Abstract
The plant-derived cannabinoids Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD) both have immunosuppressive effects; although some effects of THC are mediated by the CB2 receptor, CB2 binds CBD weakly. In examining the effects of THC and CBD on microglial proliferation, we found that these compounds potently inhibit [3H]thymidine incorporation into a murine microglial cell line with no effect on cell cycle. Treatment with THC and CBD decreased [3H]thymidine uptake into microglia, with IC50 values that match inhibition of [3H]thymidine incorporation into DNA. CBD and, less potently, THC decreased uptake of [3H]adenosine to a similar extent as [3H]thymidine in both murine microglia and RAW264.7 macrophages. Binding studies confirm that CBD binds to the equilibrative nucleoside transporter 1 with a Ki < 250 nM. Because adenosine agonists have antiinflammatory effects, and because uptake of adenosine is a primary mechanism of terminating adenosine signaling, we tested the hypothesis that CBD is immunosuppressive because it enhances endogenous adenosine signaling. In vivo treatment with a low dose of CBD decreases TNFα production in lipopolysaccharide-treated mice; this effect is reversed with an A2A adenosine receptor antagonist and abolished in A2A receptor knockout mice. These studies demonstrate that CBD has the ability to enhance adenosine signaling through inhibition of uptake and provide a non-cannabinoid receptor mechanism by which CBD can decrease inflammation.
Keywords: adenosine, lipopolysaccharide, tetrahydrocannabinol, thymidine, tumor necrosis factor-α
The marijuana-derived cannabinoids Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD) each have immunosuppressive effects (1) and are currently in clinical trials for treatment of multiple sclerosis (2). Although the CB2 cannabinoid receptor partially mediates the antiinflammatory effects of THC (3), CBD does not bind well to the known cannabinoid receptors (4); this low affinity results in the inability of CBD to produce the subjective “high” and cognitive effects that are characteristic of marijuana and THC (5, 6). To date, the mechanism by which CBD decreases inflammation is unknown, although micromolar concentrations of CBD have been shown to inhibit lipoxygenase activity (7).
In addition to multiple sclerosis, CBD has shown promise in several rodent models of inflammation. Oral treatment with CBD decreases edema and hyperalgesia in a rat paw model of carrageenan-induced inflammation (8). A single dose of CBD also decreases serum TNFα production in lipopolysaccharide (LPS)-treated mice (9). CBD improves arthritis symptoms and joint pathology in murine collagen-induced arthritis while inhibiting IFN-γ production and lymph node cell proliferation, as measured by [3H]thymidine incorporation (9). Treatment with THC also decreases proliferation of lymph node cells as well as splenocytes (10).
Given the potential of THC and CBD to decrease proliferation in immune cells, we initially examined the effects of several natural and synthetic cannabinoids on microglial cell proliferation by using [3H]thymidine incorporation. Microglia are the resident immune cells of the brain, and their proliferation has been linked to a number of neurodegenerative diseases (11). However, CBD and THC decreased [3H]thymidine incorporation into EOC-20 microglial cells with no effect on cell cycle. Further experiments demonstrate that this decrease in [3H]thymidine incorporation is due to an inhibition of [3H]thymidine uptake into cells, and that the inhibition extends to uptake of [3H]adenosine.
Release of adenosine is an endogenous mechanism of immunosuppression evoked during cellular stress and inflammation (12). Uptake of adenosine is a primary mechanism of terminating adenosine signaling; therefore, adenosine uptake inhibitors enhance endogenous activity at adenosine receptors (13). Antiinflammatory effects of adenosine agonists and uptake inhibitors are similar to the effects of plant-derived cannabinoids. Like CBD, adenosine receptor agonists and uptake inhibitors decrease serum TNFα in LPS-treated mice (13). In addition, THC and adenosine each have the ability to suppress the T helper 1 response while enhancing the T helper 2 response (14, 15).
Here, we demonstrate that THC and CBD have the ability to inhibit adenosine uptake by acting as competitive inhibitors at the equilibrative transporter. Furthermore, the subsequent enhancement of adenosine signaling by CBD in vivo is responsible for some of the drug's observed antiinflammatory effects, demonstrated by a decrease in serum TNFα evoked by LPS treatment.
Results
Cannabinoid Effects on [3H]Thymidine DNA Incorporation.
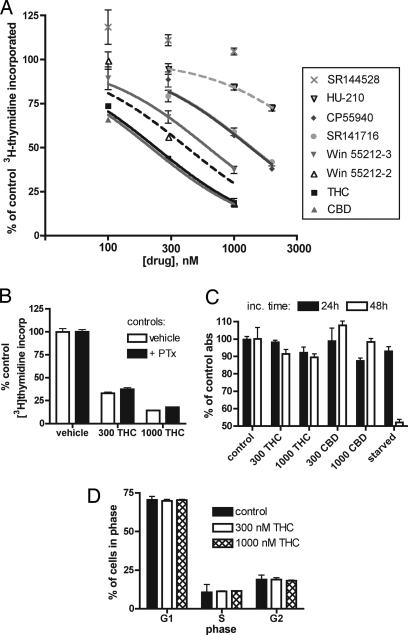
We determined the effects of the plant-derived cannabinoid THC on proliferation of the murine microglial cell line EOC-20, using [3H]thymidine incorporation to measure proliferation. In a 4-h incubation, THC inhibited [3H]thymidine incorporation into EOC-20 microglia with an IC50 of 370 nM. The cannabinoid CBD, which is also derived from marijuana but has much lower affinity for cannabinoid receptors (16), likewise inhibited EOC-20 [3H]thymidine incorporation after 4 h with potency similar to that of THC (Fig. 1A).
