Abstract
Human monoclonal antibody 2219 is a neutralizing antibody isolated from a human immunodeficiency virus type 1-infected individual. 2219 was originally selected for binding to a V3 fusion protein and can neutralize primary isolates from subtypes B, A, and F. Thus, 2219 represents a cross-reactive, human anti-V3 antibody. Fab 2219 binds to one face of the variable V3 β-hairpin, primarily contacting conserved residues on the N-terminal β-strand of V3, leaving the V3 crown or tip largely accessible. Three V3/2219 complexes reveal the antibody-bound conformations for both the N- and C-terminal regions that flank the V3 crown and illustrate how twisting of the V3 loop alters the relative dispositions and pairing of the amino acids in the adjacent V3 β-strands and how the antibody can accommodate V3 loops with different sequences.
Recent crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibodies have revealed how the immune system can utilize many different strategies to recognize this constantly evolving virus. Prototypic examples include the broadly neutralizing antibody b12, which is thought to use its long complementarity-determining region (CDR) loops to access the recessed, but conserved, CD4 binding site (63), and antibody 2G12, which capitalizes on a unique domain-swapped dimer-of-Fab configuration to create a multivalent binding surface to enhance avidity for low-affinity carbohydrate epitopes (5) on the gp120 silent face. Novel antibodies that block gp120 binding to its chemokine receptor have recently been shown to contain sulfated tyrosines in their CDR loops that likely mimic sulfated tyrosines in the chemokine receptor itself (12), whereas the anti-V3 antibody 447-52D uses a long CDR H3 loop to bind V3 in a way that makes the recognition largely sequence independent, except for interaction with the relatively conserved GPGR crown region (72). Finally, antibody 4E10, the most broadly HIV-1-neutralizing antibody known, binds to a membrane-proximal epitope on gp41 and may also use its long CDR H3 loop to interact with the membrane (6). We report here the structure of a human anti-V3 neutralizing antibody, 2219, that shows yet another way the immune system has found to evoke broad recognition of multiple HIV-1 viral isolates.
The HIV-1 viral proteins gp120 and gp41 are located on the outer membrane surface of the virus, forming a trimeric assembly of the two noncovalently associated proteins. Antibodies that neutralize the virus are directed against these envelope proteins. One of the major epitopes on gp120 is its V3 (third hypervariable) loop, a region of approximately 35 amino acids, linked by a disulfide bond at the base (Cys296-Cys331; HXB2 numbering). Although V3 is termed “hypervariable,” much of the V3 loop, including the tip or crown, is fairly well conserved, with usually just one or two chemically similar amino acid types found at each position (Table 1). The V3 region is highly immunogenic and induces a spectrum of antibodies that can either be highly specific for a particular V3 sequence (75) or be more broadly cross-reactive and neutralize many primary isolates across several HIV-1 subtypes (3, 27, 29, 30, 32). That broadly cross-reactive anti-V3 antibodies are found suggests that V3 is a key epitope to include in vaccine design.
TABLE 1.
Residues of V3 sequences by subtypea
| Subtype (no. of sequences) | Most common residue(s) (%) found at position:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 303 | 304 | 305 | 306 | 307 | 308 | 309 | 312 | 313 | 314 | 315 | 316 | 317 | 318 | 319 | 320 | |
| A (539) | T (96) | R (98) | K (58)/ T (22) | S (86)/ G (10) | V (49)/ I (46) | R (63)/ H (36) | I (94) | G (98) | P (98) | G (99) | Q (93) | A (53)/ T (37) | F (96) | Y (94) | A (92) | T (83)/ X (8) |
| B (1,912) | T (97) | R (93) | K (81)/ R (16) | S (72)/ G (20) | I (95) | H (57)/ P (16) | I (69)/ L (16) | G (96) | P (92) | G (99) | R (77)/ K (8) | A (88)/ T (6) | F (74)/ W (14) | Y (89)/ F (5) | T (50)/ A (48) | T (88)/ X (7) |
| C (443) | T (99) | R (99) | K (78)/ E (11) | S (98) | I (63)/ M (21) | R (97) | I (99) | G (99) | P (100) | G (100) | Q (99) | T (78)/ A (18) | F (98) | Y (94) | A (98) | T (93) |
| D (182) | T (73)/ I (10) | R (91) | Q (61)/ K (15) | S (41)/ G (29) | T (65)/ I (25) | H (60)/ P (16) | I (90)/ M (5) | G (99) | P (70)/ L (10) | G (97) | Q (70)/ R (25) | A (85)/ T (7) | L (55)/ F (19) | Y (70)/ F (20) | T (89)/ A (10) | X (62)/ T (31) |
| AE (356) | T (85)/ I (6) | R (95) | T (92) | S (90)/ R (4) | I (75)/ M (8) | T (3)/ R (15) | I (85)/ M (10) | G (100) | P (98) | G (100) | Q (86)/ R (10) | V (88)/ A (4) | F (93)/ L (3) | Y (97) | R (80)/ K (16) | T (97) |
| F (84) | T (96) | R (100) | K (93) | S (85)/ G (12) | I (99) | H (62)/ Q (19) | L (60)/ I (33) | G (99) | P (99) | G (99) | Q (60)/ R (40) | A (85)/ T (6) | F (99) | Y (98) | A (62)/ T (38) | T (98) |
| G (73) | T (99) | R (95) | K (90)/ R (10) | S (92) | I (96) | R (25)/ S (21) | I (38)/ F (33) | G (90)/ A (10) | P (97) | G (99) | Q (97) | A (79)/ T (15) | F (75)/ L (18) | Y (99) | A (82)/ T (18) | T (97) |
This information is compiled from more-extensive tables in Human Retroviruses and AIDS 1999: a Compilation and Analysis of Nucleic Acid and Amino Acid Sequences (46). The most commonly found residue at each position is listed followed by the percentage it is found. If the most common residue is found less than 90% of the time, the second most commonly found residue is also listed. “X” refers to a deletion at the position.
The V3 region impacts many different aspects of viral infectivity, largely because of its interaction with the gp120 coreceptor CCR5 (15, 60, 61) or CXCR4 (2) during viral cell fusion. Variation at particular V3 residues can affect coreceptor usage and lead to changes in cell tropism (34). During receptor binding and viral fusion, gp120 is thought to undergo several conformational changes, and V3 may change its conformation, location, or accessibility. Conformational flexibility in gp120 has received strong support from a recent crystal structure of an unliganded simian immunodeficiency virus gp120 core (10, 11), in which the gp120 inner domain has significant structural rearrangements compared to CD4-bound HIV gp120 (47, 48). Trimer models based on these core structures (11) suggest that the V3 region alters its location relative to the V1-V2 domain after CD4 is bound. This model is consistent with other studies where binding of soluble CD4 to intact virions enhances the accessibility of the V3 loop (54).
