot analysis of nuclear extract from COS cells transfected by Sox10 expression vector (S) and pcDNA3 control vector (C). Sox10 immunoreactivity was recognized as a single band at 64 kDa. (B) Sox10 binds to S1, S2, and S3 sites. SX is a consensus oligonucleotide for Sox10 (Table 1). S indicates the nuclear extract cotransfected with Sox10 expression vector, and C indicates the nuclear extract cotransfected with pcDNA3 empty vector. NS indicates nonspecific band. (C) A supershifted band was recognized with the Sox10 antiserum and oligonucleotide corresponding to the Sox1 site. Weak interaction was also observed for S2 and S3 (unpublished data). No oligo–protein complex recognized with mutated oligo (S1m). ssSox10, supershifted Sox10.
Discussion
Control of myelin gene expression involves concerted actions of several transcription factors (1). Here we show that Cntf expression is also controlled by transcription factors previously identified in the context of Schwann cell differentiation and myelin formation. Among these, Sox10 plays a pivotal role for Cntf up-regulation in early postnatal Schwann cells. After birth, Sox10 expression is found at P1 and strongly increases at P3 in mice. The raise in Sox10 levels is followed by a strong increase of Cntf expression at P7. Moreover, the importance of Sox10 is underlined by our observation that Cntf protein levels are significantly reduced in sciatic nerves of Sox10+/− mice, even stronger than levels of P0 protein (data not shown).
Surprisingly, Pax3 appears as a repressor of Cntf promoter activity in our cotransfection studies. Pax3 also reduced the positive regulatory effect of Sox10. Pax3 is indispensable for the differentiation of neural crest derivatives (36). Its expression in peripheral nerves of embryonic mice is high, and expression goes down around birth before Cntf expression is up-regulated. It appears tempting to speculate that Pax3 represses expression of Cntf during embryonic development and that the down-regulation of Pax3 around birth contributes to the massive up-regulation of Cntf production starting in the first postnatal week.
Interestingly, patients with deletion or mutations in the Sox10 gene reveal defects in the peripheral nervous system known as Hirschsprung disease (37, 38). In such patients, defects of the enteric ganglia and subsequently a lack of coordinated innervation of bowel and gut are prevailing, but peripheral neuropathies are also part of the disease phenotype (39). It is not clear whether the neuropathy can simply be explained as a defect in proper myelination in such patients (40). It is likely that Sox10 controls additional target genes that influence maintenance and function of the peripheral nervous system. Thus, reduced Cntf levels could contribute to this phenotype. Levels of Cntf protein in human peripheral nerves are much lower than in mice (M.S., unpublished observations), and human CNTF is less active than the mouse Cntf protein, even in human target cells (41). Thus, reduced gene expression might be more critical in humans in comparison to mice. In postnatal Cntf−/− mice, loss of motoneurons occurs (17), and degenerative alterations in Schwann cell morphology such as disintegration of paranodal networks are detectable at nodes of Ranvier (42). Furthermore, reduced Cntf levels exaggerate the disease phenotype when combined with the superoxide dismutase 1 (SOD1) (19) gene mutation resulting in more severe motoneuron disease. Thus, reduced Cntf expression might also be relevant for the pathophysiology of neuropathies caused by mutations in Sox10 or other conditions reducing Sox10 levels.
In summary, our data indicate that Sox10 up-regulates Cntf gene expression in Schwann cells and other cell types and that Cntf protein levels are reduced in Sox10 mutant mice. Dysregulation of Cntf expression could contribute to the phenotype of specific neuropathies, in particular those caused by mutations in Sox10.
Materials and Methods
Cloning and Sequencing of Murine Cntf Promoter.
A genomic clone containing a 6.5-kb insert with the complete Cntf gene was obtained by screening bacterial artificial chromosome filters (Incyte Genomics, Palo Alta, CA). Digestion of a positive bacterial artificial chromosome with EcoRV, subcloning into pBSII KS+/−, and DNA sequencing revealed that this clone also contained the 4.7-kb 5′ promoter region of Cntf including the 3′ region of the Zfp gene, which is located upstream of Cntf.
Computer Analysis of Putative Binding Sites for Transcription Factors in the Cntf Promoter.
Programs provided by the Genetics Computer Group (Madison, WI) and matinspector professional(Genomatix, Munich) (43) were used for sequence analysis of the Cntf promoter.
