Abstract
Japanese encephalitis virus (JEV), a mosquito-borne flavivirus that causes severe human disease, has been shown to block the interferon (IFN)-induced Janus kinase signal transducer and activation of transcription (Jak-Stat) signaling cascade by preventing Tyk2 tyrosine phosphorylation and Stat activation. In this study, we demonstrate that expression of the JEV nonstructural protein NS5 readily blocked IFN-stimulated Jak-Stat signaling events such as Stat1 nuclear translocation and tyrosine phosphorylation of Tyk2 and Stat1. The region of JEV NS5 responsible for Stat1 suppression was identified using various deletion clones. Deletion of 83 N-terminal residues of JEV NS5, but not the 143 C-terminal residues, abolished its ability to block IFN-stimulated Stat1 activation. The role of JEV NS5 as an IFN antagonist was further demonstrated by its ability to block the induction of interferon-stimulated genes and the antiviral effect of IFN-α against the IFN-sensitive encephalomyocarditis virus, which appears to replicate and kill cells that express NS5 even with alpha IFN treatment. Furthermore, the molecular mechanism responsible for IFN antagonism by NS5 probably involves protein tyrosine phosphatases (PTPs), as the IFN-blocking events in both JEV-infected and NS5-expressing cells were reversed by sodium orthovanadate, a broad-spectrum inhibitor of PTPs. We suggest that JEV NS5 is an IFN antagonist and that it may play a role in blocking IFN-stimulated Jak-Stat signaling via activation of PTPs during JEV infection.
Japanese encephalitis virus (JEV) is a mosquito-borne flavivirus that causes human epidemic encephalitis in Asia annually (43). The genome of JEV is a single-stranded positive-sense RNA of approximately 11 kb in length which contains a single long open reading frame encoding a polyprotein precursor. Cleavage of the polyprotein by cellular and viral proteases yields three structural proteins (core [C], precursor membrane [prM], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (31). Replication of the flavivirus is initiated by a viral RNA replicase complex through a process of RNA-dependent RNA polymerization in the perinuclear endoplasmic reticulum membranes (46, 50). Nonstructural proteins NS3 and NS5 have been identified as the major components of the viral RNA replicase complex associated with the 3′ noncoding region of genomic RNA in the initiation of viral replication (6, 47). NS5, the largest and most conserved flavivirus protein encoded in the open reading frame, contains sequences homologous to methyltransferase (MTase) and RNA-dependent RNA polymerase (RdRP); the former is involved in methylation of the 5′ RNA cap structure, and the latter is the key enzyme for viral replication (1, 11, 17, 24).
The alpha/beta interferon (IFN-α/β) response is the host's main innate immune mechanism against viral infection (39). The induction of IFNs during viral infection is mediated by the coordinate activation of multiple cellular transcription factors such as interferon regulatory factor (IRF), NF-κB, and c-Jun/ATF-2 (10, 49). IFN signaling is known to be mediated by the Janus kinase signal transducer and activation of transcription (Jak-Stat) pathway (13, 27, 39, 45), which is initiated by binding to the cell surface receptors, IFNAR1 and IFNAR2. Ligation of IFN and its receptors leads to activation of Jak1 and Tyk2 through tyrosine phosphorylation, which in turn stimulates the phosphorylation of Stats. Subsequently, phosphorylated Stat1 and Stat2 dimerize and associate with IRF-9 to form ISGF3 complexes. The formation of ISGF3 complexes in the cytoplasm results in nuclear translocation, binding to the IFN-stimulated responsive element, and consequent expression of proteins including the IFN-stimulated antiviral proteins (9). Several cellular proteins have also been identified as negative regulators of the Jak-Stat signaling pathway (42); these include the suppressor of cytokine signaling (SOCS) proteins (40, 44, 48), the protein inhibitors of activated Stats (PIAS) (32, 33), and the protein tyrosine phosphatases (PTPs) (37, 51).
Viruses have evolved various mechanisms for initiating viral replication in the host by disrupting the actions of IFN and evading IFN-stimulated antiviral responses (12, 23, 27, 41). The main strategies adapted by viruses are (i) inhibition of IFN production and secretion, (ii) competition for binding to IFN receptors through viral decoy receptors, (iii) degradation or suppression of activation of Jak-Stat components, and (iv) inhibition of the actions of IFN-induced antiviral proteins. Recently, several studies have found that flaviviruses such as JEV (30), West Nile virus (WNV) (16, 34), dengue virus serotype 2 (DEN-2) (19, 30), and tick-borne Langat virus (LGTV) (2) counteract IFN-induced Jak-Stat signaling primarily by blocking the phosphorylation of the signaling components Jak1, Tyk2, Stat1, and Stat2. Moreover, DEN-2 has also been reported to subvert the IFN response by downregulating Stat2 protein expression (22). Flavivirus proteins capable of blocking IFN signaling events include NS4B of DEN-2, WNV, and yellow fever virus (35, 36); NS2A and NS4A of DEN-2 and WNV (34, 36); and NS5 of LGTV (2).
The interplay between JEV and the IFN system has been studied recently. JEV infection results in IFN-β production through an RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation pathway (5). Even though IFN is produced by the host cells, JEV has evolved a way to block the IFN-stimulated Jak-Stat signaling by preventing activation of Tyk2 and Stats (30). In this study, we investigated the JEV protein(s) responsible for blocking IFN-induced Jak-Stat signaling. We found that expression of JEV NS5 readily blocked IFN-α-stimulated Jak-Stat signaling events such as Stat1 nuclear translocation and phosphorylation of Tyk2 and Stat1. Cells expressing JEV NS5 potentiated the IFN-sensitive encephalomyocarditis virus (EMCV) to evade IFN-mediated antiviral effects. Moreover, treatment of cells with sodium orthovanadate, a broad-spectrum inhibitor of PTPs, suppressed the abilities of JEV and NS5 to antagonize IFN. Our data indicate that JEV NS5 is a potent IFN antagonist and that it may play a pivotal role in inhibiting IFN signaling through a PTP-dependent mechanism.
MATERIALS AND METHODS
Viruses, cell lines, chemicals, and antibodies.