Until recently, all structural information for V3 has come from studies of V3 peptides or V3 peptide-antibody complexes (for a review, see reference 23). However, a recent crystal structure of a V3-containing gp120 core, in complex with CD4 and a CD4-induced monoclonal antibody (MAb), has revealed a long V3 that extends about 30 Å from its base (35) and consists of three structural domains: the base (residues 296 to 300 and 326 to 331), the stem (residues 301 to 305 and 321 to 325), and the tip or crown (residues 306 to 320). These structure-based domain names differ slightly from previously described “functional domains” for which the combined base and stem regions were called the stem (14). The base and tip/crown regions consist of antiparallel β-strands forming a β-hairpin, while the intervening stem region has a more irregular and flexible structure. In the Fab-V3 peptide structures, only the V3 tip/crown and short sections of the stem regions are visible, but their conformations generally agree with that observed in the V3-containing core gp120. V3 peptide-Fab crystal structures are available for mouse Fabs 50.1 (59, 70), 59.1 (25, 26), 58.2 (70), and 83.1 (71) and for human Fab 447-52D (72). Four of these Fabs (50.1, 59.1, 83.1, and 447-52D) recognize a conserved V3 structure, while 58.2 recognizes a V3 conformation that differs in the GPGR region of the tip/crown (26). Nuclear magnetic resonance (NMR) studies of V3 in complex with Fv fragments include a IIIb V3 peptide (RKSIRIQRGPGRAFVTIG) bound to mouse MAb 0.5β (75), where the V3 forms a hairpin with an irregular turn around GPGR. In the MN V3 complex with human Fv 447-52D (65), the peptide forms a β-hairpin with a γ-turn around GPGR, which is consistent with solid-state NMR studies (66). 447-52D has also been studied by NMR with a IIIb peptide, with the N-terminal side of the IIIb peptide adopting a conformation similar to that of the MN peptide bound to 447-52D but with a different orientation for the C-terminal strand (62). NMR studies of isolated V3 peptides show no stable structure in solution, but transient turns exist around GPGR (7-9, 17, 18, 19, 25, 33, 36, 37, 52, 64, 77, 79, 80, 84). NMR studies of peptides modified by cyclization (4, 9, 33, 38, 74, 76-78), by replacement of AlaP316 with the conformationally restricted residue α-aminoisobutyric acid (4, 25), by glycosylation (36, 37, 52), through attachment to resin beads (40), through attachment to a bacteriophage viral coat protein (39), and through attachment to carrier proteins, such as bovine pancreatic trypsin inhibitor (81) and MUC1 (22), all show an increased β-turn propensity around GPGRAF, while V3 peptides attached to filamentous bacteriophage fd viral coat protein pVIII (39) adopt a double-turn structure similar to that observed in the Fab 59.1-peptide crystal structure (25, 26).
While it is clear how broadly neutralizing MAbs that target highly conserved regions can recognize many isolates, the nature of cross-reactivity with MAbs that bind to more-variable regions is less clear. One such MAb, 447-52D, reacts with and neutralizes viruses from subtypes B, A, and F if these viruses possess a GPGR motif (3, 83), and the structural basis of this specificity has been elucidated (72).
However, anti-V3 MAb 2219, which displays cross-clade neutralizing activity (30), is less dependent on the actual motif at the V3 tip/crown. 2219, while not as broadly neutralizing as 447-52D, will neutralize a variety of primary isolates from subtypes B (BaL, SF162, JR-CSF, US1, JR-FL, 92US717, MNp, CA5, and Bx08), subtype A (CA1), and subtype F (93BR019) (28, 30). In order to investigate the structural basis of this cross-reactivity, we have determined crystal structures for antibody 2219 as its Fab fragment in complex with three different peptides (Table 2), including an unusual peptide with sequence RPRQ, rather than GPGR, at its tip/crown.
TABLE 2.
Sequences of V3 peptides cocrystallized with Fab 2219a
| Peptide name (subtype) | Residue at position:
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 301 | 309 | 312 | 325 | ||||||||||||||||||||||
| RP322 (B) | K | R | K | R | I | H | I | . | . | G | P | G | R | A | F | Y | T | T | K | ||||||
| UR29 (C) | N | N | T | K | K | S | I | K | I | . | . | R | P | R | Q | A | F | Y | A | T | N | G | I | I | G |
| UG1033 (A) | N | N | T | R | K | S | I | H | L | . | . | G | P | G | R | A | F | Y | A | T | G | D | I | I | G |
Residues in bold have ordered electron density and are included in the coordinates. The first and last residues of the UR29 and UG1033 peptides are 301 and 325. Discontinuity in the residue numbering system is found after IleP309, as the next residue is GlyP312. The HXB2 isolate, which is the basis for the standard numbering system, has a 2-residue insert between 309 and 312 that accounts for the discontinuous numbering for most other isolates. The RP322 peptide (MN) ends in NAbuC (acm) NH2.
MATERIALS AND METHODS
Construction of the V3 fusion protein.
The V3 fusion protein was cloned and expressed as previously described (44). Briefly, the V3 region of a JR-CSF viral isolate (45 amino acids; SVEINCTRPSNNTRKSIHIGPGRAFYTTGEIIGDIRQAHCNISRA) was fused to the C-terminal end of the N-terminal domain of murine leukemia virus gp70 (268 residues). This construct was expressed in CHO cells and was glycosylated at three N-linked sites, as shown by digestion with endo-β-N-acetylglucosaminidase H (44).
Fab production and purification.
Human monoclonal antibody 2219 {immunoglobulin G1(λ) [IgG1(λ)]} was produced as previously reported (30). Briefly, peripheral blood cells from HIV-1-infected individuals were transformed by Epstein-Barr virus and cultured for 3 weeks, and the supernatant was screened by enzyme-linked immunosorbent assay (ELISA) for binding to the V3 fusion protein. Cells from positive cultures were fused to the human/mouse heteromyeloma cell line SHM-D33 and cloned at limiting dilution to monoclonality. Fab fragments were prepared by cleavage of the 2219 IgG with 4% papain for 4 h, followed by purification on a mono S column (10/10 column; buffer A [50 mM sodium acetate, pH 4.5]; buffer B [50 mM sodium acetate, 1 M sodium chloride, pH 4.5]) and a Superdex 200 column (16/60 column; 20 mM Tris, 100 mM sodium chloride, pH 7.4).
Binding assays.
Eighteen V3 peptides were tested for antibody binding. Four V3 peptides, representing sequences of MN, SF2, NY/5, and CDC4, were purchased from Intracel, Inc. (Cambridge, MA). One peptide, D687, was synthesized by Genemed Biotechnologies, Inc. (South San Francisco, CA). Peptide VI191 was synthesized by Macromolecular Resources (Fort Collins, CO). The remaining peptides were synthesized by Lawrence Loomis-Price at the H. M. Jackson Foundation (Rockville, MD) by standard solid-phase methods as described previously (82).
The binding of MAbs to V3 peptides was determined by a standard ELISA as described previously (29). Briefly, ELISA plates were coated with the V3 peptides at a concentration of 1 μg/ml and left overnight at 4°C. MAbs at concentrations ranging from 0.003 to 10 μg/ml were added to the plate in duplicate, and antigen-bound MAbs were detected by using alkaline phosphatase-labeled goat anti-human IgG (γ specific) (Zymed Laboratories, South San Francisco, CA). The color was developed by adding the substrate, and the plates were read at 405 nm.
Crystallization, data collection, structure determination, refinement, and analysis.
Crystals were obtained for Fab 2219 (12 mg/ml) in complex with a 5:1 molar excess of peptide RP322 (referred to as the MN peptide; subtype B, KRKRIHIGPGRAFYTTKNAbuC(acm)NH2, where Abu refers to 2-aminobutyric acid and acm means acetamidomethylated) in 40% polyethylene glycol 400 (PEG 400), 0.2 M potassium acetate. The crystals grew in tetragonal space group P41212, with unit cell dimension a = b = 60.5 Å, and unit cell dimension c = 275.2 Å, and with one Fab-peptide complex in the asymmetric unit. Data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) beamline 9-2 to a 2.3-Å resolution, with an overall Rsym of 8.6% and with 99.8%-complete data (see Table 3 for data collection and refinement statistics for all three structures). The structure was determined by molecular replacement using as the initial model human Fab fragment NEW (Protein Data Bank [PDB] identifier [ID] 7fab), which was separated into variable and constant domains, and using the AMoRe package of programs (57).
TABLE 3.