Fig. 1.
Cannabinoids inhibit [3H]thymidine incorporation into EOC-20 microglia with no effect on cell cycle or MTT reduction. (A) [3H]Thymidine incorporation assay. Cannabinoids were present during the 4-h [3H]thymidine incubation. Results are expressed as a percent of control, and vertical lines represent SEM (n = 4); the single-site competition equation was used to determine IC50. (B) PTx treatment during [3H]thymidine incorporation assay. Cells were treated with 100 ng/ml PTx during the 24-h stimulation with LMCM; THC or vehicle was added for the final 4 h. Indicated concentrations are in nM. Results are shown as a percentage of control proliferation; respective controls are shown. Treatment with PTx alone resulted in ≈70% of normal proliferation (n = 6). (C) MTT assay. EOC-20 microglia were treated for 24 or 48 h with vehicle, THC, or CBD before assaying for reduction of MTT tetrazolium salt. Indicated concentrations are in nM; shown are the means and SEM of the resulting absorbance at 562 nm. (D) Cell cycle analysis by FACS after 4 h of treatment with THC. To exclude fragmented or fused cells, cells per cell cycle stage were calculated as a percentage of total gated cells (n = 2).
A number of synthetic cannabinoids were tested for their ability to decrease [3H]thymidine incorporation, with various effects (Fig. 1A). Specifically, the potent cannabinoid HU-210, which has an affinity for the CB1 and CB2 receptors in the picomolar range (16), was less efficacious than the low-affinity CBD in inhibiting [3H]thymidine incorporation. Whereas SR141716 and the agonist Win 55212-2 inhibited [3H]thymidine incorporation at doses slightly higher than THC or CBD, the receptor-inactive enantiomer S(−)Win 55212-3 had a potency similar to the active R(+)Win 55212-2. These data suggest an inhibitory effect independent of known cannabinoid receptors.
Both cannabinoid receptors are G protein-coupled receptors that are known to signal through Gi/o (17) and are thus inhibited by pertussis toxin (PTx). Although PTx alone reduced DNA incorporation of [3H]thymidine into EOC-20 microglia, THC continued to decrease [3H]thymidine incorporation in cells preincubated with 100 ng/ml PTx, a concentration known to inhibit signaling through Gi/o proteins (ref. 18; Fig. 1B). Furthermore, the inhibitory effects of THC and CBD were not reversed by CB1 or CB2 receptor antagonists at concentrations that block cannabinoid receptor activity (SR141716 and SR144528 added at 500 nM, data not shown) (16, 19).
Analysis of Proliferation.
Because proliferation thus far had been measured by incorporation of [3H]thymidine, we used an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay as a second measure of proliferation after cannabinoid treatment. Because this method can be less sensitive than measurement of [3H]thymidine incorporation, we examined proliferation after 24 or 48 h to ensure that possible changes could be seen. As measured by MTT assay, EOC-20 cells treated for up to 48 h with 1,000 nM THC proliferated at 90% of control cells levels; proliferation for cells treated with 1,000 nM CBD over 48 h was 98% of control (Fig. 1C). Regardless of cannabinoid effects on [3H]thymidine incorporation, THC and CBD had little to no effect on proliferation as measured by MTT reduction.
We used FACS analysis of propidium iodide-stained cells to directly measure DNA replication after THC treatment. Four hours of treatment with 300 or 1,000 nM THC did not increase the number of cells in G1 phase or decrease the number of cells in S or G2 phase (Fig. 1D), indicating that THC treatment overall does not decrease DNA synthesis or mitosis in microglia.
Inhibition of Nucleoside Uptake.
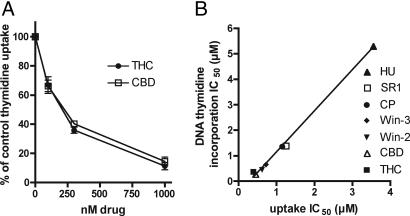
We determined the effects of cannabinoids on the transport of [3H]thymidine into EOC-20 microglia. To specifically examine inward [3H]thymidine transport, we examined uptake during the first minute after [3H]thymidine addition to the cells. THC and CBD inhibited [3H]thymidine transport and did so with almost identical potency as measured in [3H]thymidine incorporation into DNA experiments (Fig. 2). Synthetic cannabinoids also had similar inhibitory potencies in [3H]thymidine DNA incorporation and [3H]thymidine uptake experiments (r2 = 0.9954; Fig. 2B). These data indicate that CBD, THC, and other cannabinoids inhibit thymidine uptake into cells, which results in decreased [3H]thymidine incorporation into DNA.
Fig. 2.
CBD and THC inhibit uptake of [3H]thymidine into EOC-20 microglia. (A) EOC-2 cells were pretreated with cannabinoid for 30 min at 37°C, and uptake of 0.5 μCi [3H]thymidine was assayed for a period of 1 min. Nonspecific 3H counts, determined in the presence of 1 mM thymidine, were subtracted from each data point. Results are expressed as a percentage of vehicle-treated control, and vertical lines represent SEM (n = 3). (B) Comparison of cannabinoid IC50 required to inhibit DNA [3H]thymidine incorporation over 4 h, and [3H]thymidine uptake over 1 min. Nonspecific uptake was not subtracted. The linear regression line is shown; r2 = 0.995. HU, HU-210; SR1, SR144528; CP, CP55940; Win-3, Win 55212-3; Win-2, Win 55212-2.