A Taiwanese JEV strain, RP-9 (7), was used throughout this study. JEV was propagated using C6/36 cells in RPMI 1640 medium containing 5% fetal bovine serum (FBS) (Gibco). EMCV (ATCC number VR-995) was propagated in Vero cells using Eagle's minimum essential medium containing 10% FBS. BHK-21 was grown in RPMI 1640 medium containing 5% FBS. Human lung carcinoma A549 cells were maintained in F-12 medium (Gibco) supplemented with 10% FBS. Vero cells were cultured in Eagle's minimum essential medium containing 10% FBS. Human embryonic kidney 293T cells were grown in Dulbecco's modified Eagle's medium (Sigma) containing 10% FBS. Recombinant human IFN-αA/D and sodium orthovanadate were obtained from Sigma, and the protease inhibitor cocktail was obtained from Roche. Rabbit polyclonal anti-Stat1 (#9172), anti-phospho-Stat1 (Tyr701) (#9171), and anti-phospho-Tyk2 (Tyr1054/1055) (#9321) antibodies were purchased from Cell Signaling Technology. Antibodies against Tyk2 (#610173) and PKR (#610764) were obtained from BD Transduction Laboratories. Rabbit polyclonal anti-IRF-9 (sc-496) and anti-IRF-1 (sc-497) antibodies were from Santa Cruz Biotechnology. Mouse monoclonal anti-Flag M2 antibody was from Upstate Biotechnology.
Plasmids.
To construct pStat1-DsRed, the human Stat1 cDNA was amplified from IFN-α-treated A549 cells by reverse transcription-PCR using primers specific for human Stat1 (forward primer, 5′-ATGTCTCAGTGGTACGAACTTCA-3′; reverse primer, 5′-TTACACTTCAGACACAGAAATCAAC-3′) and then cloned in frame into the N terminus of a red fluorescent protein, DsRed2 (Clontech).
For the expression of JEV proteins, cDNA fragments of JEV RP-9 (GenBank accession number AF014161) (7) encoding the individual viral proteins were subcloned to a Flag-tagged pCR3.1 (Invitrogen). Initiation codons were added to these constructs at their N termini. The JEV nucleotide numbers and expected molecular sizes for each of the viral protein constructs are listed below (see Fig. 2A). For truncated NS5 plasmids, the corresponding DNA fragments were obtained from the NS5-Flag/pCR3.1 plasmid by MluI digestion for NS5 amino acids (aa) 167 to 905 or by PCR using primers specific for the other fragments and then subcloned in flame to a Flag-tagged pCR3.1. All of the constructs were handled using standard molecular cloning techniques and verified by DNA sequencing.
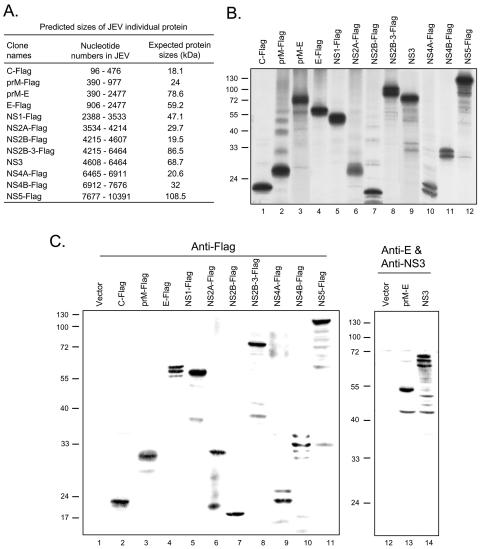
FIG. 2.
Protein expression of plasmid constructs encoding individual JEV proteins. (A) The corresponding nucleotide numbers of JEV (GenBank accession number AF014161) contained in each construct and their predicted sizes with tags but not posttranslational modifications. (B) The protein expression patterns of the C-terminally Flag-tagged pCR3.1 plasmids expressing individual JEV proteins (C, prM, E, NS1, NS2A, NS2B, NS2B-3, NS4A, NS4B, and NS5) and pCR3.1 plasmids expressing JEV prM-E and NS3 proteins in an in vitro transcription/translation system. (C) BHK-21 cells were transfected with the indicated plasmids for 24 h before the cell lysates were harvested for Western blotting with anti-Flag, anti-JEV E, and NS3 antibodies as indicated. The molecular masses (kDa) of protein standards are shown on the left side of the gel.
The SIN DNA-based vector includes a respiratory syncytial virus promoter-driving expression cassette derived from a recombinant Sindbis virus (SIN) vector (dsTE12Q) (26) and is able to express the SIN replicase complex from nonstructural genes. The replicase then turns on the subgenomic promoter to express structural and inserted genes. For the recombinant SIN constructs, the full-length and truncated NS5 DNA fragments were amplified by PCR from NS5-Flag/pCR3.1 by use of specific primers. The amplified DNA fragments with BstEII restriction sites were digested with BstEII restriction enzyme and inserted into the SIN DNA-based vector.
To construct pTY-EF-NS5-Flag for the lentivirus vector, the cDNA fragment of NS5 was amplified from NS5-Flag/pCR3.1 by PCR using specific primers. The amplified DNA fragments with BstEII restriction sites were digested with BstEII, blunted by T4 DNA polymerase, and then cloned into the EcoRI-digested, T4 DNA polymerase-blunted pTY-EF1α plasmid (4).
Generation of recombinant SIN.
Transfection of BHK-21 cells with SIN DNA-based vector was performed using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Approximately 1 μg of plasmid DNA was used to transfect 5 × 105 cells in a six-well culture plate. After 48 to 72 h of transfection, SIN was harvested from the culture supernatants, and the viral titer (PFU/ml) was determined using a plaque-forming assay on BHK-21 cells.
Lentivirus vector preparation.
The lentivirus vector system was as described previously (4, 8, 21). Briefly, 293T cells were cotransfected with pTY-EFeGFP or pTY-EF-NS5-Flag and the three helper plasmids, pNHP, pHEF-VSV-G, and pCEP4-tat, by use of Superfect transfection reagent (QIAGEN). Transfected cells were incubated at 37°C in an atmosphere of 3% CO2 for 4 to 5 h and refed with fresh medium. Cell supernatants containing the viral vectors were harvested at 24, 48, and 60 h after transfection. The virus was filtered using a 0.45-μm low-protein-binding filter and stored at −80°C. The virus supernatant was concentrated by centrifugation at 27,000 rpm at 4°C for 3.5 h using an SW28 (Beckman) rotor. Virus pellets were resuspended in 1 to 2 ml fresh medium and stored at −80°C. The lentivirus vectors encoding enhanced green fluorescent protein (eGFP) or NS5 were transduced into cells by serial dilution of the virus with 8 μg/ml Polybrene to determine the relative titers by immunofluorescence assay.
In vitro transcription and translation.
Proteins were transcribed and translated using a transcription and translation T7 quick-coupled rabbit reticulocyte lysate transcription/translational system as described by the manufacturer (Promega). Briefly, 0.5 μg of plasmid was incubated with the transcription/translation system in the presence of 35S-labeled methionine at 30°C for 90 min. The translated product was separated using sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and detected by autoradiography.