Data collection and refinement statistics
| Parameter | Result(s) for:
|
||
|---|---|---|---|
| 2219 + MN peptide on beamline SSRL 9-2 | 2219 + UR29 peptide on beamline ALS 5.0.1 | 2219 + UG1033 peptide on beamline SSRL-11-1 | |
| Wavelength (Å) | 1.033 | 1.000 | 0.984 |
| Resolution (Å)a | 2.30 (2.34-2.30) | 2.0 (2.03-2.00) | 2.35 (2.39-2.35) |
| Space group; cell dimension a, b, c (Å) | P41212; 60.5, 60.5, 275.2 | P212121; 62.9, 94.4, 96.7 | P212121; 62.7, 96.9, 97.0 |
| No. of observations | 125,481 (6,361) | 189,728 (9,361) | 93,252 (3,432) |
| No. of unique reflections | 23,835 (1,162) | 39,609 (1,963) | 24,986 (1,118) |
| Completeness (%) | 99.8 (100.0) | 99.9 (100.0) | 98.6 (92.8) |
| Rsym (%)b | 8.6 (52.5) | 6.9 (61.2) | 5.7 (48.8) |
| Avg I/σ | 22.0 (3.5) | 19.7 (2.6) | 31.9 (2.4) |
| Refinement statistics for all reflections of >0.0 σF | |||
| Resolution (Å) | 69.0-2.3 | 67.4-2.0 | 31.3-2.35 |
| No. of reflections (working) | 22,624 | 37,576 | 23,732 |
| No. of reflections (test) | 1,195 | 1,982 | 1,208 |
| Rcryst (%)c | 21.7 | 20.6 | 21.2 |
| Rfree (%)d | 26.7 | 23.8 | 24.4 |
| No. of Fab atoms | 3,315 | 3,315 | 3,315 |
| No. of peptide atoms | 137 | 137 | 128 |
| No. of water molecules | 191 | 123 | 60 |
| Avg B value (Å2) for: | |||
| Variable | 34.7 | 42.1 | 70.5 |
| Constant | 34.9 | 42.5 | 71.1 |
| Peptide | 35.5 | 43.4 | 74.0 |
| Wilson B value (Å2) | 30.8 | 33.6 | 56.0 |
| Ramachandran plots (%) for: | |||
| Most favored | 92.1 | 90.3 | 88.7 |
| Additionally allowed | 6.6 | 8.4 | 10.5 |
| Generously allowed | 0.8 | 0.5 | 0.0 |
| Disallowede | 0.5 | 0.8 | 0.8 |
| Root mean square deviations | |||
| Bond length (Å) | .013 | .020 | .013 |
| Angle (Å) | 1.46 | 1.73 | 1.33 |
Numbers in parentheses are for the highest-resolution shell of data.
Rsym = Σhkl = ∑hkl|I− <I>|/Σhkl|I|.
Rcryst = Σhkl = ∑hkl|Fobs − Fcalc|/Σhkl |Fobs|.
Rfree is the same as Rcryst except for 5% of the data excluded from the refinement.
Residue AsnL51 is in a conserved γ turn almost always found in Fab structures, SerH128 is in a disordered region of the heavy chain, and AspL151 is in clear density and is the i + 1 residue in a type II′ turn.
Fab 2219 was cocrystallized with a 5:1 molar excess of peptide UR29 (subtype C; NNTKKSIKIRPRQAFYATNGIIG) in 25% PEG monomethylether 5000, 0.2 M disodium tartrate. The crystals belong to orthorhombic space group P212121, with unit cell dimension a = 62.9 Å, b = 94.4 Å, and c = 96.7 Å. Data were collected to a 2.0-Å resolution at the Advanced Light Source (ALS) beamline 5.0.1, with an overall Rsym of 6.9% and completeness of 99.9%. The structure was determined by molecular replacement using the 2219-MN peptide structure (with peptide coordinates removed) as the initial model.
Fab 2219 was cocrystallized with a 5:1 molar excess of peptide UG1033 (subtype A; NNTRKSIHLGPGRAFYATGDIIG; 23-mer) in 13.75% PEG 20,000, 0.05 M disodium tartrate. The crystals belong to space group P212121, with unit cell dimension a = 62.7 Å, b = 96.9 Å, and c = 97.0 Å. Data were collected to a 2.35-Å resolution at the SSRL from beamline 11-1, with an overall Rsym of 5.7% and completeness of 98.6%. The structure was determined by molecular replacement using the 2219-MN peptide structure (without peptide) as a model. Data were also obtained for this particular Fab-peptide complex in several different-molecular-weight PEG-ion combinations (but with the same unit cell dimensions and space group), and although all of the crystals diffracted to similar resolutions, their structures suffered from disorder in the Fab constant domain. The constant domain in the PEG 20,000 crystal form is not disordered, but B values for the entire Fab are higher than for the other two 2219-peptide structures (Table 3). All of the structures were refined with CNS and Refmac, with maintenance of the same 5% test set of reflections throughout. Buried surface areas were calculated with the program MS (13), using a 1.7-Å probe radius and standard van der Waals radii (24). Hydrogen bonds were evaluated with the program HBPLUS (55), and van der Waals contacts were evaluated with the program Contacsym (67, 68). Shape correlation statistics (Sc) were calculated with the CCP4 program Sc, version 2.0 (49).
PDB accession numbers.
The coordinates and structure factors are deposited in the Protein Data Bank with PDB accession codes 2b0s (MN peptide complex), 2b1a (UG1033 peptide complex), and 2b1h (UR29 peptide complex).
RESULTS
Cross-reactivity of MAb 2219.
Antibody 2219 cross-reacts in an ELISA with 11 of 18 peptides representing the V3 sequences of primary HIV-1 isolates from subtypes A, B, and C (Table 4) and neutralizes viruses with the V3 sequences shown in Table 5 (28, 30). The sequences of all of the 18 peptides are different, and no pattern in the primary sequence of the peptides that could explain which peptide would or would not bind strongly to MAb 2219 was discernible, although Arg at position 308 and/or Gln at position 315 may be correlated with reduced affinity (see Table 2 for residue numbers).
TABLE 4.
Reactivities of human anti-V3 MAbs with V3 peptides tested by ELISAa
| V3 peptide (subtype) | Sequence (beginning at position 303 and ending at position 319)c | Optical density at 405 nm forb:
|
||
|---|---|---|---|---|
| 2219 | 447 | 1418 | ||
| UG1033 (A) | TRKSIHL..GPGRAFYA | 1.1 | 3.7 | 0.1 |
| VI191 (A) | TRKGIHI..GPGRAFYA | 2.5 | 3.7 | 0.1 |
| MN (B) | KRKRIHI..GPGRAFYT | 3.2 | 3.8 | 0.1 |
| SF2 (B) | TRKSIYI..GPGRAFHT | 2.4 | 3.6 | 0.1 |
| NY/5 (B) | TKKGIAI..GPGRTLYA | 3.9 | 3.9 | 0.1 |
| CDC4 (B) | TRKRVTL..GPGRVWYT | 0.9 | 3.8 | 0.1 |
| CM237 (B) | TRKSIHL..GPGKAWYT | 3.9 | 3.9 | 0.1 |
| UR29 (C) | TKKSIKI..RPRQAFYA | 3.9 | 0.1 | 0.1 |
| NOF (C) | TRKRIRV..GPGQYVYA | 0.8 | 2.9 | 0.2 |
| 12233 (C) | TRKSMRI..GPGQPFYA | 0.7 | 3.2 | 0.2 |
| MA959 (C) | TRRSIRI..GPGQVFYA | 0.6 | 3.5 | 0.1 |
| W2RW20 (A) | TRKGVRI..GPGQAFYA | 0.1 | 2.9 | 0.1 |
| CM1.CA7 (A) | TRRSIRI..GSGQTSYA | 0.1 | 0.1 | 0.1 |
| CM1.CA22 (A) | TRRSVRI..GPGQAIYA | 0.1 | 0.1 | 0.1 |
| D687 (A) | TNKNVHI..GPGQAFYA | 0.2 | 3.3 | 0.1 |
| ZAM18 (C) | TRKSIRI..GPGQAFYA | 0.2 | 3.5 | 0.1 |
| D757 (C) | TRKSIRI..GPGQTFYA | 0.1 | 3.7 | 0.1 |
| D808 (C) | TRKSTRI..GPGQTFYA | 0.1 | 1.2 | 0.1 |
Residues 303 to 319 are shown; a break in the residue numbering arises after residue 309; the next consecutive residue in these peptides is GlyP312 or ArgP312.