RNA Extraction, 5′ and 3′ RACE (Oligo Cap Methods).
Total RNA was isolated from adult mouse sciatic nerve by TRIzol (Invitrogen) according to the manufacturer’s protocol. The 5′ and 3′ RACE were performed as described in Supporting Methods, which is published as supporting information on the PNAS web site.
Plasmid Constructs, Cell Culture, Transfection, and Luciferase Assays.
DNA corresponding to various regions of the Cntf promoter was cloned by PCR with appropriate sense and antisense primers (Cz10rev2) (Table 2, which is published as supporting information on the PNAS web site). Cloning of expression and reporter constructs is described in Supporting Methods.
NIH 3T3, Cos7, Neuro2A, and IMS32 Schwann cell lines (27) and primary Schwann cells were maintained in Dulbecco–Vogt-modified Eagle’s medium with 10% FCS, 50 units/ml penicillin, and 100 μg/ml streptomycin. Transfection was performed as described in Supporting Methods and Table 1.
Nuclear Protein Extraction and Western Blotting.
Cos7 and Neuro2A cells were transfected with 37.5 μg of expression vectors for Sox10 in 100-mm dishes. In parallel, pcDNA3 vector was transfected as a control. After 24 h, cells were harvested, and nuclear extracts were collected (44). Protein concentration was measured by using a Bradford assay. Nuclear protein extracts (25 μg) were subjected to Western blotting. Anti-Sox10 antibody (45) was used at a 1:3,000 dilution. The Cntf antiserum K10 was used at a 1:5,000 dilution. Recombinant rat Cntf was used as a standard for analysis of Cntf expression levels.
EMSA.
Double-stranded 30- or 34-bp oligonucleotides (1 μg) were radiolabeled by extension of overhanging GATC 4-bp ends with Klenow fragment of DNA polymerase in the presence of [α-32P]dCTP. Labeled oligonucleotide and 4 μg of nuclear protein were incubated in 20 μl of reaction mixture. Reaction mixture for Sox10 contained 10 mM Hepes (pH 8.0), 50 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 5% glycerol, 10% BSA, and 2 μg of poly[d(I)d(C)] (Sigma). For competition experiments, unlabeled double-stranded oligonucleotides were preincubated in 250-fold excess with the protein for 20 min before labeled oligonucleotides were added. Supershifts were performed by using an antibody to Sox10 as described (45). All samples were electrophoresed on 5% native polyacrylamide gels for 1.5 h at 140 V. Gels were dried after electrophoresis and exposed to X-OMAT film (Kodak) to detect radioactivity.
Western Blot Analysis.
Sciatic nerves were dissected from P1, P3, P7, and P42 wild-type and Sox10+/− mice (8). Cell lysates from Sox10 RNA interference-transfected IMS32 cells were harvested 24 h after transfection. Protein extracts were prepared and processed for Cntf and Sox10 (46) Western blot analysis. Blots were then stripped and reacted with antibodies against actin (MAB1501R; Chemicon). Signal intensities were measured and normalized against the actin signal (aida software package; Raytest, Straubenhardt, Germany). Statistical analysis was performed by using the unpaired two-tailed t test (prism; GraphPad, San Diego).
Supplementary Material
Acknowledgments
We thank Michaela Pfister for technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (SFB581), grants from the Japan Foundation for Aging and Health (to Y.I.), and the Kanae Foundation for Life & Socio-Medical Science.