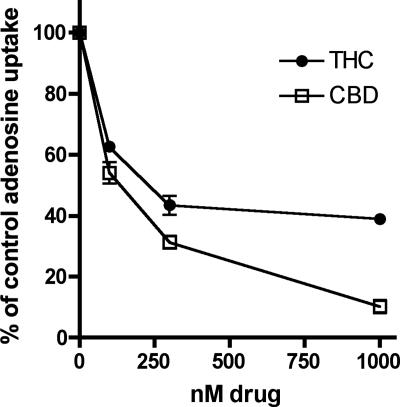
The nucleosides thymidine and adenosine can be taken into cells by the same transporter (ref. 20; see Fig. 8, which is published as supporting information on the PNAS web site). CBD inhibited transport of [3H]adenosine into EOC-20 microglia with an IC50 of 124 nM. THC also inhibited adenosine uptake in EOC-20 microglia with similar potency (Fig. 3 and Table 1). The concentration of cannabinoid required to inhibit either thymidine or adenosine uptake was similar between the two nucleosides (Table 1), suggesting a common transporter is being inhibited by these drugs.
Fig. 3.
CBD and THC inhibit [3H]adenosine uptake in EOC-20 microglia. Cells were pretreated with cannabinoid for 30 min at 37°C, and uptake of 0.5 μCi [3H]adenosine over a period of 1 min was assayed. Nonspecific uptake, determined in the presence of 1 mM adenosine, was subtracted from each data point. Results are expressed as a percentage of vehicle-treated control, and vertical lines represent SEM (n = 3).
Table 1.
Comparison of the potencies of cannabinoids to inhibit thymidine and adenosine uptake
| Compound | Thymidine uptake IC50, μM | Adenosine uptake IC50, μM |
|---|---|---|
| THC | 0.17 | 0.27 |
| CBD | 0.19 | 0.12 |
| HU-210 | 2.69 | 3.89 |
| CP55,940 | 0.76 | 1.00 |
| Win 55212-2 | 0.45 | 0.29 |
| Win 55212-3 | 0.75 | 0.45 |
| SR141716 | 0.88 | 0.95 |
| SR144528 | >30.0 | >20.0 |
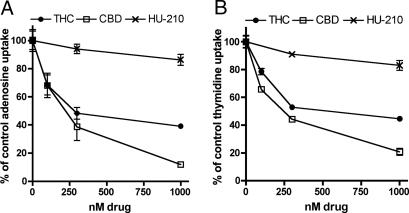
To confirm these results in different inflammatory cells, we also examined the effects of CBD and THC on adenosine uptake in the RAW264.7-transformed macrophage line. CBD inhibited adenosine and thymidine uptake in RAW264.7 cells with IC50 values of 190 and 225 nM, respectively (Fig. 4A and B). THC was slightly less potent than CBD at inhibiting uptake in RAW274.7 macrophages, with an IC50 of 334 nM for adenosine and 481 nM for thymidine (Fig. 4 A and B). As in microglia, HU-210 had no effect on nucleoside uptake in RAW264.7 cells.
Fig. 4.
Plant-derived cannabinoids inhibit adenosine and thymidine uptake in RAW264.7 macrophages. Cells were pretreated with cannabinoid for 30 min at 37°C, and uptake of radiolabeled adenosine (A) or thymidine (B) was assayed over 1 min. Nonspecific uptake, determined in the presence of 1 mM unlabeled nucleotide, was subtracted from each data point. Results are expressed as a percentage of vehicle-treated control; vertical lines represent SEM (n = 3).
Binding to the Equilibrative Nucleoside Transporter (ENT) 1.
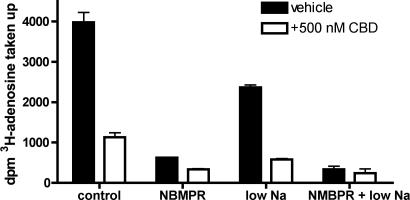
Because CBD inhibits both adenosine and thymidine uptake with similar efficacy, the drug could inhibit a common nucleoside transporter. There are two subtypes of nucleoside transporter: ENT, which are blocked by the drug S-(4-nitrobenzyl)-6-thioinosine (NBMPR), and concentrative transporters, which are dependent on the presence of extracellular sodium (20). In EOC-20 and RAW264.7 cells, the majority of adenosine and thymidine transport was NBMPR-sensitive and not affected by sodium removal, suggesting ENT are the primary transporters functioning in these cells (Fig. 5; data not shown).
Fig. 5.
Adenosine uptake in EOC-20 cells is mediated by ENT transporters. To demonstrate NBMPR sensitivity, uptake assays were carried out in microglia as described. For sodium-free experiments, the NaCl in normal BSS buffer was replaced with N-methyl-glucamine. Cannabinoids or 100 nM NBMPR was added 30 min before the addition of 3H nucleoside; uptake was measured for 1 min. Nonspecific uptake was subtracted from the total uptake. Error bars reflect SEM (n = 3).