MAbs 2219 and 447 are specific for V3. Human MAb 1418, specific for parvovirus B19, was used as a negative control.
Boldface type indicates the N-terminal half of the peptides (P303 to P309), which forms the majority of hydrogen bonds or salt bridge interactions with Fab 2219 in the crystal structures.
TABLE 5.
V3 sequences of HIV-1 strains neutralized by MAb 2219a
| Strain | Subtype | Amino acid sequence |
|---|---|---|
| BaL | B | NNTRKSIHI..GPGRAFYTT |
| SF162 | B | NNTRKSITI..GPGRAFYAT |
| JR-CSF | B | NNTRKSIHI..GPGRAFYTT |
| 92US717 | B | NNTRRSINI..GPGRAFYTT |
| MNp | B | YNKRKRIHI..GPGRAFYTT |
| CA5 | B | NNTRKGIHI..GPGRAIFTT |
| BX08 | B | NNTRKSIHI..GPGRAFYTT |
| CA1 | A | NNTRKGIHI..GPGRAIYAT |
| 93BR019 | F | NNTRKSIHI..GPGQAFYTT |
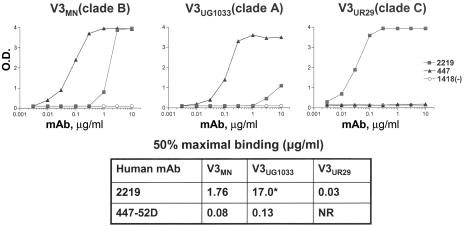
The interactions of MAb 2219 with three V3 peptides from viruses MN (subtype B), UR29 (subtype C), and UG1033 (subtype A) were chosen for further study because of the strong reactivity of the MAb with these peptides and because of the sequence diversity represented by these peptides. However, the relative binding affinities of MAb 2219 for the three peptides differ by 2 orders of magnitude, the binding affinity being highest for UR29, intermediate for MN, and lowest for UG1033 (Fig. 1). 2219 can completely neutralize MN when used at 25 μg/ml (30); however, since the viruses from which UR29 and UG1033 were derived are not available, corresponding neutralization data could not be generated. Recent data have also demonstrated the ability of MAb 2219 to potently neutralize pseudoviruses carrying the V3 consensus sequences of these subtypes A and C (42).
FIG. 1.
Reactivities of MAbs with V3 peptides by ELISA. The binding curves for anti-V3 MAbs 2219 and 447-52D are shown with filled squares and triangles, respectively. Data for the human anti-parvovirus B19 MAb 1418, used as a negative control, are shown as open circles. O.D., optical density; *, value obtained by extrapolation because the binding curve did not reach the saturation point; NR, not reactive.
Fab structure.
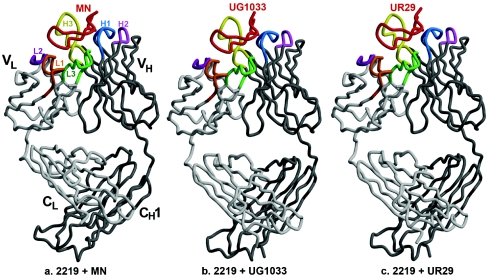
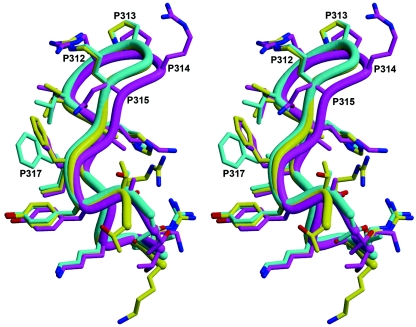
Fab 2219 [human, IgG1(λ)] was crystallized with these three V3 peptides, the sequences of which are shown in Table 2. The Fab light and heavy chains and the peptides are numbered with chain identifiers of L, H, and P, respectively. The Fab is numbered by the standard Kabat and Wu numbering system, and the peptides are numbered according to the HXB2 reference sequence (58). The three structures are very similar (Fig. 2) except for small changes in the peptide and the peptide binding site. The elbow angles for Fab 2219 bound to the MN, UR29, and UG1033 peptides are 228°, 210°, and 211°, respectively. These large (>190°) elbow angles are common for antibodies with a lambda light chain (73).
FIG. 2.
Cα traces of the Fab 2219-peptide complexes. (a) Fab 2219 plus MN peptide; (b) Fab 2219 plus UG1033 peptide; (c) Fab 2219 plus UR29 peptide. In all panels, the peptide is colored in red; Fab light and heavy chains in light and dark gray, respectively; and the Fab CDR loops L1 in orange, L2 in dark purple, L3 in green, H1 in light blue, H2 in pink, and H3 in yellow. Images were produced using Molscript (45), Bobscript (21), and Raster3D (56).
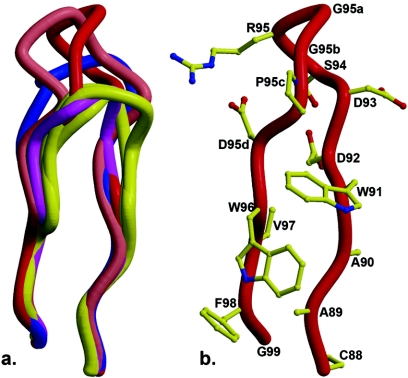
The Fab 2219 CDR loops (L1, L2, H1, H2) fall into canonical classes (1, 53) 5λ, 1, 1, and 1, respectively. 2219 CDR loop L3 represents the longest L3 loop described so far and thus forms a new canonical class (Fig. 3). The 2219 L3 has four residues inserted after L95; the next-longest L3 structure is found in human antibody 447-52D (72), with a 3-amino-acid insert. The 2219 CDR H3 is of medium length (15 amino acids), compared to those of other anti-HIV antibodies with very long H3 CDRs, such as 447-52D (20 amino acids), 2F5 (22 amino acids), 58.2 (17 amino acids), and b12 (18 amino acids), and has a “kinked” base, as predicted from its sequence (69).
FIG. 3.
New canonical structure for CDR L3. (a) The 2219 L3 loop (red) is compared to other L3 loops with three inserts (447-52D [salmon]), two inserts (2fb4 [blue], canonical class λL3-2), and one insert (1gig, 1ind [yellow], λL3-1a; 7fab [pink], λL3-1b; and 1mfa [yellow], λL3-1c). (b) The 2219 lambda chain L3 CDR loop has four inserts (95a to -d) and represents a new canonical structure.
Peptide structure and interactions.
