Abstract
μ Opioid receptors are critical for heroin dependence, and A118G SNP of the μ opioid receptor gene (OPRM1) has been linked with heroin abuse. In our population of European Caucasians (n = 118), ≈90% of 118G allelic carriers were heroin users. Postmortem brain analyses showed the OPRM1 genotype associated with transcription, translation, and processing of the human striatal opioid neuropeptide system. Whereas down-regulation of preproenkephalin and preprodynorphin genes was evident in all heroin users, the effects were exaggerated in 118G subjects and were most prominent for preproenkephalin in the nucleus accumbens shell. Reduced opioid neuropeptide transcription was accompanied by increased dynorphin and enkephalin peptide concentrations exclusively in 118G heroin subjects, suggesting that the peptide processing is associated with the OPRM1 genotype. Abnormal gene expression related to peptide convertase and ubiquitin/proteosome regulation was also evident in heroin users. Taken together, alterations in opioid neuropeptide systems might underlie enhanced opiate abuse vulnerability apparent in 118G individuals.
Keywords: drug abuse, dynorphin, enkephalin, mRNA
The use of heroin and the high fatality associated with heroin intoxication continue to escalate, as well as addiction to prescription opiate drugs. As compared with those addicted to other drugs, heroin abusers still rely heavily on an i.v. route of administration that places them at greater risk for morbidity as well as premature death due to overdose (1). The neurobiological actions of opiates have been widely studied, and it appears that the rewarding effects of heroin and its psychoactive metabolites, e.g., morphine, are strongly mediated via the μ opioid receptor (MOR). A line of compelling evidence obtained from experimental transgenic animal studies has documented that the reinforcing and locomotor behavioral properties of morphine are abolished in animals deficient in the MOR (2, 3). The localization of MORs to limbic and motor neural circuits that are tightly linked to addiction disorders (4, 5) also emphasizes the importance of the MOR in heroin dependence.
In comparison to most addictive substances, the vulnerability of heroin abuse appears to have a significant genetic load (6). Several genetic variations of the human MOR gene (OPRM1) have been identified. The most studied has been a SNP A118G in exon 1 of the OPRM1 that causes an Asn40Asp substitution in the receptor’s extracellular domain. This alters MOR function in in vitro cell systems, as evidenced by enhanced binding of β-endorphin and agonist-induced activation of G protein-coupled potassium channels (7). Although subsequent studies have failed to corroborate the original in vitro findings (8), a clear functional role has been recently documented between the 118G genotype and regulation of the OPRM1 expression (see ref. 9; mRNA and protein). Moreover, various human investigations have found the A118G SNP of OPRM1, depending on ethnicity, to be associated with opioid dependence (10) and alcoholism (11) as well as alcohol relapse in response to opiate antagonist treatment (12). Despite the growing body of literature for a functional behavioral relevance of the opioid receptor polymorphism, the underlying neurobiological correlates are unknown.
To expand our knowledge regarding the neurobiology of heroin abuse and opioid polymorphism, the current study was designed to examine the striatal endogenous opioid neuropeptide system in a large racially homogenous population of human heroin users. Most neurobiological studies of addiction have focused on the nucleus accumbens (NAc; localized within the ventral striatum), because this brain region is tightly linked with reward- and goal-directed motivated behavior (13, 14). Early experimental animal studies documented that opiate drugs are directly self-administered into the NAc (15), and that lesions of the NAc or ventral pallidum, a major output target of the NAc, impair heroin reinforcement (16). Striatal output neurons differentially express opioid neuropeptides prodynorphin and proenkephalin and contain MORs (17). We here report enhanced disturbance of the striatal opioid system, particularly for preproenkephalin (PENK) gene expression, in heroin users with the 118G SNP of the OPRM1.
Results
A118G SNP OPRM1 Genotype in Association with Heroin Use.
The distribution of the 118G allele appears to vary greatly in relation to race and ethnicity (7, 18–20), thus only Caucasian European subjects from highly homogenous populations were currently investigated. In our main population of 65 subjects (Table 1), the 118G allele was found to be in Hardy–Weinberg equilibrium. The finding of an overall allelic frequency of 11.5% for the G allele (Table 2, which is published as supporting information on the PNAS web site) is in line with previous reports of 10–14% in populations of European descent (7, 11, 18, 19). Subjects homozygous for the 118G are termed G/G, heterozygous subjects are termed A/G, and homozygous A118 subjects are termed A/A. There was an overall effect of genotype (likelihood ratio χ2(1) = 6.238; P = 0.044). The A/G genotype was significantly more frequent in the heroin than in the control group (χ2(1) = 6.153, P = 0.013). The frequency of the A/G genotype among our control subjects was 3.8% (1 of 26), but it was 25.6% (10 of 39) among heroin individuals (Table 2). Thus, 91% of the A/G genotype individuals were heroin subjects. Only one control and one heroin subject were homozygous 118G carriers. To further substantiate the OPRM1 finding, an additional 53 subjects (14 controls and 39 heroin subjects) were genotyped, and the results of the total population corroborated the first findings. The frequency of the A/G genotype was significantly higher in the heroin group (χ2(1) = 4.741, P = 0.030), and 88% of all A/G genotype individuals were heroin subjects.
Table 1.