Western immunoblot analysis.
Cell lysates were prepared in SDS sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mM dithiothreitol, 0.1% bromophenol blue) containing a cocktail of protease inhibitors (Roche). Similar amounts of proteins were loaded into the gel, separated by SDS-polyacrylamide gel electrophoresis, and transferred to a nitrocellulose membrane (Hybond-C Super; Amersham). The nitrocellulose membrane was blocked with 5% skim milk in TBS-T (25 mM Tris, 0.8% NaCl, 2.68 mM KCl, pH 7.4, with 0.1% Tween 20) and subsequently incubated overnight with the primary antibody. The blots were then treated with a horseradish peroxidase-conjugated secondary antibody (Amersham) and developed using an ECL system (Amersham). For reblotting, the membrane was washed with 1× ReBlot plus strong antibody-stripping solution (Chemicon) for 15 min at room temperature. The membrane was then blocked twice with 5% skim milk in TBS-T for 5 min; this procedure was followed by reprobing with the antibody as described above.
Immunofluorescence.
BHK-21 cells were cotransfected with pStat1-DsRed and various plasmids encoding JEV proteins in serum-free medium with Lipofectamine (Invitrogen). Transfected cells were left untreated (control) or were treated with IFN-αA/D (2,500 U/ml), after which they were fixed and permeabilized in 2% formaldehyde at the indicated times. The cells were probed with anti-Flag M2 antibody and subsequently with Alexa-Fluor 488 goat anti-mouse immunoglobulin G (Molecular Probes). Subsequently, the expression and location of JEV proteins (green) and Stat1-DsRed (red) were observed under a fluorescence microscope.
IFN antiviral assay.
Briefly, the parental A549 cells and those transduced with lentivirus vectors expressing eGFP or JEV NS5 were seeded in six-well plates and incubated with or without recombinant human IFN-αA/D (500 U/ml) for 16 h. Cells were then adsorbed with mouse EMCV (multiplicity of infection [MOI] = 0.1) for 1 h. The cells were washed twice in serum-free F-12 medium and then maintained in F-12 medium supplemented with 10% FBS. Culture media were harvested for the plaque-forming assay at 32 h postinfection, and cells were fixed and stained with crystal violet for the cytopathic effect (CPE) assay at 48 h postinfection.
RESULTS
JEV NS5 effectively inhibits IFN-α-induced Stat1 nuclear translocation.
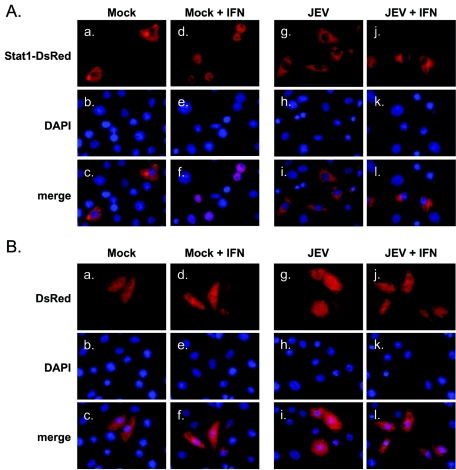
To monitor the IFN-stimulated Jak-Stat signaling, we constructed pStat1-DsRed, which encodes the human Stat1 fused in frame with a red fluorescent protein, DsRed. The Stat1-DsRed fusion protein was predominantly located in the cytoplasmic compartment of the untreated BHK-21 cells (Fig. 1A, panels a through c). After IFN-α treatment (1, 8, 12, and 15 h), the DsRed-tagged Stat1 colocalized with DAPI (4′,6′-diamidino-2-phenylindole) staining (Fig. 1A, panels d through f; also data not shown), indicating that the Stat1-DsRed fusion protein translocated to and remained in the nucleus in response to IFN-α. In contrast, JEV infection resulted in cytoplasmic retention of Stat1-DsRed upon IFN-α treatment (Fig. 1A, panels g through l), which is consistent with our previous finding that JEV is capable of counteracting IFN-α signal transduction (30). The cellular distribution of DsRed alone was localized in both the cytoplasm and the nucleus of mock- and JEV-infected cells irrespective of IFN-α treatment (Fig. 1B). These results demonstrate that the intracellular status of Jak-Stat signaling can be visualized using the Stat1-DsRed fusion protein.
FIG. 1.
Blocking of IFN-stimulated nuclear translocation of Stat1-DsRed fusion protein in JEV-infected BHK-21 cells. BHK-21 cells adsorbed with JEV (MOI = 10) or medium only (Mock) for 1 h were transfected with pStat1-DsRed for 6 h. Cells were then treated with 2,500 U/ml of IFN-αA/D or left untreated for 15 h and subsequently fixed and permeabilized for immunofluorescence assay. Cellular localization of Stat1-DsRed (A) or control DsRed (B) was observed under a fluorescence microscope (red). The nuclei were stained with DAPI (blue). Representative cells from the same field and the merged images for each experimental group are shown.
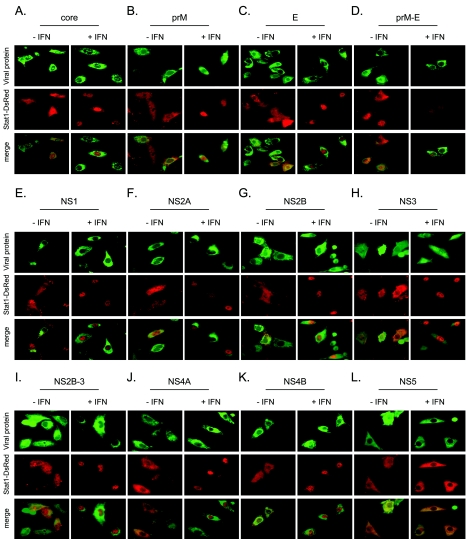
Viral proteins acting as IFN antagonists to inhibit IFN signal transduction pathway have been identified for several viruses (39, 41). Our results indicate that inhibition of IFN-α signaling by JEV likely requires viral protein synthesis, suggesting that a certain JEV protein(s) acts as an IFN antagonist (30). To identify the JEV protein(s) responsible for blocking IFN signaling, a series of JEV expression plasmids were generated to individually express Flag-tagged C, prM, E, NS1, NS2A, NS2B, NS2B-3, NS4A, NS4B, and NS5, as well as untagged prM-E and NS3. The leader sequences for the three viral glycoproteins, prM, E, and NS1, have been preserved in the corresponding constructs. To ensure proper folding of proteins or to maintain the intrinsic enzymatic activity, we also made the constructs expressing prM plus E (prM-E) (18) and NS2B plus NS3 (NS2B-3) (3). These constructs all expressed proteins of the expected molecular sizes as verified by an in vitro transcription/translation system (Fig. 2A and B), and by Western blotting in the transfected cells (Fig. 2C). We cotransfected pStat1-DsRed with plasmids expressing various JEV proteins in BHK-21 cells, which were then stimulated with IFN-α. Cells expressing viral proteins were identified by immunofluorescence staining with anti-Flag, anti-JEV E, or NS3 antibody. The localization of Stat1-DsRed was observed under a fluorescence microscope. As shown in Fig. 3, BHK-21 cells expressing NS5 (panel L) clearly showed cytoplasmic retention of Stat1-DsRed in response to treatment with a high dose of IFN-α (2,500 U/ml), whereas in cells expressing other viral proteins (panels A through K), Stat1-DsRed was readily translocated to the nucleus upon IFN-α treatment. In the transfected cells, it was seen that without IFN treatment, Stat1-DsRed mainly remained in the cytoplasm (Fig. 3). These results indicate that JEV NS5 potently inhibits IFN-α signaling by blocking IFN-induced nuclear translocation of Stat1.