Abbreviations
- Cntf
ciliary neurotrophic factor
- IMS32
immortalized Schwann cell line 32
- S1, S2 and S3
Sox10 high mobility group type DNA-binding motifs 1, 2 and 3
- S1m, S2m and S3m
mutated S1, S2 and S3
- Pn
postnatal day n.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wegner M. Glia. 2000;29:118–123. [PubMed] [Google Scholar]
- 2.Kioussi C., Gross M. K., Gruss P. Neuron. 1995;15:553–562. doi: 10.1016/0896-6273(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 3.Monuki E. S., Weinmaster G., Kuhn R., Lemke G. Neuron. 1989;3:783–793. doi: 10.1016/0896-6273(89)90247-x. [DOI] [PubMed] [Google Scholar]
- 4.Jaegle M., Mandemakers W., Broos L., Zwart R., Karis A., Visser P., Grosveld F., Meijer D. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 5.Bermingham J. R., Jr, Scherer S. S., O’Connell S., Arroyo E., Kalla K. A., Powell F. L., Rosenfeld M. G. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 6.Arroyo E. J., Bermingham J. R., Jr, Rosenfeld M. G., Scherer S. S. J. Neurosci. 1998;18:7891–7902. doi: 10.1523/JNEUROSCI.18-19-07891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlbrodt K., Schmidt C., Sock E., Pingault V., Bondurand N., Goossens M., Wegner M. J. Biol. Chem. 1998;273:23033–23038. doi: 10.1074/jbc.273.36.23033. [DOI] [PubMed] [Google Scholar]
- 8.Britsch S., Goerich D. E., Riethmacher D., Peirano R. I., Rossner M., Nave K. A., Birchmeier C., Wegner M. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stöckli K. A., Lottspeich F., Sendtner M., Masiakowski P., Carroll P., Götz R., Lindholm D., Thoenen H. Nature. 1989;342:920–923. doi: 10.1038/342920a0. [DOI] [PubMed] [Google Scholar]
- 10.Stöckli K. A., Lillien L. E., Näher-Noe M., Breitfeld G., Hughes R. A., Thoenen H., Sendtner M. J. Cell Biol. 1991;115:447–459. doi: 10.1083/jcb.115.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahl N., Yancopoulos G. D. J. Neurobiol. 1994;25:1454–1466. doi: 10.1002/neu.480251111. [DOI] [PubMed] [Google Scholar]
- 12.Ip N. Y., Yancopoulos G. D. Annu. Rev. Neurosci. 1996;19:491–515. doi: 10.1146/annurev.ne.19.030196.002423. [DOI] [PubMed] [Google Scholar]
- 13.Bonni A., Sun Y., Nadal-Vicens M., Bhatt A., Frank D. A., Rozovsky I., Stahl N., Yancopoulos G. D., Greenberg M. E. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 14.Sendtner M., Kreutzberg G. W., Thoenen H. Nature. 1990;345:440–441. doi: 10.1038/345440a0. [DOI] [PubMed] [Google Scholar]
- 15.Sendtner M., Stöckli K. A., Thoenen H. J. Cell Biol. 1992;118:139–148. doi: 10.1083/jcb.118.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sendtner M., Götz R., Holtmann B., Thoenen H. J. Neurosci. 1997;17:6999–7006. doi: 10.1523/JNEUROSCI.17-18-06999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masu Y., Wolf E., Holtmann B., Sendtner M., Brem G., Thoenen H. Nature. 1993;365:27–32. doi: 10.1038/365027a0. [DOI] [PubMed] [Google Scholar]
- 18.DeChiara T. M., Vejsada R., Poueymirou W. T., Acheson A., Suri C., Conover J. C., Friedman B., McClain J., Pan L., Stahl N., et al. Cell. 1995;83:313–322. doi: 10.1016/0092-8674(95)90172-8. [DOI] [PubMed] [Google Scholar]
- 19.Giess R., Holtmann B., Braga M., Grimm T., Muller-Myhsok B., Toyka K. V., Sendtner M. Am. J. Hum. Genet. 2002;70:1277–1286. doi: 10.1086/340427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giess R., Maurer M., Linker R., Gold R., Warmuth-Metz M., Toyka K. V., Sendtner M., Rieckmann P. Arch. Neurol. (Chicago) 2002;59:407–409. doi: 10.1001/archneur.59.3.407. [DOI] [PubMed] [Google Scholar]
- 21.Linker R. A., Maurer M., Gaupp S., Martini R., Holtmann B., Giess R., Rieckmann P., Lassmann H., Toyka K. V., Sendtner M., et al. Nat. Med. 2002;8:620–624. doi: 10.1038/nm0602-620. [DOI] [PubMed] [Google Scholar]