Demographic data on the population (n = 65) used for genotyping
| Group | Control | Heroin |
|---|---|---|
| Number | n = 26 | n = 39 |
| Age, yr | 34.73 ± 2.27 (15–58) | 26.33 ± 0.85 (19–46) |
| Gender | M = 23, F = 3 | M = 33, F = 6 |
| PMI, h | 22.08 ± 1.09 | 23.58 ± 0.28 |
| Storage time, weeks | 195.2 ± 22.5 (51.1–410.7) | 195.4 ± 16.1 (98–547) |
| pH | 6.74 ± 0.03 (6.15–6.98) | 6.53 ± 0.03 (6.06–6.85) |
| Ethanol (blood) | n = 3 | n = 6 |
| Cause of death | Alcohol intoxication (n = 2), electric shock (n = 2), pulmonary emboli (n = 1), myocardial infarct (n = 17), pneumonia (n = 2), sudden death (n = 2) | Heroin overdose |
| Inclusion criteria | Negative toxicology of opiate/other drugs | Positive opiate toxicology |
| No history of opiate abuse | History of opiate abuse | |
| No history of abuse of other drugs | No history of abuse of other drugs | |
| No physical body-needle tracks | Physical body-needle tracks | |
| No history of psychiatric disorder |
A representative subset (n = 53; n = 19, controls; n = 34, heroin user) from the population was used for molecular and biochemical analyses. Values are presented as mean ± SEM; ranges are in parentheses. yr, year; M, male; F, female; PMI, postmortem interval.
In addition to the MOR, heroin exerts its actions through delta opioid receptors that also mediate drug reward (21). A potentially functional SNP in the OPRD1 gene (T80G) has been described that predicts an amino acid change from phenylalanine to cysteine (22). The T80G SNP was detected in only one heterozygous control subject in the main population of 65 subjects, giving an allelic frequency of 0.8% that did not allow for further investigation.
OPMR1 Genotype in Relation to Heroin-Related Effects on Opioid Neuropeptide mRNA Expression.
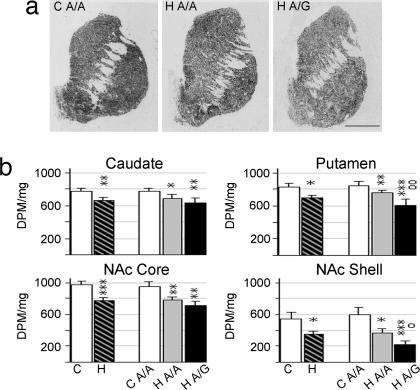
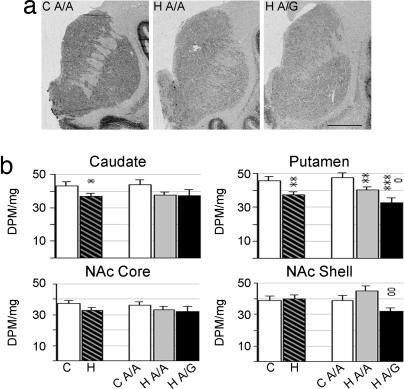
Brain PENK and preprodynorphin (PDYN) mRNA expression levels were examined in a subset of the same control and heroin users as the OPMR1 polymorphism study (Table 1). In situ hybridization histochemistry was used to detect mRNA expression levels in discrete anatomical subregions of the human striatum. Consistent with previous studies (23), PENK mRNA expression was highly abundant in the striatum of controls with highest levels detected in the dorsal subregions (Fig. 1a). Heroin subjects, as compared with controls, had reduced PENK mRNA levels throughout the dorsal and ventral striatum (Fig. 1). mRNA levels were significantly lower in the caudate nucleus (F2,46 = 5.678; P = 0.003, covariate age), putamen (F3,46 = 5.602; P = 0.036, covariates age, pH), NAc core (F1,39 = 13,418; P = 0.0007), and shell (F1,39 = 4.445; P = 0.042). Age was negatively and pH positively correlated with mRNA expression levels; however, there was no significant age or pH × group interaction.
Fig. 1.
PENK mRNA expression in striatal regions of control and heroin subjects. (a) Representative autoradiograms of coronal cryosections hybridized with PENK antisense riboprobe. (Scale bar, 1 cm.) (b) Semiquantification of PENK mRNA expression levels. Values are expressed as DPM per milligram (mean ± SEM). C, control; H, heroin users, including both A/A and A/G genotypes; H A/A, heroin users of A/A genotype; H A/G, heroin users of A/G genotype. ∗, difference from control; o, difference between heroin A/A and A/G genotypes; ∗, o, P < 0.05; ∗∗, oo, P < 0.01; ∗∗∗, P < 0.001.
To determine whether the A118G OPMR1 polymorphism influenced gene expression levels in heroin abusers, the measurements were also analyzed in consideration of genotype. Because of the small number (n = 2) of A/G and G/G genotypes in the control group, only control A/A subjects could be evaluated. The single heroin subject with a G/G genotype was included in the heroin A/G subgroup. There was a significant influence of genotype on mRNA expression. Post hoc analysis revealed that PENK mRNA levels in the putamen (P = 0.008) and NAc shell (P = 0.045) of heroin users were decreased 20.4% and 41.4%, respectively, in subjects carrying the G allele as compared with the A/A subjects (Fig. 1b).