FIG. 3.
Inhibition of IFN-stimulated nuclear translocation of Stat1-DsRed by JEV NS5 in BHK-21 cells. BHK-21 cells were cotransfected with plasmids expressing the indicated JEV proteins and pStat1-DsRed. Twenty-four hours after transfection, cells were stimulated with or without IFN-αA/D (2,500 U/ml) for 16 h. Cells were then fixed and permeabilized for immunofluorescence assay. JEV proteins (C, prM, E, NS1, NS2A, NS2B, NS2B3, NS4A, NS4B, and NS5) were detected using a mouse anti-Flag antibody and Alexa-Fluor 488 goat anti-mouse antibody (green). JEV prM-E and NS3 were detected using anti-JEV E and NS3 antibodies, respectively, and then stained with Alexa-Fluor 488 goat anti-mouse antibody (green). Nuclear translocation of Stat1-DsRed (red) was observed under a fluorescence microscope. Representative cells from the same field for each experimental group are shown.
Stat1 and Tyk2 phosphorylation are blocked by JEV NS5.
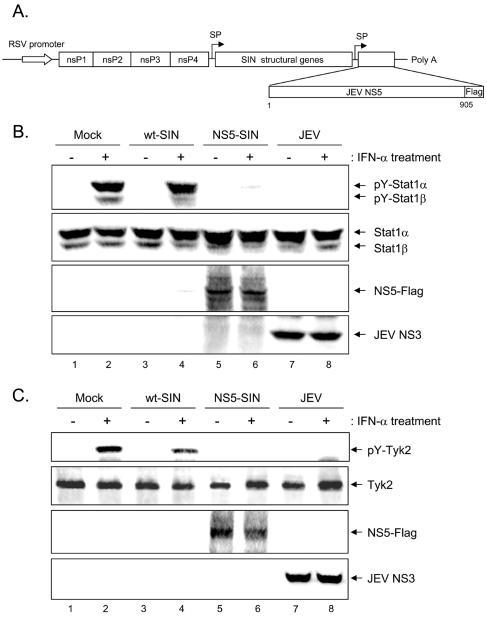
To verify the potential IFN-antagonistic activity of NS5 on endogenous proteins, we generated a recombinant SIN to overexpress a Flag-tagged JEV NS5 under a SIN subgenomic promoter as outlined in Fig. 4A. The recombinant NS5-SIN was harvested from the transfected BHK-21 cells to a titer similar to that of wild-type SIN (wt-SIN) (data not shown). Using a high multiplicity of infection (MOI = 10), all of the cells were infected with SIN and expressed JEV NS5. The wt-SIN infection did not impair the ability of Vero cells to respond to IFN-α, as evidenced by a high level of Stat1 tyrosine phosphorylation in mock-infected and wt-SIN-infected cells (Fig. 4B, lanes 2 and 4). In contrast, NS5-SIN infection rendered the cells unresponsive to IFN-α treatment, as determined by an analysis of Stat1 phosphorylation on Western blotting, which is similar to what was seen in the JEV-infected Vero cells (Fig. 4B, lanes 6 and 8). Since we previously observed that JEV inhibited IFN-α signaling by blocking the activation of IFN receptor-associated kinase Tyk2 (30), we next examined whether Tyk2 was also suppressed by the NS5 protein. Even though a slight repression of IFN-stimulated Tyk2 phosphorylation was noticed in wt-SIN-infected cells (Fig. 4C, lanes 2 and 4), probably due to a somewhat negative effect of Tyk2 activation by SIN infection, IFN-α-stimulated Tyk2 tyrosine phosphorylation was completely blocked in Vero cells overexpressing JEV NS5 (Fig. 4C, lane 6), as was seen with JEV infection (Fig. 4C, lane 8). These results clearly indicate that JEV NS5 not only blocked nuclear translocation of the Stat1-DsRed fusion protein but also inhibited tyrosine phosphorylation of endogenous Stat1 and Tyk2 upon high-dose IFN-α stimulation.
FIG. 4.
Reduction of IFN-α-stimulated Stat1 and Tyk2 tyrosine phosphorylation in Vero cells infected with a recombinant Sindbis virus expressing JEV NS5. (A) Schematic representation of a recombinant SIN expressing JEV NS5 protein. The construct, including a respiratory syncytial virus (RSV) promoter-driving expression cassette, was able to transcribe and translate the SIN replicase complex from nonstructural genes (nsP1, nsP2, nsP3, and nsP4). The replicase produced then activates the subgenomic promoter (SP) to drive the expression of SIN structural genes and JEV NS5. (B) Vero cells were mock infected or infected with wt-SIN, recombinant NS5-SIN, or JEV (MOI = 10) for 6 h. The cells were then stimulated with IFN-αA/D (1,000 U/ml) for 30 min or left unstimulated. The cell lysates were harvested for Western blotting with anti-phospho-Stat1 (Tyr701), anti-Stat1, anti-Flag, and anti-JEV NS3 antibodies, as indicated on the right sides of the gels. (C) Vero cells were prepared as described for panel B except that cells were stimulated with IFN-αA/D for 15 min before harvesting for Western blotting with anti-phospho-Tyk2 (Tyr1054/1055), anti-Tyk2, anti-Flag, and anti-JEV NS3 antibodies as indicated.
Determination of the region of JEV NS5 required for IFN-α antagonism.