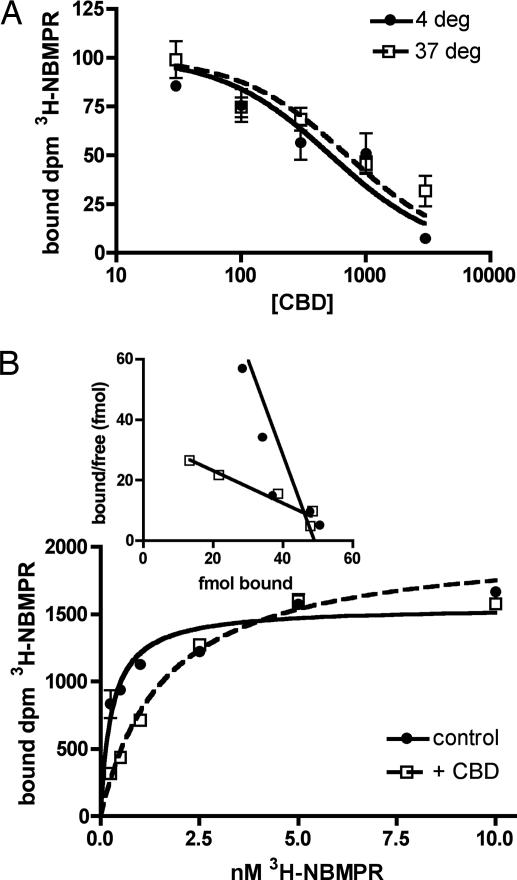
We examined CBD binding affinity for ENT using competition studies with [3H]NBMPR. NBMPR binds ENT1 with high affinity at the exofacial substrate recognition site (21) and is not known to be taken up into cells; however, to minimize possible transport, we also examined binding at 4°C. CBD inhibited [3H]NBMPR binding to EOC-20 cells with a Ki of 209 ± 89 nM at 4°C and 237 ± 94 nM at 37°C (Fig. 6A), consistent with IC50 values obtained in nucleoside uptake. CBD acts as a competitive inhibitor of [3H]NBMPR, because preincubation with 500 nM CBD increases the Kd for [3H]NBMPR by ≈6-fold, with no effect on the Bmax (Fig. 6B). Control Kd was 0.374 ± 0.071 nM, whereas the Kd in CBD-treated cells was 2.522 ± 0.674 nM (P = 0.013 by t test); control Bmax was 36.30 ± 5.56 fmol, whereas the Bmax in CBD-treated cells was 38.89 ± 2.30 fmol (P = 0.721).
Fig. 6.
CBD is a competitive inhibitor at the ENT1 transporter in EOC-20 cells. (A) Cells were preincubated for 30 min with indicated concentrations of CBD before incubation with 1 nM [3H]NBMPR for 30 min at 4°C or 37°C. Nonspecific binding, determined in the presence of 10 μM nitrobenzylthioguanosine, was subtracted; the combined results of three experiments are shown. IC50 values, derived from the solution of a single-site competition equation in individual experiments (each n = 3), were used to calculate the dissociation constant for CBD. (B) Kinetics of [3H]NBMPR binding. EOC-20 cells were preincubated with vehicle or 500 nM CBD before [3H]NBMPR was added for a 30-min incubation at 37°C. Nonspecific binding was subtracted. Shown is one representative experiment of three; the lines represent the solution of the one-site binding equation (n = 3). A Scatchard plot of the data is shown (Inset).
Adenosine Mediation of CBD Effects on TNFα.
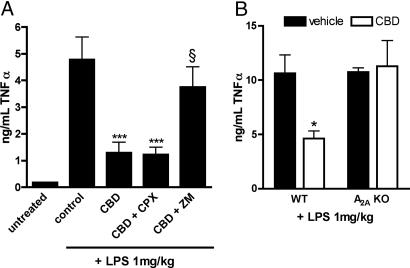
Adenosine transport inhibitors decrease TNFα in LPS-treated mice by increasing the amount of endogenous adenosine available to bind the A2A receptor (13). Because CBD inhibits adenosine transport, we hypothesized that the A2A receptor mediates the decrease in TNFα seen with CBD treatment (9). CBD (1 mg/kg) before LPS injection was sufficient to significantly decrease TNFα, compared with vehicle-treated mice (Fig. 7). Although this decrease remained unchanged upon coadministration of the A1 adenosine receptor antagonist 8-cyclopentyl-1,3-dipropylxanthine (3 mg/kg, i.p.), pretreatment with the A2A receptor antagonist ZM 241385 (10 mg/kg, i.p.) reversed the effects of CBD on TNFα (Fig. 7A). Neither antagonist alone significantly altered the effect of LPS to increase TNFα levels (data not shown). Finally, mice null for the adenosine A2A receptor were insensitive to the effects of CBD on the TNFα response to LPS, compared with wild-type mice of the same strain (Fig. 7B).
Fig. 7.