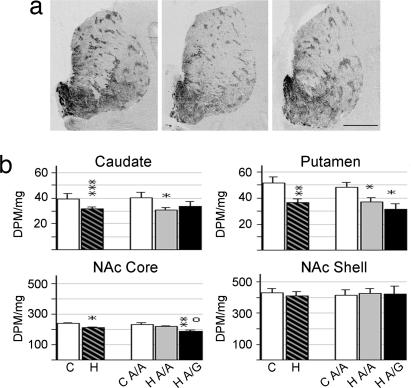
The pattern of PDYN mRNA expression was similar to previous reports, with expression most abundant in the limbic-related regions of the striatum, namely the NAc, and in particular the shell subregion (Fig. 2a), as well as the patch (striosome) compartment in the putamen and caudate nucleus (23). Similar results were obtained from the putamen and caudate nucleus motor (Fig. 2b) and associative (Table 3, which is published as supporting information on the PNAS web site) subregions. As compared with controls, heroin subjects had lower PDYN mRNA levels in the caudate nucleus (motor region, F2,43 = 9.725, P = 0.0005, covariate age; associative region, F2,43 = 9.425, P = 0.002, covariate age), putamen (motor region, F1,45 = 9.075 P = 0.004; associative region, F2,39 = 5.195; P = 0.067, covariate sex), and NAc core (F1,36 = 6.010; P = 0.019). There was no PDYN mRNA alteration in the NAc shell. Assessment of the PDYN mRNA levels expressed in the patch and matrix compartments of the dorsal striatum revealed the same direction of change as that observed in the total area (data not shown). Age was negatively correlated to the PDYN expression levels, and there were no age × group interactions. Analysis of the A118G polymorphism revealed genotype effects primarily on PDYN mRNA levels in the NAc core; the A/G heroin subgroup showed a 19.7% lower expression than the A/A heroin subjects (P = 0.024; Fig. 2b).
Fig. 2.
PDYN mRNA expression in striatal regions of control and heroin subjects. (a) Representative autoradiograms of coronal cryosections hybridized with PDYN antisense riboprobe. (Scale bar, 1 cm.) (b) Semiquantification of PDYN mRNA expression levels in the motor caudate nucleus, motor putamen, NAc core, and NAc shell. Values are expressed as DPM per milligram (mean ± SEM). ∗, difference from control, o, difference between heroin A/A and A/G genotype; ∗, o, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. For abbreviations, see Fig. 1 legend.
Toxicological measures of heroin metabolites in the urine and blood in the heroin subjects were analyzed for correlation to opioid gene expression. Urine and blood morphine levels were 1.75 ± 0.44 and 0.39 ± 0.08 μg/ml, respectively, with no significant difference between the A/G and A/A groups. A significant positive correlation was evident between PENK mRNA levels and urine morphine in the NAc (shell, r = 0.652, P = 0.022; core, r = 0.881, P = 0.009). Urine morphine concentrations were negatively correlated with PDYN mRNA expression in the putamen (associative region, r = −0.631, P = 0.028; motor region, r = −0.569, P = 0.042). The significant correlations between toxicology and PENK and PDYN were contributed by the A/A subjects. The morphine/codeine concentration ratio was 16.2 ± 7.6 and 3.6 ± 1.2 μg/ml in the urine and blood, respectively; >1 morphine/codeine concentration ratio normally indicates heroin usage rather than other medication with codeine (24). Only a few subjects had positive toxicology for 6-monoacetylmorphine, the rapid metabolite of heroin, and no significant correlation was found with the opioid neuropeptide mRNA expression levels.
OPMR1 Genotype in Relation to Heroin-Related Effects on Opioid Neuropeptide Levels.
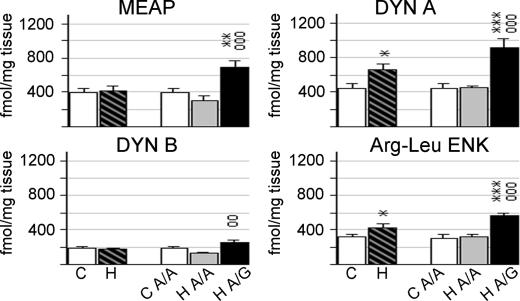
To assess the relationship between gene expression and their peptide products, radioimmunoassay was used to measure opioid peptide levels in tissue taken from the caudate nucleus. Met-enkephalin-Arg6-Phe7 (MEAP) is specifically derived from the PENK gene. Despite significant alterations in PENK mRNA expression, there was no significant difference in MEAP peptide levels between all heroin and control subjects (Fig. 3). There were, however, significant group effects in consideration of genotype (F2,39 = 9.588, P = 0.0012; covariate age). Heroin subjects with A/G genotype had 2.3-fold higher levels of MEAP than heroin A/A individuals (P = 0.0002).
Fig. 3.
Enkephalin and dynorphin peptides levels in the caudate nucleus of control and heroin subjects. Values are expressed as fmol/mg protein (mean ± SEM). ∗, difference from control; o, difference between heroin A/A and A/G genotype; ∗, o, P < 0.05; ∗∗, oo, P < 0.01; ∗∗∗, ooo, P < 0.001. For abbreviations, see Fig. 1 legend.
Dynorphin A, dynorphin B, and Arg-Leu enkephalin are derived from the PDYN gene. Dynorphin A (F1,37 = 6.903; P = 0.012) and Arg-Leu enkephalin (F1,39 = 4.362; P = 0.043) were significantly increased in heroin subjects (Fig. 3). Heroin subjects with A/G genotype had ≈2-fold higher levels of dynorphin A (heroin A/G vs. heroin A/A, P = 0.0002; heroin A/G vs. control, P = 0.0001) and Arg-Leu enkephalin (heroin A/G vs. heroin A/A; P = 0.0008, heroin A/G vs. control, P = 0.00005) than either A/A heroin or controls subjects. Heroin subjects with A/G genotype had significantly higher levels of dynorphin B than heroin A/A individuals (P = 0.001). There was a trend for increased dynorphin B levels in heroin subjects with the G allele vs. controls (P = 0.061), whereas the heroin subjects of genotype A/A showed a trend for decreased levels (P = 0.070) as compared with the control group. Peptide levels had no significant correlation to morphine or ethanol toxicology.