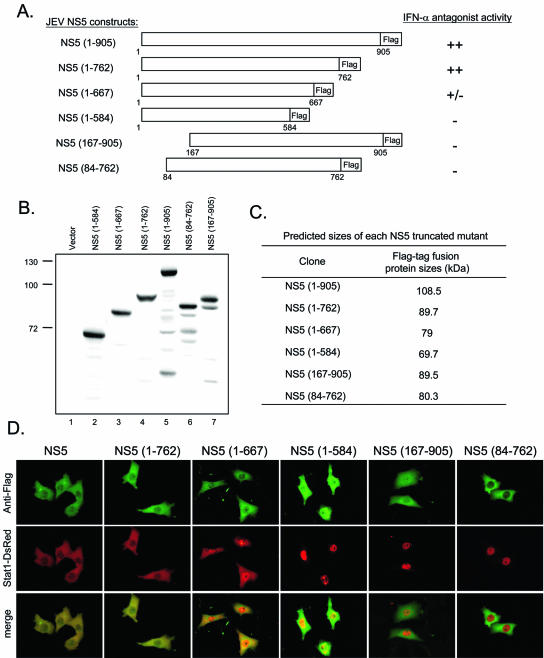
To define the minimum sequence of JEV NS5 required to block IFN signaling, a series of N-terminally and C-terminally truncated mutants of NS5 were constructed (Fig. 5A). The expression of full-length and truncated NS5 proteins with expected molecular sizes was verified by Western blotting (Fig. 5B and C). The ability of these truncated NS5 proteins to block IFN-α signaling was then tested by cotransfection with pStat1-DsRed in BHK-21 cells. Cells expressing the various NS5 proteins were identified by immunofluorescent staining with anti-Flag antibody. Expression of the 762 N-terminal residues of NS5 gave a response similar to that of the full-length NS5: Stat1-DsRed was completely sequestered in the cytoplasm, even in the presence of a high dose of IFN-α (Fig. 5D). However, in cells that expressed the 667 N-terminal amino acids of NS5, Stat1-DsRed was detected in both the cytoplasm and the nuclei. Further deletion of the C terminus up to residue 584 completely abolished the ability of the resulting protein to block IFN-α-induced Stat1 nuclear translocation. Removal of the 83 or 166 N-terminal residues of JEV NS5 also resulted in a loss of IFN-antagonistic activity. Our results show that a large proportion of JEV NS5 proteins are involved in exerting this newly attributed function, probably reflecting the fact that a certain conformation of NS5 may be required for it to exert this effect.
FIG. 5.
Analysis of the region of JEV NS5 that determines its IFN antagonist activity. (A) Schematic diagram of the full-length and truncated NS5-Flag-tagged fusion constructs and a summary of their IFN antagonist activities. (B) Protein expression patterns of NS5 deletion constructs. BHK-21 cells transfected with the indicated NS5 constructs for 24 h were harvested and analyzed by Western blotting with anti-Flag antibody. (C) Predicted molecular sizes of NS5 truncated mutants. (D) BHK-21 cells were cotransfected with pStat1-DsRed and various plasmids expressing the full-length or truncated NS5 proteins. At 24 h after transfection, cells were stimulated with IFN-αA/D (2,500 U/ml) for 16 h and then fixed and permeabilized for immunofluorescence assay. NS5 proteins were detected by using anti-Flag antibody and Alexa-Fluor 488 goat anti-mouse antibody (green). The locations of Stat1-DsRed (red) in the Flag-positive cells were observed under a fluorescence microscope. Representative cells from the same field for each experimental group are shown.
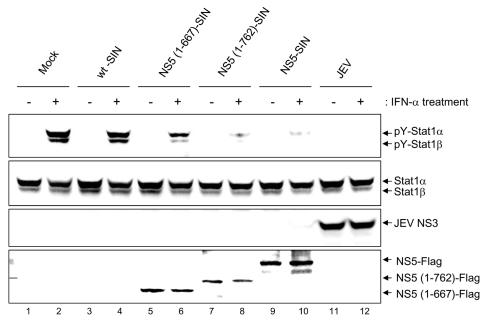
The ability of truncated NS5 to interfere with IFN signaling was supported by results obtained from cells infected with recombinant SINs expressing residues 1 to 762 or 1 to 667 of JEV NS5, which completely or partially blocked IFN-α-induced Stat1-DsRed nuclear translocation, respectively (Fig. 5D). As shown in Fig. 6, endogenous Stat1 phosphorylation induced by IFN-α was almost completely inhibited by the full-length and the 762 N-terminal residues of JEV NS5 (Fig. 6, lanes 8 and 10), whereas it was partially inhibited by the 667 N-terminal residues of JEV NS5 (Fig. 6, lane 6). We suggest that deletion of the 143 C-terminal residues did not affect the antagonistic activity of JEV NS5 against IFN.
FIG. 6.
Deletion of 143 C-terminal residues does not affect the ability of NS5 to block IFN-α-stimulated Stat1 tyrosine phosphorylation. Vero cells were mock infected or infected with wt-SIN, various recombinant SIN constructs, or JEV (MOI = 10) as indicated at the top of the lanes for 6 h. The cells were then stimulated with IFN-αA/D (1,000 U/ml) for 30 min or left unstimulated before the cell lysates were harvested for Western blotting with anti-phospho-Stat1 (Tyr701), anti-Stat1, anti-JEV NS3, and anti-Flag antibodies as shown on the right sides of the gels.
JEV NS5 suppresses the antiviral activities of IFN against EMCV in A549 cells.