CBD decreases TNFα via activation of A2A adenosine receptors. (A) Male ICR mice were pretreated for 1 h before LPS injection with a single dose of CBD (1 mg/kg, i.p.) or vehicle. Thirty minutes later, mice were given vehicle, 8-cyclopentyl-1,3-dipropylxanthine (CPX, 3 mg/kg, i.p), or ZM 241385 (ZM, 10 mg/kg, i.p.). Mice were treated with LPS (1 mg/kg, i.v.) 1 h after CBD injection. One hour after LPS treatment, mice were killed, and serum was collected. TNFα levels were determined by ELISA (n = 6). ∗∗∗, P < 0.001 compared with vehicle control (one-way ANOVA followed by Bonferroni's posttest); §, P > 0.05 compared with control and P < 0.05 compared with CBD alone (one-way ANOVA followed by Bonferroni's posttest). (B) Wild-type or A2A-null (KO) C57BL/6 mice were pretreated for 1 h with 1 mg/kg CBD or vehicle. Mice were treated with LPS, serum was collected after 1 h, and TNFα was determined by ELISA (n = 4). ∗, P = 0.014 compared with vehicle-treated by unpaired t test.
Discussion
We began these studies to characterize the effects of plant-derived cannabinoids on microglial proliferation, as measured by incorporation of [3H]thymidine into DNA. THC and CBD dose-dependently decreased DNA incorporation of [3H]thymidine in EOC-20 microglia without affecting cell cycle or total number of cells. Instead, the predominant effect of these plant-derived cannabinoids is to decrease nucleoside transport into microglial and macrophage cells. Early reports describe an inhibitory effect of 100 μM THC on nucleoside uptake in lymphocytes, but the results were attributed to nonspecific membrane effects, which is very possible given the high concentrations used (22, 23). CBD also has been shown, at micromolar concentrations, to inhibit norepinephrine, dopamine, and serotonin uptake in synaptosomes and anandamide uptake in cultured cells (reviewed in ref. 7). We found that THC, CBD, and Win 55212-2 each inhibit thymidine and adenosine transport, with IC50 values in the nanomolar range. Interestingly, although THC and CBD had similar effects on thymidine transport, THC was less efficacious in inhibiting adenosine transport in both microglia and macrophages, with a maximal inhibition of ≈60%. Given these data, we suggest that the [3H]thymidine incorporation assay be used cautiously when examining cannabinoid effects on proliferation; the extent of interference due to inhibition of [3H]thymidine uptake or accumulation of extracellular adenosine will doubtless depend on the specific transporters available in the subject cells. Although we saw no significant effects in our MTT experiments, THC and higher concentrations of CBD can decrease proliferation of some inflammatory (10) and cancer (24) cells, as measured by tetrazolium reduction.
Our data indicate that CBD is a competitive inhibitor of adenosine uptake by an ENT. The experimentally derived Kd for [3H]NBMPR in untreated cells was ≈0.5 nM, which increased 5-fold in the presence of 500 nM CBD. Because the ENT1 subtype is the only transporter with a low nanomolar affinity for NBMPR, this suggests that CBD acts as a competitive inhibitor at the ENT1 transporter. In vivo, this inhibition increases the availability of endogenous adenosine to produce immunosuppression because the effects of CBD on TNFα were blocked by an A2A receptor antagonist and absent in A2A receptor knockout mice. This mechanism gives rationale to the antiinflammatory activity of CBD identified in other mouse models (8, 9). However, we cannot rule out the possibility that, in addition to binding to the ENT transporter, CBD could also bind and activate the A2A receptor.
We have calculated, using the Cheng and Prusoff equation (40), that the Ki for CBD to inhibit adenosine transport is ≈0.12 μM. Because CBD inhibition is competitive, the potency of CBD to inhibit adenosine uptake in vivo will be dependent on the concentration of adenosine. Assuming a Kd value of adenosine for the ENT1 transporter of ≈40 μM (25, 26) and concentrations of adenosine ranging from 0.8 μM in normal human serum to 8 μM after septic shock (27) and local concentrations as high as 100 μM (28), we determined that the IC50 of CBD under normal physiological conditions is 0.122 μM, which rises to 0.144 μM given an adenosine concentration of 8 μM and peaks at 0.42 μM at adenosine concentrations of 100 μM.
There are previous reports of interactions between THC and the adenosine system: antisense toward A1 receptors blocks THC-induced ataxia (29), and long-term treatment with THC is known to desensitize cerebellar A1 receptors (30). There is also some evidence of dependence on adenosine receptor signaling for cannabinoid effects; for instance, A2A receptors are required for physical dependence on THC, and THC-induced rewarding and aversive effects are significantly reduced in A2A-null mice (31). Inhibition of adenosine uptake by THC could account for these observed interactions.
CBD is an attractive medical alternative to smoked marijuana or plant extract because of its lack of psychoactive and cognitive effects (5), but little is known of its specific effects. The implications for this study are many. First, drugs that enhance adenosine signaling are of clinical interest in treatment of inflammation, the reduction of infarct size following myocardial or cerebral ischemia (20, 32), and as anti-seizure therapy (33). Nucleoside transporter inhibitors are also intriguing as adjunct therapy in cancer patients to potentiate the effectiveness of antifolate drugs (34). CBD is already known to be well tolerated in humans (35), which has previously been a problem for other nucleoside inhibitor drugs (36, 37). However, in daily use of CBD or THC, there is a chance for adenosine receptor desensitization; chronic consumption of ethanol, another adenosine transporter inhibitor, results in cell tolerance to adenosine through heterologous desensitization of receptors (38). For this reason, it is important to determine the long-term effects of marijuana or CBD use on adenosine receptors.
Materials and Methods
Reagents.