Correlation analyses were carried out between the opioid peptide and respective mRNAs. In consideration that mRNA expression measurements of the caudate nucleus could be compromised because of the tissue taken from this region for peptide analysis, correlation analyses were performed with mRNA measurements obtained from the putamen, because this region was intact and representative of the dorsal striatum. Significant correlations were most evident in the control group. MEAP and PENK mRNA had a weak positive correlation of r = 0.550 (P = 0.051). Of the dynorphin-related peptides, significant correlations were apparent only for dynorphin B (motor region, r = 0.744, P = 0.006; associative region, r = 0.783, P = 0.003).
Genes Related to Intracellular Processing Are Altered in Relation to Heroin Use and OPMR1 Polymorphism.
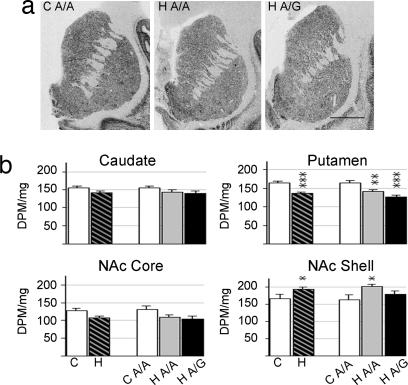
In an initial attempt to understand whether OPMR1 genotype was associated with rate-limiting components of opioid peptide processing, the expression of prohormone convertase-2 (PC2) (25, 26) was evaluated. ProPC2 mRNA expression had a relative homogenous distribution in the striatum (Fig. 4), with the exception of some control and heroin subjects with strongly labeled cell clusters in the NAc and high-expressing patches in the putamen. The mRNA levels of proPC2 were significantly increased (F1,31 = 4.163, P = 0.050) in the NAc shell of the heroin group (Fig. 4). Post hoc analysis in relation to genotype revealed that the increase was due to higher expression levels in the heroin A/A subjects. In contrast to the NAc shell, proPC2 mRNA levels were decreased in the putamen of the heroin subjects (F2,42 = 13.749, P < 0.0001), and there was a trend for a stronger reduction in the A/G heroin subgroup as compared with the A/A heroin individuals (P = 0.061). There was no difference in proPC2 expression between control and heroin groups in the caudate nucleus and NAc core. Toxicological evaluation revealed that proPC2 mRNA expression had a strong positive correlation with 6-monoacetylmorphine (caudate nucleus, r = 0.933, P = 0.007; putamen, r = 0.852, P = 0.031).
Fig. 4.
ProPC2 mRNA expression in striatal regions of control and heroin subjects. (a) Representative autoradiograms of coronal cryosections hybridized with proPC2 antisense riboprobe. (Scale bar, 1 cm.) (b) Semiquantification of proPC2 mRNA expression levels. Values are expressed as DPM per milligram (mean ± SEM). ∗, difference from control; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. For abbreviations, see Fig. 1 legend.
Agonist regulation of the MOR has been shown to be mediated via the ubiquitin/proteosomal system (27). E3 ubiquitin ligases are critical enzymes in ubiquitination, and a pilot gene screening experiment showed a homologous to E6-AP C terminus (HECT) type of E3 ubiquitin ligase HECT and RCC1 domain protein (HERC1), to be relevant to heroin exposure (our unpublished data). ProHERC1 mRNA had a homogenous expression throughout the human striatum (Fig. 5). A significant group effect was detected for proHERC1 mRNA levels expressed in the caudate nucleus (F1,43 = 4.356; P = 0.043) and putamen (F1,43 = 7.659, P = 0.008; Fig. 5b). ProHERC1 levels in the putamen were decreased to a greater extent in the A/G heroin subgroup as compared with the A/A heroin subjects (P = 0.016). Significantly lower HERC1 levels were also detected in the NAc shell of the heroin A/G subgroup as compared with the heroin A/A individuals (P = 0.005). Correlation analyses revealed that significant association between proHERC1 mRNA expression and blood morphine was predominantly contributed by the A/G subjects (caudate nucleus, r = −0.99, P < 0.0001; NAc shell, r = −0.900, P = 0.037).
Fig. 5.
ProHERC1 mRNA expression in striatal regions of control and heroin subjects. (a) Representative autoradiograms of coronal cryosections hybridized with ProHERC1 antisense riboprobe. (Scale bar, 1 cm.) (b) Semiquantification of ProHERC1 mRNA expression levels. Values are expressed as DPM per milligram (mean ± SEM). ∗, difference from control, o, difference between heroin A/A and A/G genotypes; ∗, o, P < 0.05; ∗∗, oo, P < 0.01; ∗∗∗, P ≤ 0.001. For abbreviations, see Fig. 1 legend.