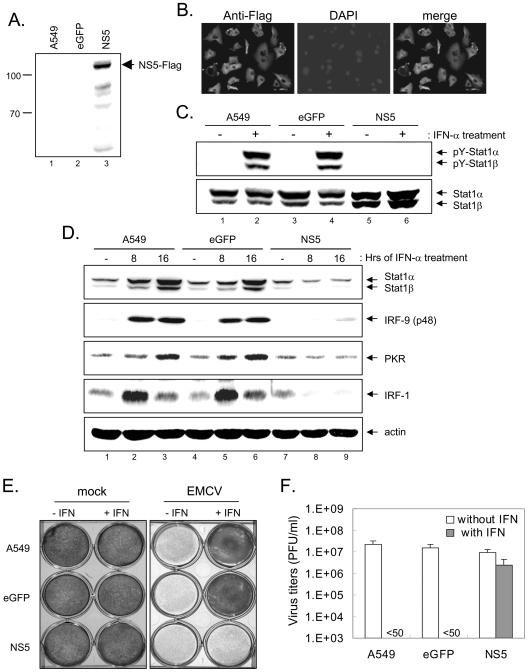
The antiviral effect of IFN is mainly mediated through the Jak-Stat signaling pathway. Our finding that JEV NS5 blocked this signaling event prompted us to test whether JEV NS5 could indeed counteract the antiviral effect that IFN-α exerts against IFN-sensitive viruses. As SIN infection induced severe cell death and is not suitable for this type of experiment, we used a lentivirus expression system to overexpress JEV NS5. A549 cells were transduced with lentivirus vectors expressing JEV NS5 or a control eGFP. The expression of JEV NS5 was confirmed by Western blotting (Fig. 7A) and immunofluorescent staining with anti-Flag antibody (Fig. 7B). These lentivirus-transduced cells all grew at rates similar to that of the parental A549 cells, and no cytotoxicity was noticed. Stat1 tyrosine phosphorylation stimulated by IFN-α was blocked in A549 cells expressing NS5 but not in the parental A549 cells or in those expressing eGFP (Fig. 7C). Furthermore, induction of several interferon-stimulated gene products, such as Stat1, IRF-1, IRF-9, and PKR, was impaired in A549 cells expressing NS5 (Fig. 7D, lanes 7 through 9), which greatly contrasted with what was seen for the parental A549 cells and those expressing eGFP upon IFN treatment (Fig. 7D, lanes 1 through 6). The antiviral activities of IFN-α against EMCV, an IFN-sensitive virus, in the parental A549 cells and in cells expressing eGFP or JEV NS5 were then compared based on the extents of the CPE and viral production. As shown in Fig. 7E, IFN-α potently inhibited the EMCV-induced CPE in parental cells and in A549 cells that expressed eGFP but not in those expressing JEV NS5. EMCV viral production was greatly suppressed by IFN-α in parental cells and A549 cells that expressed eGFP, whereas it remained virtually unchanged in cells that overexpressed JEV NS5 (Fig. 7F). Our results demonstrate that the antiviral effect of IFN-α against EMCV is substantially attenuated by JEV NS5.
FIG. 7.
Enhancement of EMCV replication in response to IFN-α in A549 cells expressing JEV NS5 through a lentivirus vector. Western blotting (A) and immunofluorescence staining (B) with anti-Flag antibody show that JEV NS5 protein was expressed in A549 cells transduced with a lentivirus vector encoding Flag-tagged NS5 but not in the parental A549 cells or in cells transduced with eGFP control. (C) Cells described above were left unstimulated or were stimulated with IFN-αA/D (1,000 U/ml) for 30 min before the cell lysates were harvested for Western blotting with anti-phospho-Stat1 (Tyr701) and anti-Stat1. (D) Protein expression patterns of interferon-stimulated genes. Cells as indicated on the top were left unstimulated or were stimulated with IFN-αA/D (1,000 U/ml) for 8 or 16 h. The cell lysates were harvested and analyzed by Western blotting using the antibodies indicated on the right sides of the gels. (E) Cells described in the legends for panels A and B were left untreated or were pretreated with IFN-αA/D (500 U/ml) for 16 h before mock or EMCV (MOI = 0.1) infection. At 48 h postinfection, the EMCV-induced CPEs were determined by staining with crystal violet. (F) At 32 h postinfection, culture supernatants were harvested for plaque-forming assays; virus titers are expressed as PFU per ml.
Blocking of IFN signaling by JEV and NS5 is mediated through PTPs.
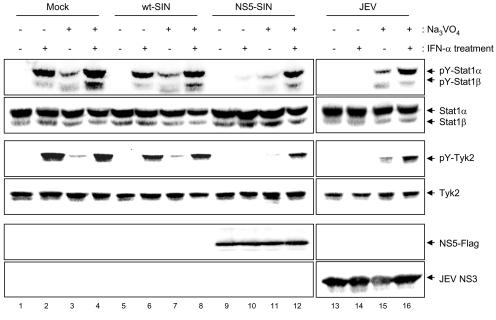
Next, we addressed the molecular mechanism responsible for blocking IFN signaling by JEV and NS5. We hypothesized that the inhibition of IFN-induced Tyk2 and Stat1 phosphorylation by JEV and NS5 might be mediated (i) by a direct physical interaction of NS5 with the receptor-associated kinases resulting in a hindrance to their activation, as suggested by papillomavirus E6 (28), or (ii) through activation of specific intracellular inhibition signals. A mammalian two-hybrid system (Stratagene) was used to examine protein-protein interactions, and NS5 did not exhibit any direct interaction with Tyk2 or Jak1 (data not shown). Cellular proteins which suppress the Jak-Stat pathway, such as SOCS, the protein inhibitors of activated Stats, and PTPs, have been described previously (42). Of these proteins, PTPs suppress the IFN-stimulated Jak-Stat pathway by dephosphorylation of receptors, receptor-associated kinases, and Stat tyrosine residues. To determine the role that PTPs play in the blocking of IFN signaling by JEV, we investigated whether pretreatment of cells with sodium orthovanadate, a broad-spectrum inhibitor of PTPs (14), could suppress the effect of JEV and NS5 on Jak-Stat signaling (Fig. 8). Although the background levels of Stat1 and Tyk2 phosphorylation were slightly elevated in all of the cells treated with sodium orthovanadate (Fig. 8, lanes 3, 7, 11, and 15), the blocking of Stat1 and Tyk2 phosphorylation observed in the JEV- and NS5-SIN-infected Vero cells (Fig. 8, lanes 10 and 14) was completely abolished by sodium orthovanadate (Fig. 8, lanes 12 and 16). We attempted to determine the identity of the PTPs involved in this molecular process by using specific PTP inhibitors such as inhibitors of CD45 (4 μM; Calbiochem) and PTP1B (25 μM; Calbiochem), but none resulted in effective blocking such as that observed for sodium orthovanadate (data not shown). Thus, our results suggest that blocking of IFN-α signaling by JEV and NS5 probably involves the activation of certain unidentified cellular PTPs.
FIG. 8.
Treatment with sodium orthovanadate reverses the blocking effect of IFN-stimulated Stat1 and Tyk2 phosphorylation in JEV- and NS5-SIN-infected cells. Vero cells pretreated with or without sodium orthovanadate (25 μM) for 16 h were mock infected or infected with wt-SIN, NS5-SIN, or JEV (MOI = 10) in the presence or absence of sodium orthovanadate (25 μM) for 6 h as indicated above the lanes. The cells were then stimulated with IFN-αA/D (1,000 U/ml) for 30 min (or 15 min for Tyk2) or left unstimulated before the cell lysates were harvested for Western blot analyses using anti-phospho-Stat1 (Tyr701), anti-Stat1, anti-phospho-Tyk2 (Tyr1054/1055), anti-Tyk2, anti-Flag, and anti-JEV NS3 antibodies as shown on the right sides of the gels.