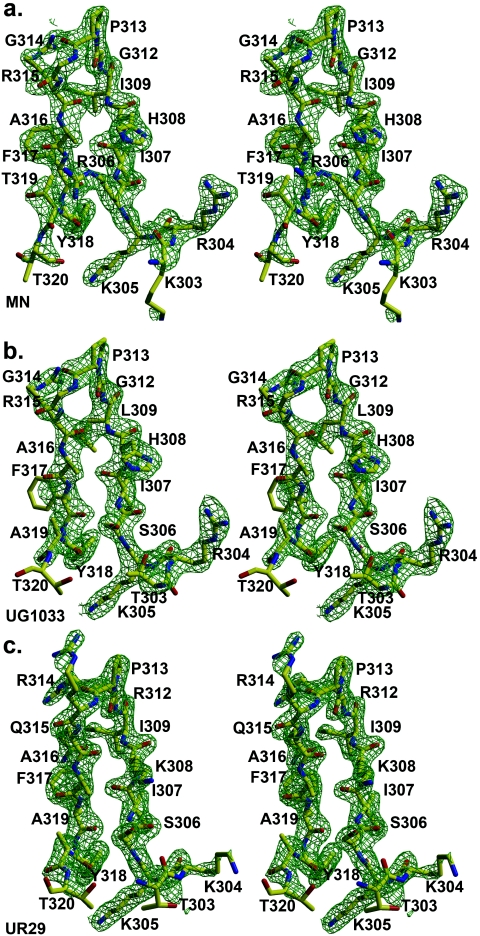
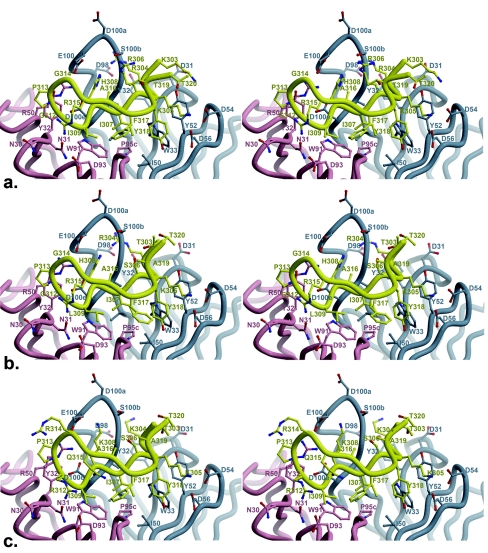
The three peptides bound to Fab 2219 (19, 23, and 23 amino acids long for the MN, UR29, and UG1033 peptides, respectively) all exhibit good electron density for 16 residues (Fig. 4), with all peptides forming a β-hairpin with a type II β-turn around residues P312 to P315. This well-ordered electron density represents the longest stretch of interpretable V3 peptide electron density seen thus far in crystal structures of Fab-V3 complexes, which have so far ranged from 9 to 11 ordered amino acids (for peptides varying in length from 16 to 24 amino acids). All of the hydrogen bonds or salt bridge interactions (Table 6) from peptide to Fab are made by the N-terminal half of the peptide (P303 to P309), with one additional hydrogen bond from ArgP315 (MN and UG1033 peptides) or ArgP312 (UR29 peptide). Although the C-terminal half of the peptide (P314 to P320) is also well defined, it makes less contact with the Fab (Table 7). C-terminal peptide residues P314, P316, P319, and P320 make no contact with Fab in any of the three structures, while PheP317 and TyrP318 make only side chain interactions. The P315 side chain makes just three contacts (one hydrogen bond) in the MN and UG1033 complexes and no contacts in the UR29 complex. In the UR29 and UG1033 crystal structures, the C-terminal portion of the peptide contacts a symmetry-related Fab, with residues L204 to L206 forming an extended β-sheet with the peptide; however, the C-terminal peptide conformation is the same as in the MN peptide complex, which makes no corresponding crystal contacts. The buried molecular surface areas on Fab and peptide are 783 Å2 and 708 Å2 (MN), 770 Å2 and 667 Å2 (UG1033), and 765 Å2 and 630 Å2 (UR29), with a total of 213, 172, and 160 contacts made between Fab and peptides MN, UG1033, and UR29, respectively (Table 7). The buried molecular surface on the Fab (for the MN, UG1033, and UR29 complexes) is contributed from both heavy (62, 62, and 61%, respectively) and light (38, 38, 37%) chains, using CDR loops L1 (15, 16, 15%), L2 (2, 3, 2%), L3 (20, 19, 22%), H1 (18,15, 16%), H2 (17, 15, 15%), and H3 (28, 33, 30%). The corresponding buried peptide surface involves all peptide residues except for P314, P316, P319, and P320, with the largest individual contribution from P304 (17%, 15%, 18%) (Table 7). The peptide-Fab interface has good shape correlation statistics (Sc) (49) of 0.83, 0.78, and 0.78 for the MN, UG1033, and UR29 complexes, similar to those for other Fab-V3 peptide complexes, such as 447-52D (1q1j, 0.77), 59.1 (1acy, 0.80), 50.1 (1ggi, 0.79), 58.2 (1f58, 0.80), and 83.1 (1nak, 0.78). At the peptide-Fab interfaces of all three structures, there is one conserved, interfacial water molecule, stabilized by hydrogen bonds to P304O, H95O, and H33N.
FIG. 4.
Fobs-Fcalc electron density for V3 peptides bound to Fab 2219. (a) MN peptide; (b) UG1033 peptide; (c) UR29 peptide. Electron density is contoured at 1.5σ.
TABLE 6.
Hydrogen bonding and salt bridge distances between Fab 2219 and different V3 peptides and intrapeptide hydrogen bonds within each V3 peptidea
| Atom | Residues in peptides and peptide atom | Distance (Å)
|
||
|---|---|---|---|---|
| MN | UG1033 | UR29 | ||
| Fab atom | ||||
| AsnL31OD1 | R/R/QP315NH2 | 2.14 | 2.28 | |
| AsnL31OD1 | G/G/RP312NH1 | 2.35 | ||
| TyrL32N | I/L/IP309O | 2.93 | 2.77 | 3.02 |
| TyrH32OH | R/R/KP304NZ | 3.31 | ||
| TrpH33NE1 | K/K/KP305O | 2.77 | 2.78 | 2.82 |
| TyrH52OH | K/T/TP303NZ | 3.18 | ||
| AspH54OD1 | K/K/KP305NZ | 2.68 | 2.75 | 2.91 |
| AspH56OD2 | K/K/KP305NZ | 2.62 | 2.45 | 2.76 |
| GluH100OE1 | H/H/KP308NZ | 2.76 | ||
| SerH100bO | H/H/KP308NZ | 2.35 | ||
| GluH100O | R/R/KP304NH1 | 2.87 | 2.81 | |
| GluH100O | R/R/KP304NH2 | 3.47 | 3.17 | |
| GlyH100cN | K/T/TP303O | 3.30 | 2.83 | |
| GlyH100cN | K/T/TP303OG1 | 3.25 | ||
| GlyH100cO | R/R/KP304NH1 | 3.04 | 2.86 | |
| AlaH100dN | R/S/SP306O | 2.79 | 2.76 | 2.82 |
| AlaH100dO | H/H/KP308N | 3.02 | 2.91 | 3.08 |
| Peptide atom | ||||
| I/I/IP307N | F/F/FP317O | 2.81 | 2.87 | 2.97 |
| I/L/IP309N | R/R/QP315O | 3.12 | 3.30 | |
| G/G/RP312N | R/R/QP315O | 3.47 | 3.47 | |
| R/R/QP315N | G/G/RP312O | 3.04 | 3.27 | 3.29 |
| F/F/FP317N | I/I/IP307O | 2.90 | 2.92 | 2.90 |
| R/R/KP304N | K/T/TP303OG1 | 2.15 | 2.25 | |
| R/S/SP306OG | K/T/TP303O | 3.42 | ||
| I/L/IP309N | R/R/QP315OE1 | 3.10 | ||
TABLE 7.