Discussion
Various investigations have evaluated the OPRM1 polymorphism in regard to drug abuse vulnerability (7, 10–12, 18–20). As compared with other studies, our results revealed a more profound (≈90%) association between heroin use and the A/G genotype. This could be due to the rather homogenous, although small, racial and ethnic population genotyped. Bart et al. (10) also found a high frequency (65%) of the 118G allele in Swedish heroin abusers from a relatively homogenous population. The higher frequency of the 118G allele in the Swedish heroin population could also reflect susceptibility to heroin “overdose,” thus the relationship between the 118G SNP and opioid function in respiratory brain regions is currently being explored. Moreover, as a postmortem investigation, the current drug users were not characterized based on an antemortem clinical diagnosis of heroin dependence, although most subjects had documented evidence of heroin abuse. Nevertheless, the focus and novel aspect of this investigation, as compared with other genotype studies, are the direct examination of the human brain that enabled detection of discrete neuronal disturbances in relation to OPRM1 polymorphism.
Although molecular and biochemical studies of the human brain are challenging because of the multiple caveats of controlling postmortem factors, the present study clearly documents an impaired opioid neuropeptide system in heroin abusers that is exaggerated in A/G individuals. These alterations would most likely be linked to the downstream effects of MOR, because the A118G SNP directly influences the expression of MOR, at both the mRNA and protein levels (8, 9), which is localized to PENK- and PDYN-expressing striatal cells. The most significant molecular alterations were evident for the PENK mRNA in which down-regulation was apparent throughout the striatum in the heroin users, indicating widespread effects on limbic and motor functions. Of the striatal regions, PENK mRNA expression was reduced to a greater extent in the NAc shell with an additional 40% reduction in A/G as compared with A/A heroin users. The PENK system, in contrast to PDYN, is associated with positive reinforcement (28), and the NAc shell is the striatal subregion most linked with the rewarding effects of drugs in animal studies (14). Thus the current findings could suggest greater alteration of reward processing in relation to A118G OPRM1 polymorphism. NAc enkephalin is central to hedonic responses not only to drugs of abuse but also for natural rewards such as palatable food (13). It is important to note that, despite the strong alterations of PENK mRNA expression in both the dorsal and ventral striatum of heroin users, morphine toxicology was significantly associated only with expression levels in the NAc shell. This suggests a more dynamic response of the limbic PENK NAc populations to heroin intake that may relate to the rapid positive mood-altering effects of the drug.
Reduced PDYN mRNA was also detected in the NAc, but the alterations were confined to the NAc core and were present only in A/G subjects. The NAc core is a central motor component of the limbic motive neurocircuit mediating, e.g., goal-directed behavior and impulse control (29). Similar to PENK, heroin-related reduction of PDYN mRNA expression levels was also evident in the dorsal striatum. The down-regulation of the opioid neuropeptide genes in heroin users may directly reflect the drug stimulation of MORs coupled to inhibitory G proteins (30). Although this could be feasible for the PDYN transcript in light of the reduced mRNA expression with increased morphine concentrations, there was a positive correlation between morphine toxicology and PENK transcription. Thus, the reduction of PENK mRNA expression in the NAc would seem to reflect the long-term chronic state of heroin use. Multiple rat studies have reported similar reduced striatal PENK mRNA levels after repeated morphine exposure (31, 32). The repeated presence of heroin in the brain is expected to function as “endogenous enkephalin” and lead to a counterbalance of decreased PENK expression. The opposing effects of acute heroin intake on the opioid neuropeptide transcription levels in our study indicate a differential regulation of PENK and PDYN striatal gene expression by heroin. However, MORs are situated on both PDYN and PENK medium spiny projection striatal neurons (17, 33), suggesting that other neural systems such as dopamine, which differentially regulates striatal PDYN and PENK, might contribute to the differential opioid neuropeptide effects (34).
In contrast to the reduced transcription of the PDYN and PENK mRNAs, heroin subjects carrying the G allele had elevated levels of peptides derived from the opioid neuropeptide genes. Most of the striatal peptide levels measured are expected to arise from neurites (35) and striatal axon collaterals of the PENK and PDYN projection neurons (36); striatal afferents from the cerebral cortex contribute very little to the dynorphin and enkephalin levels. Thus it appears that transcription and translation of the opioid neuropeptides are not as directly linked in the abuse population as they are in controls. Tissue levels of peptides reflect the balance of various factors, synthesis, processing, release, and degradation. Decreased release over time, in combination with reduced synthesis, could contribute to increased peptide tissue concentrations. Disturbance of peptide processing and degradation may also exist. The current proPC2 mRNA findings do not, however, provide an answer for the genotype differences in the peptide concentrations. The neuronal systems underlying processing and degradation of the opioid peptides are complex and will require in-depth investigations. The proPC2 findings were nevertheless intriguing, considering that the elevation of the proPC2 mRNA in the NAc shell was contributed only by A/A heroin subjects. Moreover, of the neural markers currently examined, the proPC2 gene expression appeared most sensitive to the rapid effects of heroin intake, as evidenced by the strong correlation with 6-monoacetylmorphine proPC2 (and HERC1) mRNA expression, which was increased in the NAc but decreased in the putamen, clearly demonstrates subregional striatal differences in intracellular processing in heroin abusers. Tissue levels were currently measured only in the dorsal striatum, so it remains to be confirmed whether such subregional differences would indeed be reflected at the peptide level, and multiple enzymes (mRNA and protein) associated with peptide processing need to be investigated for an understanding of the neural systems that could contribute to abnormal peptide levels in A/G heroin users.