DISCUSSION
In this report, we demonstrated that JEV NS5, but not the other viral proteins, strongly inhibited IFN-α signaling by preventing IFN-induced nuclear translocation of a Stat1-DsRed fusion protein (Fig. 3). Moreover, the endogenous Stat1 and Tyk2 phosphorylation in response to IFN-α was also blocked in cells infected with a recombinant SIN expressing JEV NS5, but not in wt-SIN-infected cells (Fig. 4). The ability of JEV NS5 to interfere with the antiviral effect of IFN was further demonstrated in cells transduced with a lentivirus vector expressing JEV NS5, in which EMCV replicated to a high level even in the presence of IFN-α (Fig. 7). Recently, NS5 of LGTV, a tick-borne flavivirus, was found to be an IFN antagonist, as expression of LGTV NS5 alone inhibited the Stat1 phosphorylation in response to IFN (2). As previous studies of other mosquito-borne flaviviruses did not denote NS5 as a potential IFN antagonist (34, 36), Best et al. (2) suggested that there may be a divergence in the IFN evasion strategies used by tick- and mosquito-borne flaviviruses. Our finding that JEV NS5 is also a potent IFN antagonist may reflect a much more complicated and independent evolutionary process of individual flaviviruses to their host IFN systems.
Several flaviviruses encode more than one nonstructural protein as IFN-signaling inhibitors. Expression of DEN-2 NS4B, and to a lesser extent, NS2A and NS4A, was capable of blocking IFN signaling (36). Kunjin virus (KUN), a subtype of WNV, encoded nonstructural proteins NS2A, NS2B, NS3, NS4A, and NS4B but did not encode NS1 and NS5, which inhibit IFN-α signaling (34). These findings suggest that DEN-2 and KUN encode various nonstructural proteins which may cooperatively contribute to the inhibition of IFN signaling during infections. In particular, the NS4B of flaviviruses, including that of DEN-2, yellow fever virus, KUN, and WNV, possesses a conserved function in inhibiting IFN-α signaling (34-36). The result that we did not identify JEV NS4B as an IFN antagonist (Fig. 3) may be due to the fact that we used a mature NS4B instead of an NS4B plus its 2K signal peptide segment. The presence of this 2K segment (29) or an unrelated signal peptide at the N terminus of NS4B has been shown to be required for its IFN antagonism (35). That the NS5 of certain flaviviruses but not that of others serves as an IFN antagonist may imply that different flaviviruses have adapted different proteins or mechanisms to interfere with IFN-α signaling and evade host immune defenses. This proposition is not unlikely, considering that even DEN-2 has been reported to utilize different mechanisms for interfering with IFN-α signaling, viz., by reducing either Tyk2 activation or Stat2 protein expression (19, 22).
NS5 is the largest and most conserved flavivirus protein (905 aa long in the case of JEV) and contains the enzymatic activities of MTase (aa 54 to 228) and RdRP (aa 253 to 901) at its N-terminal and C-terminal domains, respectively (31). The C-terminal RdRP domain of NS5 contains eight highly conserved sequence motifs that have been recognized in many RdRPs (25). As IFN antagonism is a novel function for flavivirus NS5, it was of interest to identify its responding domain. Deletion of C-terminal residues 763 to 905, which includes part of the RdRP domain, did not impair the antagonistic activity of NS5 against IFN (Fig. 5 and 6). Deletion of C-terminal residues 585 to 905 or of N-terminal residues 1 to 83 or 1 to 166 abolished the ability of NS5 to counteract the IFN-induced Jak-Stat pathway. Thus, our results suggest that neither the enzymatic activity of MTase nor that of RdRP alone is capable of conferring IFN antagonism of NS5. A large proportion of the sequences of NS5 (approximately 80%) is required for this newly identified function, and details of the sequence requirement await elucidation by site-specific mutagenesis studies. The identification of the sequences in NS5 responsible for blocking IFN-α signaling is of importance for identifying the determinants of virus virulence and for providing defined targets for antiviral drugs and vaccine designs.
At the present time, the precise mechanism(s) responsible for the blocking of IFN signaling by flaviviruses remains to be determined. Human papillomavirus-18 E6 protein physically interacted with Tyk2 through a domain that normally binds to the cytoplasmic portion of the IFN receptor IFNAR1 and therefore impairs the activation of Tyk2 upon IFN-α stimulation (28). LGTV NS5 was found to interact with IFN-α/β and IFN-γ receptor complexes but neither with the receptor-associated kinases Jak1 and Tyk2 nor with Stat1 (2). Using a mammalian two-hybrid system, we also failed to demonstrate a direct physical interaction between JEV NS5 and Tyk2 or Jak1 (data not shown). However, whether JEV NS5 is associated with the IFN receptor complex remains to be determined.
Several negative regulators of the Jak-Stat pathway, such as PTPs and SOCS proteins, have been identified. The induction of SOCS proteins has been shown to regulate the Jak-Stat pathway in a negative manner (40, 44, 48). We found previously that cellular transcription or de novo induction of cellular protein may not be essential for JEV-mediated suppression of IFN signaling (30). Furthermore, the expression levels of SOCS1 and SOCS3 measured by reverse transcription-PCR and Western blotting analysis were not significantly different between the mock- and JEV-infected cells (data not shown). These data suggest that SOCS proteins may not be involved in the blocking of IFN signaling by JEV. The mechanism by which JEV blunts the IFN-signaling events probably involves certain PTPs, because JEV and NS5 lost their abilities to block IFN-induced Jak-Stat signaling in the presence of the PTP inhibitor sodium orthovanadate (Fig. 8). PTPs participate in the negative regulation of Jak-Stat signal transduction through dephosphorylation of phosphorylated Jak and/or Stat components (42). Of the cellular PTPs, both PTP1B and CD45 have been reported to negatively regulate IFN-α signaling by tyrosine dephosphorylation of Tyk2 (20, 37). Our finding that specific inhibitors against CD45 and PTP1B failed to block the effect of JEV and NS5 on IFN signaling (data not shown) suggests that other PTPs, such as SHP1, SHP2, or other unidentified cellular PTPs, may be involved. As vaccinia virus encodes an H1 protein (VH1) that functions as a protein tyrosine phosphatase to inhibit IFN-γ signaling through Stat1 dephosphorylation (38), an alternative explanation for our finding shown in Fig. 8 is that JEV NS5 might itself function as a PTP. Numerous members of the PTP family and the vaccinia virus-encoded PTP (VH1) contain the signature sequence HCXAGXXR at their active sites (15). Sequence alignment of JEV NS5 and other JEV proteins showed that the signature sequence of PTPs is not found, implying that JEV proteins may not possess PTP activity. However, it is important to identify the activated cellular PTP(s) responsible for the blocking of IFN-α signaling by JEV and NS5.