Total van der Waals and hydrogen bond contact with Fab 2219 and buried surface areas of the V3 peptide
| Peptide residue position (amino acids)a | Molecular surface area (Å2) of indicated atom buried inb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MN
|
UG1033
|
UR29
|
|||||||
| Main | Side | Area | Main | Side | Area | Main | Side | Area | |
| P303 (K, T, T) | 1 | 9 | 68.2 | 1 | 3 | 41.1 | 5 | 2 | 39.1 |
| P304 (R, R, K) | 5 | 33 | 118.7 | 3 | 38 | 121.2 | 2 | 17 | 96.0 |
| P305 (K, K, K) | 7 | 27 | 74.5 | 6 | 28 | 75.0 | 7 | 25 | 70.5 |
| P306 (R, S, S) | 6 | 3 | 41.9 | 5 | 1 | 25.5 | 4 | 1 | 25.3 |
| P307 (I, I, I) | 2 | 16 | 63.4 | 2 | 8 | 65.1 | 2 | 7 | 54.4 |
| P308 (H, H, K) | 6 | 15 | 83.4 | 6 | 14 | 89.9 | 5 | 12 | 87.6 |
| P309 (I, L, I) | 14 | 10 | 70.4 | 12 | 2 | 73.1 | 13 | 9 | 63.7 |
| P312 (G, G, R) | 14 | 0 | 23.3 | 11 | 0 | 21.5 | 9 | 7 | 61.1 |
| P313 (P, P, P) | 4 | 4 | 31.4 | 3 | 4 | 27.7 | 3 | 4 | 27.8 |
| P314 (G, G, R) | 0 | 0 | 1.4 | 0 | 0 | 1.2 | 0 | 0 | 1.5 |
| P315 (R, R, Q) | 0 | 3 | 30.6 | 0 | 3 | 31.2 | 0 | 0 | 0.0 |
| P316 (A, A, A) | 0 | 0 | 0.0 | 0 | 0 | 0.0 | 0 | 0 | 0.0 |
| P317 (F, F, F) | 0 | 21 | 47.6 | 0 | 12 | 43.3 | 0 | 16 | 47.2 |
| P318 (Y, Y, Y) | 0 | 12 | 53.1 | 0 | 9 | 51.5 | 0 | 9 | 54.3 |
| P319 (T, A, A) | 0 | 0 | 0.0 | 0 | 0 | 0.0 | 0 | 0 | 0.0 |
| P320 (T, T, T) | 0 | 0 | 0.0 | 0 | 0 | 0.0 | 0 | 0 | 1.1 |
The amino acids found at each position for the MN, UG1033, and UR29 peptides, in that order, are shown in parentheses after the residue number.
Numbers represent contact with main or side chain atoms of the peptide and the molecular surface area (Å2) buried by each peptide residue.
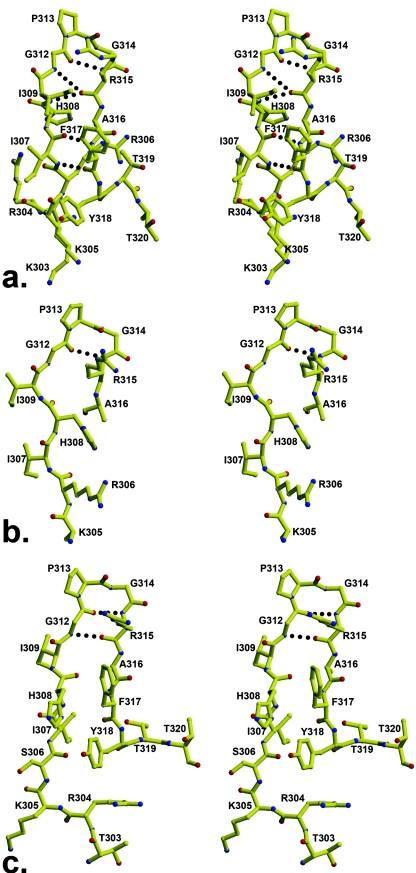
The peptides in the three different complexes share almost identical backbone conformations, but the UR29 peptide with its unusual ArgP312ProP313ArgP314GlnP315 crown sequence uses different residue positions to contact Fab than the other peptides (Fig. 5), causing the UR29 backbone to deviate slightly in this region. UR29 ArgP312 forms a hydrogen bond with AsnL31 from Fab, while ArgP315 in the MN and UG1033 peptides makes the comparable hydrogen bond (Fig. 6). ArgP314 in UR29 projects out into the solvent, and GlnP315 points towards PheP317 but has no interaction with Fab.
FIG. 5.
Stereoview of three peptides bound to Fab 2219. The peptides (MN [yellow], UR29 [pink], and UG1033 [cyan]) were superimposed using only their respective Fab coordinates. ArgP315 (MN and UG1033) and ArgP312 make comparable interactions with the Fab fragment. Note the slight movement of PheP317 in response to the Ile→Leu mutation at P309 in the UG1033 structure.
FIG. 6.
Specific binding interactions between V3 peptides and Fab 2219. (a) 2219 with MN peptide; (b) 2219 with UG1033 peptide; (c) 2219 with UR29 peptide. The UR29 peptide has the unusual RPRQ crown sequence and uses the ArgP312 side chain flexibility to make hydrogen bond interactions comparable to those of ArgP315 in the other two complexes. The UG1033 peptide has an IleP309Leu mutation, and the longer Leu side chain pushes the PheP317 side chain from its position in the other two complexes to an unfavored rotamer with weak electron density.
A more subtle conformational change is seen in the UG1033 peptide, where a seemingly conservative change from Ile to Leu at position P309 causes the PheP317 side chain to alter its preferred rotamer conformation by about 50° (the χ1 torsion angle equals −66° and −57° in the MN and UR29 complexes and −110° in the UG1033 complex) to avoid close contact with the longer Leu side chain (Fig. 4 and 6). Electron density is weak for the UG1033 Phe side chain, suggesting that it may also be somewhat disordered.
Conserved peptide conformation.
The peptides bound to 2219 share structural homology with V3 peptides bound to mouse Fabs 50.1, 59.1, and 83.1; human Fab 447-52D; and the V3 region in the V3-containing gp120 core. The isolated N-terminal (amino acids 303 to 312) or C-terminal (amino acids 313 to 320) regions of 2219-bound V3 peptides overlap well with the corresponding regions from all peptides and V3-containing gp120 core. However, a difference in the ψ angle for IleP309 in the 2219-bound peptides causes the relative disposition of the N- and C-terminal halves of the 2219 peptides to be distorted or twisted, compared to the disposition of the 447-52D and V3-containing gp120 core structures (Fig. 7). Thus, the isolated N- and C-terminal halves overlap well, but the peptide in its entirety does not. A comparison of the 2219 MN peptide dihedral angles and those of a V3 peptide bound to 447-52D (PDB ID 1q1j) and the V3-containing gp120 core (PDB ID 2b4c) show that, with the exception of deviations at the extreme N and C termini and the twist at IleP309, the peptides are very similar (Table 8). The strand twist results in an ∼180° flip of the HisP308 carbonyl oxygen and all residues N-terminal to that oxygen. The change in relative orientations of the N and C strands also alters the intrastrand hydrogen bonding ladder. In the V3-containing gp120 core, two hydrogen bonds are made in the V3 tip or crown (GlyP312N to ArgP315O and GlyP312O to ArgP315N), while in the 2219-bound V3 peptide, the GlyP312O to ArgP315N hydrogen bond is retained, but new hydrogen bonds are formed between IleP307N to PheP317O, IleP307O to PheP317N, and IleP309N to ArgP315O (Fig. 7).
FIG.7.
V3 peptides bound to 2219 and 447-52D and the corresponding region from the V3-containing gp120 core. The 2219 V3 peptide (MN) (a), 447-52D peptide (MN) (b), and V3 tip from the V3-containing gp120 core (JR-FL) (c) are shown in approximately equivalent orientations, with hydrogen bonds shown as dotted black lines. A twist in the main-chain torsion angle around IleP309 results in different relative orientations of the N- and C-terminal strands in the 2219 structure (a) so that in the 2219-bound V3, residues IleP309, IleP307, PheP317, and TyrP318 occupy the same face of the β-hairpin, but in the other two V3 loop structures bound to 447-52D and in the V3-containing gp120 core, these residues would be on opposite faces (Table 8).
TABLE 8.