Intracellular disturbances in relation to heroin use were also evident for the striatal expression of the HECT-related (HERC1) gene. HERC1 can function as an E3 ubiquitin–protein ligase (37) and can influence multiple cellular processes, including protein degradation, receptor trafficking and degradation, and signal transduction (38). The ubiquitin/proteosome pathway plays a prominent role in morphine-induced regulation of MOR (27), and heroin users, irrespective of genotype, had down-regulated HERC1 mRNA expression in the putamen. The observation that HERC1expression in A/G individuals was positively correlated with morphine toxicology suggests an impact of genotype on the acute HERC1 response. Genotype masked the overall heroin effect in the NAc shell, because a significant reduction of the proHERC1 mRNA was apparent only in A/G subjects. Based on the involvement of the ubiquitin/proteosome system with MOR function, the greater impairment of the proHERC1 mRNA expression in the A/G heroin users could relate to the dysfunction of MOR regulation that is a characteristic feature of 118G individuals (9).
It is important to point out that it was not possible to determine the neurobiological consequences of the G allele in the normal human brain, because very few control subjects carried the 118G allele. This again emphasizes the possible vulnerability of A/G individuals in regard to heroin. Inconsistent reports regarding the A118G SNP with heroin abuse and important consideration of ethnicity suggest that heroin vulnerability may include other genes in disequilibrium with the A118G SNP. Nonetheless, that significant disturbances are now identified for the opioid neuropeptide as well as for neural systems linked to intracellular processing in A/G heroin users provides an important first step in understanding the neurobiological correlates of the A118G OPRM1 polymorphism in human addiction disorders.
Methods
Human Brains.
Human brains from apparent heroin overdose and normal control Caucasian subjects without head trauma were collected at autopsy within 24 h after death at the Department of Forensic Medicine, Semmelweis University. Postmortem brain material was also obtained from the National Institute of Forensic Medicine (Karolinska Institutet). All material was obtained under approved local ethical guidelines. The cause and manner of death were determined by a forensic pathologist after evaluating the circumstances of death, toxicology data, autopsy results, and police reports, as well as family interviews and medical records, when available. All cases were assessed for common drugs of abuse (including alcohol) and for therapeutic agents. The general characteristics of inclusion criteria for control and heroin subjects are described in Table 1. The heroin group represented a unique drug abuse population, because this group consisted of predominantly heroin users with no methadone treatment.
The total population of subjects consisted of 118 subjects from the Hungarian (heroin users, n = 64; control, n = 38) and Swedish (heroin users, n = 14; control, n = 2) collections. OPMR1 and OPDR1 genotyping was initially performed on 65 cases (Table 1), and molecular and peptide measurements were assessed on a subset (n = 53) of these subjects. An additional 53 cases (14 arising from the Swedish Caucasian population) were genotyped for the A118G SNP in the OPMR1. For the molecular studies, immediately after autopsy, brains were cut coronally in 1.5-cm slabs, frozen, and kept at −70°C. Blocks (≈5 × 7 cm) were cut over the rostral striatum; cryosections (20 μm, Microm HM560; Microm International, Walldorf, Germany) were quickly mounted on Superfrost Plus glass (BRL, Newton, MA) and kept at −30°C until analysis. Before sectioning, tissue punches were taken from the dorsal caudate nucleus for peptide measurements. All molecular and biochemical procedures were carried out blinded to the subject group.
Allelic Analysis of OPMR1 and OPDR1.
DNA was purified from cerebellar tissue of controls and heroin subjects (Table 1) using DNeasy columns (Qiagen, Valencia, CA). The OPMR1 A118G SNP was genotyped from PCR according to Gelernter et al. (19) by using primers ArtUp and R2-D2 (Table 4, which is published as supporting information on the PNAS web site). The OPDR1 T80G was genotyped by using primers ex1F27Cu and exF27Cd (see ref. 22; Table 4).
In Situ Hybridization Histochemistry.
The PENK riboprobe was an EcoRI/Pvu 792-bp fragment complementary to the full coding region of the PENK human gene that was subcloned in a psp65 plasmid (39). The PDYNFL1 probe was synthesized from a pGEM3Z plasmid with the 5′ region of the gene (GenBank accession no. NM_024411, bases −215/−72) (40). Templates for the PC2 (GenBank accession no. NM_002594, bases 452–776) and HERC1 (GenBank accession no. NM_003922, bases 264–572) probes were synthesized from a human cDNA library using nested PCR (Table 4). Riboprobes were generated by in vitro transcription using SP6 polymerase and [α-35S]UTP (Amersham Pharmacia Biosciences). In situ hybridization was performed as described (see ref. 41 and Supporting Text, which is published as supporting information on the PNAS web site).
Film Analysis.
Optical density values were measured (scion image; Wayne Rasband, National Institute of Mental Health, Bethesda) from digitalized images with a resolution of 300 dots per inch (scanned by scanmaker iii; Microtek Electronics, Düsseldorf, Germany). Optical density values were converted to disintegrations per minute (DPM) per milligram by reference to coexposed C14 standards (ARC, St. Louis). Measurements were taken from distinct regions of interest in the dorsal striatum (caudate and putamen) and NAc where it is dissociated into core and shell subdivisions (Fig. 6, which is published as supporting information on the PNAS web site). Distinction of motor and associative subregions of the caudate and putamen is based on the functional organization of the primate striatum (36). Because of the heterogeneous expression of PDYN in the dorsal striatum, in addition to measurements of the total area, separate analysis of expression in the patch (striosome; islands of high PDYN expression) and matrix compartments was also performed by using threshold analysis. Background was subtracted from adjacent white matter areas, and the DPM/mg values from duplicate slides were averaged.