In summary, our results clearly show that the JEV nonstructural protein NS5 blocks IFN-α signaling by inhibiting Tyk2 and Stat1 activation. Using a standard IFN sensitivity assay, we demonstrated that cells expressing NS5 through a lentivirus vector subverted the IFN-mediated antiviral effect and resulted in increased EMCV replication and a greater cytopathic effect in response to IFN-α treatment. Results from a series of NS5 truncated mutants suggest that NS5 probably requires an intact N terminus, but not the 143 C-terminal residues, to mediate its IFN-antagonistic activity. Furthermore, the inhibition of IFN-α signaling by JEV and NS5 likely involved the activities of cellular PTPs. Thus, we suggest that JEV NS5 not only acts as an important enzymatic component of flavivirus RNA replication machinery but also contributes to the blocking of IFN-mediated innate immunity during JEV infection.
Acknowledgments
We thank J. M. Hardwick for dsTE12Q and L. J. Chang for the lentivirus vector system.
This work was supported by grants awarded to Y.-L.L. from the National Health Research Institute (NHRI-EX94-9433SI) and Academia Sinica, Taiwan, Republic of China.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers, T. J., A. Grakoui, and C. M. Rice. 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 65:6042-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, L. J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6:715-728. [DOI] [PubMed] [Google Scholar]
- 5.Chang, T. H., C. L. Liao, and Y. L. Lin. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect. 8:157-171. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. J., M. D. Kuo, L. J. Chien, S. L. Hsu, Y. M. Wang, and J. H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L. K., Y. L. Lin, C. L. Liao, C. G. Lin, Y. L. Huang, C. T. Yeh, S. C. Lai, J. T. Jan, and C. Chin. 1996. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology 223:79-88. [DOI] [PubMed] [Google Scholar]
- 8.Cui, Y., T. Iwakuma, and L. J. Chang. 1999. Contributions of viral splice sites and cis-regulatory elements to lentivirus vector function. J. Virol. 73:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doly, J., A. Civas, S. Navarro, and G. Uze. 1998. Type I interferons: expression and signalization. Cell. Mol. Life Sci. 54:1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 13.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, J. A. 1991. Use of vanadate as protein-phosphotyrosine phosphatase inhibitor. Methods Enzymol. 201:477-482. [DOI] [PubMed] [Google Scholar]
- 15.Guan, K. L., S. S. Broyles, and J. E. Dixon. 1991. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature 350:359-362. [DOI] [PubMed] [Google Scholar]
- 16.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 18.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 19.Ho, L. J., L. F. Hung, C. Y. Weng, W. L. Wu, P. Chou, Y. L. Lin, D. M. Chang, T. Y. Tai, and J. H. Lai. 2005. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J. Immunol. 174:8163-8172. [DOI] [PubMed] [Google Scholar]
- 20.Irie-Sasaki, J., T. Sasaki, W. Matsumoto, A. Opavsky, M. Cheng, G. Welstead, E. Griffiths, C. Krawczyk, C. D. Richardson, K. Aitken, N. Iscove, G. Koretzky, P. Johnson, P. Liu, D. M. Rothstein, and J. M. Penninger. 2001. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature 409:349-354. [DOI] [PubMed] [Google Scholar]
- 21.Iwakuma, T., Y. Cui, and L. J. Chang. 1999. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology 261:120-132. [DOI] [PubMed] [Google Scholar]
- 22.Jones, M., A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79:5414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 24.Koonin, E. V. 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74:733-740. [DOI] [PubMed] [Google Scholar]
- 25.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 26.Levine, B., J. E. Goldman, H. H. Jiang, D. E. Griffin, and J. M. Hardwick. 1996. Bcl2 protects mice against fatal alphavirus encephalitis. Proc. Natl. Acad. Sci. USA 93:4810-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 28.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 29.Lin, C., S. M. Amberg, T. J. Chambers, and C. M. Rice. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 67:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., and C. M. Rice. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23-61. [DOI] [PubMed] [Google Scholar]
- 32.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, B., S. Mink, K. A. Wong, N. Stein, C. Getman, P. W. Dempsey, H. Wu, and K. Shuai. 2004. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 5:891-898. [DOI] [PubMed] [Google Scholar]
- 34.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers, M. P., J. N. Andersen, A. Cheng, M. L. Tremblay, C. M. Horvath, J. P. Parisien, A. Salmeen, D. Barford, and N. K. Tonks. 2001. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J. Biol. Chem. 276:47771-47774. [DOI] [PubMed] [Google Scholar]
- 38.Najarro, P., P. Traktman, and J. A. Lewis. 2001. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J. Virol. 75:3185-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki, A., H. Yasukawa, A. Suzuki, S. Kamizono, T. Syoda, I. Kinjyo, M. Sasaki, J. A. Johnston, and A. Yoshimura. 1999. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 4:339-351. [DOI] [PubMed] [Google Scholar]
- 41.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 42.Shuai, K., and B. Liu. 2003. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3:900-911. [DOI] [PubMed] [Google Scholar]
- 43.Solomon, T. 2003. Recent advances in Japanese encephalitis. J. Neurovirol. 9:274-283. [DOI] [PubMed] [Google Scholar]
- 44.Song, M. M., and K. Shuai. 1998. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J. Biol. Chem. 273:35056-35062. [DOI] [PubMed] [Google Scholar]
- 45.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 46.Uchil, P. D., and V. Satchidanandam. 2003. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 278:24388-24398. [DOI] [PubMed] [Google Scholar]
- 47.Uchil, P. D., and V. Satchidanandam. 2003. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology 307:358-371. [DOI] [PubMed] [Google Scholar]
- 48.Vlotides, G., A. S. Sorensen, F. Kopp, K. Zitzmann, N. Cengic, S. Brand, R. Zachoval, and C. J. Auernhammer. 2004. SOCS-1 and SOCS-3 inhibit IFN-alpha-induced expression of the antiviral proteins 2,5-OAS and MxA. Biochem. Biophys. Res. Commun. 320:1007-1014. [DOI] [PubMed] [Google Scholar]
- 49.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 50.Westaway, E. G., J. M. Mackenzie, and A. A. Khromykh. 2003. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 59:99-140. [DOI] [PubMed] [Google Scholar]
- 51.Yetter, A., S. Uddin, J. J. Krolewski, H. Jiao, T. Yi, and L. C. Platanias. 1995. Association of the interferon-dependent tyrosine kinase Tyk-2 with the hematopoietic cell phosphatase. J. Biol. Chem. 270:18179-18182. [DOI] [PubMed] [Google Scholar]