THC, CBD, SR144528, and SR141716 were obtained through the National Institute on Drug Abuse drug supply program, and CP55,940 was donated by Pfizer Central Research (Groton, CT). HU-210 was a generous gift from R. Mechoulam (Hebrew University, Jerusalem). All other materials, drugs, and radioisotopes used in these studies were obtained from standard commercial sources.
Cell Culture.
EOC-20 microglia and RAW264.7 macrophages were obtained from American Type Culture Collection (Manassas, VA) and maintained according to manufacturer's instructions. For EOC-20 cells, 20% LADMAC-conditioned medium (LMCM) was used as a source of macrophage colony-stimulating factor. LADMAC (American Type Culture Collection) is a bone marrow-derived cell line that secretes macrophage colony-stimulating factor (39), and the medium obtained from these cells was titrated for potency in inducing proliferation.
[3H]Thymidine Incorporation Assays.
A [3H]thymidine incorporation assay was used to measure proliferation as described (19), with minor modifications. EOC-20 cells were grown to 60% confluency in complete medium and synchronized by replacing the medium with DMEM. After 24 h of starvation, LMCM was added back to the medium to stimulate cell growth. Cannabinoids (in 1 μl of DMSO) and [3H]thymidine (Amersham Pharmacia Biosciences) were added for a 4-h incubation. Cells were washed, macromolecules were precipitated and solubilized, and incorporated [3H]thymidine was determined by scintillation counting.
Cell Cycle Analysis.
Cell cycle analysis was done by FACS after staining DNA with propidium iodide. Microglia in 60-mm dishes were starved for 24 h in DMEM and stimulated for 24 h with LMCM. During the final 4 h of stimulation, cells were treated with THC or DMSO vehicle. After incubation, cells were washed twice in cold PBS, scraped in 100 μl of PBS per dish, pelleted by centrifugation, and resuspended in 100 μl of PBS. Cold 95% ethanol (280 μl per tube) was added dropwise to each sample while vortexing gently, and samples were incubated for 1 h at 4°C to permeabilize. Pelleted cells were resuspended in 125 μl of ribonuclease (500 units/ml in PBS), and samples were incubated for 15 min at 37°C before adding 625 μl of propidium iodide (50 μg/ml in PBS) per tube. Fluorescence was analyzed on a FACS Scan Flow Cytometer (Becton Dickinson). Cells were gated to exclude cell fragments or fused cells, and cells in each stage were determined as a percentage of total gated cells.
MTT Assay.
Microglial cells were grown in 12-well plates to 60% confluency and synchronized by replacing the normal medium with DMEM. After 24 h, drug treatments and LMCM were added. Some wells were designated “starved controls” and denied LMCM. After 23 or 47 h of treatment, 100 μl of 2.5 mg/ml MTT tetrazolium salt was added to each well, and the plate was returned to a 37° CO2 incubator for 1 h. Medium was aspirated, 500 μl of acidified isopropanol (500 μl HCl per 75 ml isopropanol) was added to each well, and the plate was shaken for 5 min. Absorbance at 562 nm was determined in a 200-μl aliquot.
Nucleoside Uptake Assays.
Unless indicated, assays were carried out at 37°C in Earle's normal balanced salt solution (BSS). Cells grown in 12-well plates were washed once in BSS and preincubated 30 min at 37°C with drug or DMSO vehicle, added in 1 μl. Uptake began after addition of 0.5 μCi [3H]adenosine or [3H]thymidine (1 Ci = 37 GBq); after 1 min buffer was rapidly aspirated, and cells were washed once with ice-cold PBS. Cells were solubilized in 0.2 M NaOH/1% SDS, and radioactivity was determined. Nonspecific uptake was defined as uptake in the presence of 1 mM adenosine or thymidine, which was added 1 min before 3H nucleotide. For low-sodium medium, NaCl in BSS was replaced with N-methylglucamine at equal molarity.
[3H]NBMPR Binding.
EOC-20 cells in 12-well plates were washed once, and the medium was replaced with BSS. CBD was added in 1 μl of DMSO, and nitrobenzylthioguanosine (10 μM) was used to determine nonspecific binding. After a 30-min preincubation, 0.25–10 nM [3H]NBMPR was added; plates were shaken either in a 37°C water bath or on ice for 30 min then washed with 1 ml of ice-cold PBS. Radioactivity was determined after solubilizing cells in NaOH/SDS.
LPS Treatment of Mice.
All animal studies were carried out in accordance with the National Institutes of Health Guide for the Use and Care of Laboratory Animals. Male ICR or C57BL/6 mice were pretreated for 1 h with 1 mg/kg CBD or vehicle (DMSO/emulphor/saline in a 1:1:18 ratio) i.p. After 30 min, mice were given 8-cyclopentyl-1,3-dipropylxanthin, ZM 241285, or vehicle i.p. For ZM 241385, it was necessary to decrease vehicle ratio to 1:1:8; TNFα levels from mice treated with this vehicle were indistinguishable from mice treated with 1:1:18 vehicle (data not shown). After drug pretreatment, mice were given LPS in a single 50-μl tail vein injection. After 1 h, mice were killed by swift decapitation after anesthesia with isoflurane, and blood was collected. Serum was kept at 4°C overnight and assayed for TNFα the next day.
TNFα Assay.
Serum TNFα levels were determined by using an ELISA kit from Assay Designs (Ann Arbor, MI). Serum samples were diluted 1:4 (ICR mice) or 1:9 (C57BL/6 mice) with buffer, and absorbance values were compared with a standard curve generated by using the recombinant TNFα provided.