Peptide Measurements.
Opioid peptide concentrations from the caudate nucleus were determined by using radioimmunoassays for MEAP and dynorphin peptides, as described (see ref. 42 and Supporting Text).
Statistical Analysis.
Normal distribution of the data was analyzed by using Shapiro–Wilk’s W test of normality. If not normally distributed, data sets were normalized using natural logarithm. General linear stepwise regression analysis was used to evaluate group differences and identify covariates: age, brain pH, postmortem interval (dichotomized <12 and <24 h), sex, blood ethanol, and brain freezer storage time. Statistical outliers were excluded. Post hoc analysis regarding heroin A/A and A/G genotype subgroups was performed by using Fisher’s least-square difference. Significance was set at P < 0.05 and trends considered for P < 0.10. Statistical evaluations were carried out by using the jmp (SAS Institute, Cary, NC) and statistica (StatSoft Scandinavia, Uppsala, Sweden) software packages.
Supplementary Material
Acknowledgments
We thank Mrs. Alexandra Tylec for technical assistance and Elisabeth Berg (Department of Learning, Informatics, Management, and Ethics, Medical Statistics Division, Karolinska Institutet) for statistical guidance. This study was supported by National Institutes of Health Grant NIDA DA15446; Hungarian Scientific Research Fund Grant T32227; the Swedish National Drug Policy Coordinator (Y.L.H., G.B., and I.N.); and Swedish Science Council Grants 11252 (to Y.L.H.), 14681 (to G.B.), and 12588 (to I.N.). M.C.H. was supported by the Swedish Institute and Hungarian Scholarship Board.
Abbreviations
- MEAP
Met-enkephalin-Arg6-Phe7
- MOR
μ opioid receptor
- NAc
nucleus accumbens
- PC2
prohormone convertase-2
- PDYN
preprodynorphin
- PENK
preproenkephalin
- DPM
disintegrations per minute.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hulse G. K., English D. R., Milne E., Holman C. D. Addiction. 1999;94:221–229. doi: 10.1046/j.1360-0443.1999.9422216.x. [DOI] [PubMed] [Google Scholar]
- 2.Matthes H. W., Maldonado R., Simonin F., Valverde O., Slowe S., Kitchen I., Befort K., Dierich A., Le Meur M., Dolle P., et al. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 3.Sora I., Elmer G., Funada M., Pieper J., Li X. F., Hall F. S., Uhl G. R. Neuropsychopharmacology. 2001;25:41–54. doi: 10.1016/S0893-133X(00)00252-9. [DOI] [PubMed] [Google Scholar]
- 4.Daunais J. B., Letchworth S. R., Sim-Selley L. J., Smith H. R., Childers S. R., Porrino L. J. J. Comp. Neurol. 2001;433:471–485. doi: 10.1002/cne.1154. [DOI] [PubMed] [Google Scholar]
- 5.Mansour A., Fox C. A., Thompson R. C., Akil H., Watson S. J. Brain Res. 1994;643:245–265. doi: 10.1016/0006-8993(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 6.Tsuang M. T., Lyons M. J., Meyer J. M., Doyle T., Eisen S. A., Goldberg J., True W., Lin N., Toomey R., Eaves L. Arch. Gen. Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 7.Bond C., LaForge K. S., Tian M., Melia D., Zhang S., Borg L., Gong J., Schluger J., Strong J. A., Leal S. M., et al. Proc. Natl. Acad. Sci. USA. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyer A., Koch T., Schroder H., Schulz S., Hollt V. J. Neurochem. 2004;89:553–560. doi: 10.1111/j.1471-4159.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Wang D., Johnson A. D., Papp A. C., Sadee W. J. Biol. Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 10.Bart G., Heilig M., LaForge K. S., Pollak L., Leal S. M., Ott J., Kreek M. J. Mol. Psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bart G., Kreek M. J., Ott J., LaForge K. S., Proudnikov D., Pollak L., Heilig M. Neuropsychopharmacology. 2005;30:417–422. doi: 10.1038/sj.npp.1300598. [DOI] [PubMed] [Google Scholar]
- 12.Oslin D. W., Berrettini W., Kranzler H. R., Pettinati H., Gelernter J., Volpicelli J. R., O’Brien C. P. Neuropsychopharmacology. 2003;28:1546–1552. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- 13.Kelley A. E., Bakshi V. P., Haber S. N., Steininger T. L., Will M. J., Zhang M. Physiol. Behav. 2002;76:365–377. doi: 10.1016/s0031-9384(02)00751-5. [DOI] [PubMed] [Google Scholar]
- 14.Koob G. F. Ann. N.Y. Acad. Sci. 1992;654:171–191. doi: 10.1111/j.1749-6632.1992.tb25966.x. [DOI] [PubMed] [Google Scholar]