Main chain torsion angles for V3 peptides bound to Fabs 2219 and 447-52D and the V3 region in the V3-containing gp120 core
| Peptide residue | Angle | Torsion angle for:
|
Avg deviationa | ||
|---|---|---|---|---|---|
| 2219MN | 447-52D | gp120 | |||
| P303 | φ | −58 | 0 | ||
| ψ | −162 | −75 | 44 | ||
| P304 | φ | −109 | −108 | 1 | |
| ψ | 24 | 148 | 62 | ||
| P305 | φ | −75 | −124 | 25 | |
| ψ | −32 | 144 | −12 | 74 | |
| P306 | φ | −61 | −158 | −147 | 41 |
| ψ | 131 | 143 | 146 | 6 | |
| P307 | φ | −110 | −116 | −118 | 3 |
| ψ | 123 | 132 | 160 | 14 | |
| P308 | φ | −94 | −116 | −129 | 13 |
| ψ | 106 | 122 | 121 | 7 | |
| P309 | φ | −93 | −113 | −84 | 11 |
| ψ | −27 | 99 | 140 | 65 | |
| P312 | φ | −157 | 176 | −158 | 12 |
| ψ | −178 | 169 | 131 | 20 | |
| P313 | φ | −55 | −65 | −59 | 4 |
| ψ | 122 | 130 | 99 | 12 | |
| P314 | φ | 70 | 74 | 100 | 12 |
| ψ | 8 | 5 | 8 | 1 | |
| P315 | φ | −123 | −88 | −172 | 30 |
| ψ | 129 | −166 | 149 | 24 | |
| P316 | φ | −147 | −111 | −131 | 12 |
| ψ | 150 | 167 | 9 | ||
| P317 | φ | −117 | −133 | 8 | |
| ψ | 165 | 153 | 6 | ||
| P318 | φ | 53 | 60 | 4 | |
| ψ | 41 | 65 | 12 | ||
| P319 | φ | −80 | −150 | 35 | |
| ψ | 4 | −2 | 3 | ||
| P320 | φ | −62 | −101 | 20 | |
| ψ | 47 | 0 | |||
The average deviation from the mean of the three torsion angles (from 2219, 447-52D, and gp120).
The C-terminal portion of the 2219-bound peptide contacts the Fab fragment through two side chains (PheP317 and TyrP318) but makes no backbone contacts. This strand has structural homology to the corresponding region on the intact gp120-V3 structure (35) (Table 8 and Fig. 7) and also is consistent with the weak electron density observed for the 447-52D C terminus (described in reference 72 but not included in the PDB submission). Interestingly, none of the V3 conformations that bound to 2219 or 447-52D and/or that are bound in gp120-V3 have the double turn seen in 59.1 (25, 26), although V3 bound to mouse Fab 83.1 has what appears to correspond to the start site of this double turn (71).
2219 binds to the UR29 peptide with an affinity 2 orders of magnitude higher than to the MN or UG1033 peptides (Fig. 1). Analyses of the structures do not give a clear rationale for this difference in binding affinity, as the buried molecular surface area, the buried hydrophobic molecular surface area, the shape complementarity to the surface, and the numbers of contacts and hydrogen bonds are all very similar for the three Fab-peptide complexes. Actually, slightly fewer contacts, smaller surface areas, and a slightly lower shape correlation statistic are observed for the more strongly binding UR29 peptide complex. There are some differences in intrapeptide hydrogen bonding due to sequence differences in the three peptides (Table 6), the most striking around GlnP315 in UR29. Small differences in the UR29 backbone conformation (Fig. 6) cause the loss of two intrapeptide hydrogen bonds (P309N to P315O and P312N to P315O) found in MN and UG1033, but GlnP315 then makes two different intrapeptide hydrogen bonds not found in the two other peptides. GlnP315OE1 makes a hydrogen bond to IleP309N (3.10 Å), and GlnP315NE2 is about 3.5 Å from the center of the PheP317 phenyl ring, a reasonable distance for an aromatic hydrogen bond (Fig. 6) (50). Thus, although the numbers of intrapeptide hydrogen bonds in the three peptides are similar (5, 6, and 6 in the MN, UG1033, and UR29 peptides, respectively) it is possible that different intrapeptide hydrogen bonding interactions may serve to better stabilize the UR29 peptide in solution, thus lowering the entropic penalty for binding. However, as seen in many other systems, it is often difficult to directly correlate the structural results with binding affinities (18, 20, 51).
DISCUSSION
The human neutralizing MAb 2219 was studied to investigate the structural basis for cross-reactivity with a variable epitope, the V3 loop of the HIV-1 envelope glycoprotein gp120. 2219 binds to different V3 peptides (Table 4) and neutralizes viruses from HIV-1 subtypes B, A, and F (Table 5) (30). These results demonstrate that a single antibody, raised against a variable region of the virus, can bind to peptides with different viral sequences and can neutralize different viral isolates, suggesting that vaccine constructs that can elicit such antibodies may be able to induce a broadly effective immune response.
Examination of the interactions made between the three different V3 peptides and 2219 help to explain the reactivity of the Fab fragment for some of the different peptides tested by ELISA in Table 4. For example, V3 residues P303 to P306 make a significant number of hydrogen bond and van der Waals contacts but are solvent exposed, so that alteration of these side chains may not abolish binding. In contrast, residues P307 and P309 are buried in hydrophobic pockets in the antigen combining site and are probably restricted to small, hydrophobic residues, such as Ile, Leu, or Val, to attain high affinity. Residues most often found at position 307 include Ile, Val, Thr, and Met (Table 1), while at position 309 Ile, Leu, Phe, and Met are found most frequently (Table 1). One peptide (12233, subtype C) that reacts with MAb 2219 (Table 4) has the larger, Met residue at P307; computer graphics modeling in which IleP307 is replaced with a Met shows that the Met side chain can be accommodated with little or no adjustment to the Fab complex. Fab 2219 reactivity is decreased for peptides with Arg at position 308 and Gln at position 315 (Table 4). HisP308 points out of the combining site in the 2219-peptide structures and interacts with several residues from the long H3 CDR loop. As good reactivity is observed with peptides containing His, Tyr, Ala, and Lys at position 308, it is not obvious why replacement with Arg (about the same length and charge as a Lys) would severely reduce affinity, although the bulkier Arg side chain might be a tight fit in the 2219 combining site. At position 315, ArgP315 in the MN and UG1033 peptide complex structures makes a hydrogen bond to AsnL31 (Table 6). Computer modeling in which ArgP315 is replaced with a Gln suggests that Gln could easily make the corresponding hydrogen bond, albeit a longer one (3.0 Å compared to ∼2.2 Å), with little rearrangement of the peptide or Fab, but it is possible that a longer, hence weaker, hydrogen bond at this position would reduce affinity.
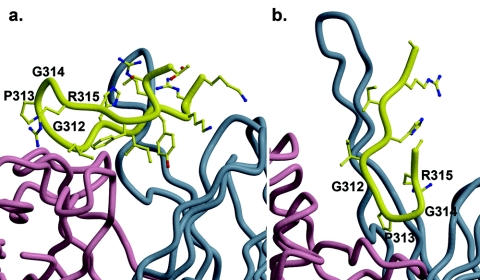
2219 is able to recognize the unusual V3 crown sequence of UR29 largely because of its mode of binding, where the antibody binds primarily to one face of V3, with the crown largely free from contact (Fig. 8). In addition, the UR29 crown sequence (RPRQ) allows the antibody to maintain its interaction between AsnL31 and an Arg from the peptide, in which the Arg corresponds to P315 in the MN and UG1033 peptides but to P312 in UR29. In contrast, the broadly reactive antibody 447-52D cannot bind peptide UR29 (Fig. 1). Examination of the interactions between 447-52D and its bound V3 peptide with an MN sequence (72) shows that conversion of GPGR to RPRQ would likely cause ArgP314 to clash with 447-52D ArgH50 or AspH58. V3 binds 447-52D with its crown region buried in a pocket but with the remaining side chain positions accessible to solvent (Fig. 8). This mode of binding allows 447-52D to recognize many different V3 sequences (83) but would prevent it from accommodating large side chains at position 314. The V3 face binding mode used by 2219 has not been seen previously for the other anti-V3 antibodies, most of which (59.1, 58.2, 83.1) bind around the crown/tip region. The most similar binding mode is that of the MN-specific antibody 50.1, which binds the same face of V3, but only to the N-terminal strand.