- 22.Sendtner M., Schmalbruch H., Stöckli K. A., Carroll P., Kreutzberg G. W., Thoenen H. Nature. 1992;358:502–504. doi: 10.1038/358502a0. [DOI] [PubMed] [Google Scholar]
- 23.Mitsumoto H., Ikeda K., Holmlund T., Greene T., Cedarbaum J. M., Wong V., Lindsay R. M. Ann. Neurol. 1994;36:142–148. doi: 10.1002/ana.410360205. [DOI] [PubMed] [Google Scholar]
- 24.Dobrea G. M., Unnerstall J. R., Rao M. S. Dev. Brain Res. 1992;66:209–219. doi: 10.1016/0165-3806(92)90082-8. [DOI] [PubMed] [Google Scholar]
- 25.Herbarth B., Pingault V., Bondurand N., Kuhlbrodt K., Hermans-Borgmeyer I., Puliti A., Lemort N., Goossens M., Wegner M. Proc. Natl. Acad. Sci. USA. 1998;95:5161–5165. doi: 10.1073/pnas.95.9.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southard-Smith E. M., Kos L., Pavan W. J. Nat. Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 27.Watabe K., Fukuda T., Tanaka J., Honda H., Toyohara K., Sakai O. J. Neurosci. Res. 1995;41:279–290. doi: 10.1002/jnr.490410215. [DOI] [PubMed] [Google Scholar]
- 28.Saotome Y., Winter C. G., Hirsh D. Gene. 1995;152:233–238. doi: 10.1016/0378-1119(94)00717-7. [DOI] [PubMed] [Google Scholar]
- 29.Carroll P., Sendtner M., Meyer M., Thoenen H. Glia. 1993;9:176–187. doi: 10.1002/glia.440090303. [DOI] [PubMed] [Google Scholar]
- 30.Pritchard C., Grosveld G., Hollenbach A. D. Gene. 2003;305:61–69. doi: 10.1016/s0378-1119(02)01186-1. [DOI] [PubMed] [Google Scholar]
- 31.Faus I., Hsu H. J., Fuchs E. Mol. Cell. Biol. 1994;14:3263–3275. doi: 10.1128/mcb.14.5.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlierf B., Ludwig A., Klenovsek K., Wegner M. Nucleic Acids Res. 2002;30:5509–5516. doi: 10.1093/nar/gkf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peirano R. I., Goerich D. E., Riethmacher D., Wegner M. Mol. Cell. Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watabe K., Sakamoto T., Kawazoe Y., Michikawa M., Miyamoto K., Yamamura T., Saya H., Araki N. Neuropathology. 2003;23:68–78. doi: 10.1046/j.1440-1789.2003.00478.x. [DOI] [PubMed] [Google Scholar]
- 35.Wegner M. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goulding M. D., Chalepakis G., Deutsch U., Erselius J. R., Gruss P. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tassabehji M., Read A. P., Newton V. E., Harris R., Balling R., Gruss P., Strachan T. Nature. 1992;355:635–636. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- 38.Hoth C. F., Milunsky A., Lipsky N., Sheffer R., Clarren S. K., Baldwin C. T. Am. J. Hum. Genet. 1993;52:455–462. [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng W., Au D. K., Knowles C. H., Anand P., Tam P. K. J. Pediatr. Surg. 2001;36:296–300. doi: 10.1053/jpsu.2001.20695. [DOI] [PubMed] [Google Scholar]
- 40.Inoue K., Shilo K., Boerkoel C. F., Crowe C., Sawady J., Lupski J. R., Agamanolis D. P. Ann. Neurol. 2002;52:836–842. doi: 10.1002/ana.10404. [DOI] [PubMed] [Google Scholar]
- 41.Wong V., Pearsall D., Arriaga R., Ip N. Y., Stahl N., Lindsay R. M. J. Biol. Chem. 1995;270:313–318. doi: 10.1074/jbc.270.1.313. [DOI] [PubMed] [Google Scholar]
- 42.Gatzinsky K. P., Holtmann B., Daraie B., Berthold C. H., Sendtner M. Glia. 2003;42:340–349. doi: 10.1002/glia.10221. [DOI] [PubMed] [Google Scholar]
- 43.Quandt K., Frech K., Karas H., Wingender E., Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sock E., Leger H., Kuhlbrodt K., Schreiber J., Enderich J., Richter-Landsberg C., Wegner M. J. Neurochem. 1997;68:1911–1919. doi: 10.1046/j.1471-4159.1997.68051911.x. [DOI] [PubMed] [Google Scholar]
- 45.Kuhlbrodt K., Herbarth B., Sock E., Enderich J., Hermans-Borgmeyer I., Wegner M. J. Biol. Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 46.Stolt C. C., Lommes P., Sock E., Chaboissier M. C., Schedl A., Wegner M. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.