- 15.Olds M. E. Brain Res. 1982;237:429–440. doi: 10.1016/0006-8993(82)90454-1. [DOI] [PubMed] [Google Scholar]
- 16.Hubner C., Koob G. Brain Res. 1990;508:20–29. doi: 10.1016/0006-8993(90)91112-t. [DOI] [PubMed] [Google Scholar]
- 17.Guttenberg N. D., Klop H., Minami M., Satoh M., Voorn P. NeuroReport. 1996;7:2119–2124. doi: 10.1097/00001756-199609020-00011. [DOI] [PubMed] [Google Scholar]
- 18.Bergen A. W., Kokoszka J., Peterson R., Long J. C., Virkkunen M., Linnoila M., Goldman D. Mol. Psychiatry. 1997;2:490–494. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- 19.Gelernter J., Kranzler H., Cubells J. Mol. Psychiatry. 1999;4:476–483. doi: 10.1038/sj.mp.4000556. [DOI] [PubMed] [Google Scholar]
- 20.Luo X., Kranzler H. R., Zhao H., Gelernter J. Am. J. Med. Genet. B. 2003;120:97–108. doi: 10.1002/ajmg.b.20034. [DOI] [PubMed] [Google Scholar]
- 21.Martin T. J., Kim S. A., Cannon D. G., Sizemore G. M., Bian D., Porreca F., Smith J. E. J. Pharmacol Exp. Ther. 2000;294:975–982. [PubMed] [Google Scholar]
- 22.Gelernter J., Kranzler H. R. Hum. Genet. 2000;107:86–88. doi: 10.1007/s004390000340. [DOI] [PubMed] [Google Scholar]
- 23.Hurd Y. L., Herkenham M. Neuroscience. 1995;64:571–586. doi: 10.1016/0306-4522(94)00417-4. [DOI] [PubMed] [Google Scholar]
- 24.Ceder G., Jones A. W. Clin. Chem. 2001;47:1980–1984. [PubMed] [Google Scholar]
- 25.Miller R., Toneff T., Vishnuvardhan D., Beinfeld M., Hook V. Y. Neuropeptides. 2003;37:140–148. doi: 10.1016/s0143-4179(03)00027-1. [DOI] [PubMed] [Google Scholar]
- 26.Berman Y., Mzhavia N., Polonskaia A., Furuta M., Steiner D. F., Pintar J. E., Devi L. A. J. Neurochem. 2000;75:1763–1770. doi: 10.1046/j.1471-4159.2000.0751763.x. [DOI] [PubMed] [Google Scholar]
- 27.Chaturvedi K., Bandari P., Chinen N., Howells R. D. J. Biol. Chem. 2001;276:12345–12355. doi: 10.1074/jbc.M008054200. [DOI] [PubMed] [Google Scholar]
- 28.Bals-Kubik R., Shippenberg T. S., Herz A. Eur. J. Pharmacol. 1990;175:63–69. doi: 10.1016/0014-2999(90)90153-w. [DOI] [PubMed] [Google Scholar]
- 29.Cardinal R. N., Pennicott D. R., Sugathapala C. L., Robbins T. W., Everitt B. J. Science. 2001;292:2499–2501. doi: 10.1126/science.1060818. [DOI] [PubMed] [Google Scholar]
- 30.Williams J. T., Christie M. J., Manzoni O. Physiol. Rev. 2001;81:299–343. doi: 10.1152/physrev.2001.81.1.299. [DOI] [PubMed] [Google Scholar]
- 31.Georges F., Stinus L., Bloch B., Le Moine C. Eur. J. Neurosci. 1999;11:481–490. doi: 10.1046/j.1460-9568.1999.00462.x. [DOI] [PubMed] [Google Scholar]
- 32.Romualdi P., Lesa G., Ferri S. Brain Res. 1991;563:132–136. doi: 10.1016/0006-8993(91)91525-6. [DOI] [PubMed] [Google Scholar]
- 33.Olive M. F., Anton B., Micevych P., Evans C. J., Maidment N. T. J. Neurosci. 1997;17:7471–7479. doi: 10.1523/JNEUROSCI.17-19-07471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerfen C. R., Enber T. M., Susel Z., Chase T. N., Monsma F. J., Mahan L. C., Sibley D. R. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- 35.Van Bockstaele E. J., Sesack S. R., Pickel V. M. J. Comp. Neurol. 1994;341:1–15. doi: 10.1002/cne.903410102. [DOI] [PubMed] [Google Scholar]
- 36.Parent A., Côté P.-Y., Lavoie B. Prog. Neurobiol. 1995;46:131–197. [PubMed] [Google Scholar]
- 37.Cruz C., Paladugu A., Ventura F., Bartrons R., Aldaz M., Rosa J. L. Cytogenet. Cell Genet. 1999;86:68–69. doi: 10.1159/000015414. [DOI] [PubMed] [Google Scholar]
- 38.Hochrainer K., Mayer H., Baranyi U., Binder B., Lipp J., Kroismayr R. Genomics. 2005;85:153–164. doi: 10.1016/j.ygeno.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Noda M., Teranishi Y., Takahashi H., Toyuosato M., Notake M., Nakanishi S., Numa S. Nature. 1982;297:431–434. doi: 10.1038/297431a0. [DOI] [PubMed] [Google Scholar]
- 40.Nikoshkov A., Hurd Y. L., Yakovleva T., Bazov I., Marinova Z., Cebers G., Pasikova N., Gharibyan A., Terenius L., Bakalkin G. FASEB J. 2005;19:1543–1545. doi: 10.1096/fj.05-3743fje. [DOI] [PubMed] [Google Scholar]
- 41.Fagergren P., Smith H. R., Daunais J. B., Nader M. A., Porrino L. J., Hurd Y. L. Eur. J. Neurosci. 2003;17:2212–2218. doi: 10.1046/j.1460-9568.2003.02636.x. [DOI] [PubMed] [Google Scholar]
- 42.Nylander I., Stenfors C., Tan-No K., Mathé A. A., Terenius L. Neuropeptides. 1997;31:257–365. doi: 10.1016/s0143-4179(97)90072-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.