Statistical Analysis.
All data were analyzed by using prism software (Graphpad, San Diego, CA), and means with standard error (SEM) were determined for collected samples. The Ki values for competing ligands were determined from IC50 values by using the formula of Cheng and Prusoff (40): Ki = IC50/[1 + (radioligand)/Kd], where the Kd is the dissociation constant of the radioligand. IC50 values for the competitors were determined from nonlinear regression fitting of the concentration response data to the equation: % inhibition = 100/[1 + 10(log x − log IC50)], where x is the concentration of inhibitor.
The Kd and Bmax values were determined from nonlinear regression fitting of the concentration response data to the equation: bound = Bmax × x/(Kd + x), where x is the concentration of ligand.
To determine statistical significance of samples among multiple groups, one-way ANOVA followed by Bonferroni's posttest was used. When determining statistical differences between two individual groups, an unpaired t test was performed and the exact P value given.
Supplementary Material
Acknowledgments
We thank Dr. Jian-Fan Chen (Boston University, Boston) for creating and providing the A2A-null mice, Dr. Eugene Ponomarev for assistance in FACS analysis, Dr. David Rademacher and Gregory McHugh for their assistance in i.v. injections, and Dr. Christopher Kearn for preliminary experiments. We also thank Drs. Garrett Gross and Jason Peart for helpful discussions. This work was supported by National Institutes of Health Grants DA09155, DA16967, and NS041314 (to C.J.H.) and HL077707 (to J.A.A.).
Abbreviations
- PTx
pertussis toxin
- LPS
lipopolysaccharide
- BSS
Earle's buffered salt solution
- CBD
cannabidiol
- ENT
equilibrative nucleoside transporter
- NBMPR
S-(4-nitrobenzyl)-6-thioinosine
- THC
Δ9-tetrahydrocannabinol
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. R.M. is a guest editor invited by the Editorial Board.
References
- 1.Croxford J. L., Yamamura T. J. Neuroimmunol. 2005;166:3–18. doi: 10.1016/j.jneuroim.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 2.Wade D. T., Makela P., Robson P., House H., Bateman C. Mult. Scler. 2004;10:434–441. doi: 10.1191/1352458504ms1082oa. [DOI] [PubMed] [Google Scholar]
- 3.Buckley N. E., McCoy K. L., Mezey E., Bonner T., Zimmer A., Felder C. C., Glass M. Eur. J. Pharmacol. 2000;396:141–149. doi: 10.1016/s0014-2999(00)00211-9. [DOI] [PubMed] [Google Scholar]
- 4.Mechoulam R., Parker L. A., Gallily R. J. Clin. Pharmacol. 2002;42:11S–19S. doi: 10.1002/j.1552-4604.2002.tb05998.x. [DOI] [PubMed] [Google Scholar]
- 5.Belgrave B. E., Bird K. D., Chesher G. B., Jackson D. M., Lubbe K. E., Starmer G. A., Teo R. K. Psychopharmacology (Berlin) 1979;64:243–246. doi: 10.1007/BF00496070. [DOI] [PubMed] [Google Scholar]
- 6.Razdan R. K. Pharmacol. Rev. 1986;38:75–149. [PubMed] [Google Scholar]
- 7.Pertwee R. G. In: Cannabinoids. Di Marzo V., editor. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 32–83. [Google Scholar]
- 8.Costa B., Colleoni M., Conti S., Parolaro D., Franke C., Trovato A. E., Giagnoni G. Naunyn-Schmiedeberg's Arch. Pharmacol. 2004;369:294–299. doi: 10.1007/s00210-004-0871-3. [DOI] [PubMed] [Google Scholar]
- 9.Malfait A. M., Gallily R., Sumariwalla P. F., Malik A. S., Andreakos E., Mechoulam R., Feldmann M. Proc. Natl. Acad. Sci. USA. 2000;97:9561–9566. doi: 10.1073/pnas.160105897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steffens S., Veillard N. R., Arnaud C., Pelli G., Burger F., Staub C., Karsak M., Zimmer A., Frossard J. L., Mach F. Nature. 2005;434:782–786. doi: 10.1038/nature03389. [DOI] [PubMed] [Google Scholar]
- 11.Streit W. J., Walter S. A., Pennell N. A. Prog. Neurobiol. 1999;57:563–581. doi: 10.1016/s0301-0082(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 12.Hasko G., Cronstein B. N. Trends Immunol. 2004;25:33–39. doi: 10.1016/j.it.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Noji T., Takayama M., Mizutani M., Okamura Y., Takai H., Karasawa A., Kusaka H. J. Pharmacol. Exp. Ther. 2002;300:200–205. doi: 10.1124/jpet.300.1.200. [DOI] [PubMed] [Google Scholar]
- 14.Erdmann A. A., Gao Z.-G., Jung U., Foley J., Borenstein T., Jacobson K. A., Fowler D. H. Blood. 2005;105:4707–4714. doi: 10.1182/blood-2004-04-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein T. W., Newton C., Larsen K., Lu L., Perkins I., Nong L., Friedman H. J. Leukocyte Biol. 2003;74:486–496. doi: 10.1189/jlb.0303101. [DOI] [PubMed] [Google Scholar]
- 16.Howlett A. C., Barth F., Bonner T. I., Cabral G., Casellas P., Devane W. A., Felder C. C., Herkenham M., Mackie K., Martin B. R., et al. Pharmacol. Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- 17.Glass M., Northup J. K. Mol. Pharmacol. 1999;56:1362–1369. doi: 10.1124/mol.56.6.1362. [DOI] [PubMed] [Google Scholar]
- 18.Howlett A. C., Qualy J. M., Khachatrian L. L. Mol. Pharmacol. 1986;29:307–313. [PubMed] [Google Scholar]
- 19.Carrier E. J., Kearn C. S., Barkmeier A. J., Breese N. M., Yang W., Nithipatikom K., Pfister S. L., Campbell W. B., Hillard C. J. Mol. Pharmacol. 2004;65:999–1007. doi: 10.1124/mol.65.4.999. [DOI] [PubMed] [Google Scholar]
- 20.Kong W., Engel K., Wang J. Curr. Drug Metab. 2004;5:63–84. doi: 10.2174/1389200043489162. [DOI] [PubMed] [Google Scholar]
- 21.Hyde R. J., Cass C. E., Young J. D., Baldwin S. A. Mol. Membr. Biol. 2001;18:53–63. [PubMed] [Google Scholar]
- 22.Desoize B., Leger C., Jardillier J. C., Nahas G. Adv. Biosci. 1978;22–23:145–159. doi: 10.1016/b978-0-08-023759-6.50017-2. [DOI] [PubMed] [Google Scholar]
- 23.Desoize B., Leger C., Nahas G. C. R. Hebd. Seances Acad. Sci. D. 1978;287:1441–1444. [PubMed] [Google Scholar]
- 24.Massi P., Vaccani A., Ceruti S., Colombo A., Abbracchio M. P., Parolaro D. J. Pharmacol. Exp. Ther. 2004;308:838–845. doi: 10.1124/jpet.103.061002. [DOI] [PubMed] [Google Scholar]
- 25.Kiss A., Farah K., Kim J., Garriock R. J., Drysdale T. A., Hammond J. R. Biochem. J. 2000;352:363–372. [PMC free article] [PubMed] [Google Scholar]
- 26.Hong M., Schlichter L., Bendayan R. J. Pharmacol. Exp. Ther. 2000;292:366–374. [PubMed] [Google Scholar]
- 27.Martin C., Leone M., Viviand X., Ayem M. L., Guieu R. Crit. Care Med. 2000;28:3198–3202. doi: 10.1097/00003246-200009000-00014. [DOI] [PubMed] [Google Scholar]
- 28.Sottofattori E., Anzaldi M., Ottonello L. J. Pharm. Biomed. Anal. 2001;24:1143–1146. doi: 10.1016/s0731-7085(00)00574-4. [DOI] [PubMed] [Google Scholar]
- 29.Dar M. S., Mustafa S. J. Brain Res. 2002;957:53–60. doi: 10.1016/s0006-8993(02)03599-0. [DOI] [PubMed] [Google Scholar]
- 30.Selley D. E., Cassidy M. P., Martin B. R., Sim-Selley L. J. Mol. Pharmacol. 2004;66:1275–1284. doi: 10.1124/mol.104.000604. [DOI] [PubMed] [Google Scholar]
- 31.Soria G., Castane A., Berrendero F., Ledent C., Parmentier M., Maldonado R., Valverde O. Eur. J. Neurosci. 2004;20:2203–2213. doi: 10.1111/j.1460-9568.2004.03682.x. [DOI] [PubMed] [Google Scholar]
- 32.Hirai K., Ashraf M. J. Mol. Cell. Cardiol. 1998;30:1803–1815. doi: 10.1006/jmcc.1998.0745. [DOI] [PubMed] [Google Scholar]
- 33.Boison D. Neuroscientist. 2005;11:25–36. doi: 10.1177/1073858404269112. [DOI] [PubMed] [Google Scholar]
- 34.Smith P. G., Thomas H. D., Barlow H. C., Griffin R. J., Golding B. T., Calvert H., Newell D. R., Curtin N. J. Clin. Cancer Res. 2001;7:2105–2113. [PubMed] [Google Scholar]
- 35.Cunha J. M., Carlini E. A., Pereira A. E., Ramos O. L., Pimentel C., Gagliardi R., Sanvito W. L., Lander N., Mechoulam R. Pharmacology. 1980;21:175–185. doi: 10.1159/000137430. [DOI] [PubMed] [Google Scholar]
- 36.Baldwin S. J., Mackey J. R., Cass C. E., Young J. D. Mol. Med. Today. 1999;5:216–224. doi: 10.1016/S1357-4310(99)01459-8. [DOI] [PubMed] [Google Scholar]
- 37.Mackey J. R., Baldwin S. A., Young J. D., Cass C. E. Drug Resist. Updat. 1998;1:310–324. doi: 10.1016/s1368-7646(98)80047-2. [DOI] [PubMed] [Google Scholar]
- 38.Gordon A. S., Nagy L., Mochly-Rosen D., Diamond I. Biochem. Soc. Symp. 1990;56:117–136. [PubMed] [Google Scholar]
- 39.Sklar M. D., Tereba A., Chen B. D., Walker W. S. J. Cell. Physiol. 1985;125:403–412. doi: 10.1002/jcp.1041250307. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Y. C., Prusoff W. H. Biochem. Pharmacol. 1973;22:3099–3110. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.