FIG. 8.
Comparison of modes of binding of the two V3 antibodies 2219 and 447-52D. (a) 2219 bound to V3 peptide (MN), with the peptide in yellow, the light chain in pink, and the heavy chain in blue. The N- and C-terminal regions of V3 contact Fab, but crown residues GlyP314 and ArgP315 are largely free from contact with Fab. (b) 447-52D bound to V3 peptide (MN) with the color scheme shown in panel a. In this complex, V3 binds with its crown region buried in the antibody combining site and with the N-terminal side bound to the H3 loop via main-chain hydrogen bonds, leaving the N-terminal side chains accessible to solvent. Thus, 447-52D can tolerate changes in the N-terminal side chain positions but not in the GPGR region.
While most of the binding and neutralization data can be illuminated by the crystal structure results, there still remain some unanswered questions. For example, 447-52D neutralizes GPGR- but not GPGQ-containing viral isolates (83), but the data in Table 4 indicate that 447-52D can still bind to several GPGQ-containing V3 peptides, often better than 2219. Yet, the crystal structure shows that 447-52D binds to GPGR via a salt bridge interaction from AspH95 to ArgP315, while 2219 recognizes the same ArgP315, but with a hydrogen bond, suggesting that 2219 would be tolerant of noncharged residues (such as Gln) (Table 4). These results suggest that the V3 regions of GPGR viral isolates may differ subtly in conformation from those of the GPGQ viral isolates, and this notion is supported by the fact that viruses bearing V3 loops containing the GPGQ motif induce a profile of anti-V3 antibodies in humans different from that of viruses bearing GPGR (31, 43).
Small structural changes in the V3 region, such as the change in a single torsion angle in the 2219-bound V3, can lead to large overall differences when the V3 structures are compared. Although all seven of the crystallographically determined V3 region structures share significant structural homology, the individual N- and C-terminal strands that look very similar may differ when analyzed in combination with each other (Fig. 7). It may be that V3 maintains the secondary structure of its N- and C-terminal strands, but by twisting the loop these strands can move with respect to one another, thus placing different sets of residues on the faces of the V3 β-hairpin. Nevertheless, given the similarity of many of the structures derived from crystallographic, NMR, and functional studies, the picture that emerges is one of a dynamic V3 structure with a conserved base and tip but with a flexible stem that assumes preferred alternative conformations that are influenced by its sequence, its environment in the context of gp120, and its role in the interaction with coreceptor CCR5 or CXCR4.
The selection of neutralizing MAbs may be more effective with a V3 fusion protein than with linear V3 peptides (30, 44). The V3 region in the fusion peptide is likely more ordered or restricted than in linear V3 peptides, and given the presence of the disulfide bond at its base and the correctly placed carbohydrate moieties, the fusion protein may better represent the native V3 structure (41). Undoubtedly, a successful V3-based vaccine will have to mimic one or a range of V3 conformations found on intact virus and also induce antibodies that can tolerate some sequence variability within the V3 loop. The 447-52D and 2219 structures have shown two distinctly different ways to address this problem, through primary main-chain interactions between V3 and the CDR H3 of antibody (447-52D) or by binding to a conserved face of the β-hairpin that allows variation in the crown. As more neutralizing and nonneutralizing V3 antibody structures are determined, the definition of the requirements for broad neutralization by V3 antibodies can be improved and applied to vaccine design efforts.
Acknowledgments
We thank Sharon Ferguson and Constance Williams for excellent technical assistance and SSRL Beamline 11-1 and ALS Beamline 5.0.2 for synchotron beamtime.
We gratefully acknowledge support by NIH grants GM-46192 (I.A.W. and R.L.S.), AI36085, AI27742, and HL 59725 (S.Z.-P. and M.K.G.); a grant from the Department of Veterans Affairs (S.Z.-P.); and a grant from the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium (I.A.W.).
This is manuscript 17762-MB from the Scripps Research Institute.
REFERENCES
- 1.Al-Lazikani, B., A. M. Lesk, and C. Chothia. 1997. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 273:927-948. [DOI] [PubMed] [Google Scholar]
- 2.Basmaciogullari, S., G. J. Babcock, D. Van Ryk, W. Wojtowicz, and J. Sodroski. 2002. Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein of importance for CXCR4 binding. J. Virol. 76:10791-10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabezas, E., M. Wang, P. W. Parren, R. L. Stanfield, and A. C. Satterthwait. 2000. A structure-based approach to a synthetic vaccine for HIV-1. Biochemistry 39:14377-14391. [DOI] [PubMed] [Google Scholar]
- 5.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 6.Cardoso, R. M., M. B. Zwick, R. L. Stanfield, R. Kunert, J. M. Binley, H. Katinger, D. R. Burton, and I. A. Wilson. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22:163-173. [DOI] [PubMed] [Google Scholar]
- 7.Catasti, P., E. M. Bradbury, and G. Gupta. 1996. Structure and polymorphism of HIV-1 third variable loops. J. Biol. Chem. 271:8236-8242. [DOI] [PubMed] [Google Scholar]
- 8.Catasti, P., J. D. Fontenot, E. M. Bradbury, and G. Gupta. 1995. Local and global structural properties of the HIV-MN V3 loop. J. Biol. Chem. 270:2224-2232. [DOI] [PubMed] [Google Scholar]
- 9.Chandrasekhar, K., A. T. Profy, and H. J. Dyson. 1991. Solution conformational preferences of immunogenic peptides derived from the principal neutralizing determinant of the HIV-1 envelope glycoprotein gp120. Biochemistry 30:9187-9194. [DOI] [PubMed] [Google Scholar]
- 10.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Determining the structure of an unliganded and fully glycosylated SIV gp120 envelope glycoprotein. Structure 13:197-211. [DOI] [PubMed] [Google Scholar]
- 11.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 12.Choe, H., W. Li, P. L. Wright, N. Vasilieva, M. Venturi, C. C. Huang, C. Grundner, T. Dorfman, M. B. Zwick, L. Wang, E. S. Rosenberg, P. D. Kwong, D. R. Burton, J. E. Robinson, J. G. Sodroski, and M. Farzan. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161-170. [DOI] [PubMed] [Google Scholar]
- 13.Connolly, M. L. 1993. The molecular surface package. J. Mol. Graphics 11:139-141. [DOI] [PubMed] [Google Scholar]
- 14.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degano, M., K. C. Garcia, V. Apostolopoulos, M. G. Rudolph, L. Teyton, and I. A. Wilson. 2000. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity 12:251-261. [DOI] [PubMed] [Google Scholar]
- 17.de Lorimier, R., M. A. Moody, B. F. Haynes, and L. D. Spicer. 1994. NMR-derived solution conformations of a hybrid synthetic peptide containing multiple epitopes of envelope protein gp120 from the RF strain of human immunodeficiency virus. Biochemistry 33:2055-2062. [DOI] [PubMed] [Google Scholar]
- 18.Dettin, M., A. De Rossi, M. Autiero, J. Guardiola, L. Chieco-Bianchi, and C. Di Bello. 1993. Structural studies on synthetic peptides from the principal neutralizing domain of HIV-1 gp120 that bind to CD4 and enhance HIV-1 infection. Biochem. Biophys. Res. Commun. 191:364-370. [DOI] [PubMed] [Google Scholar]
- 19.Dettin, M., R. Roncon, M. Simonetti, S. Tormene, L. Falcigno, L. Paolillo, and C. Di Bello. 1997. Synthesis, characterization and conformational analysis of gp 120-derived synthetic peptides that specifically enhance HIV-1 infectivity. J. Pept. Sci. 3:15-30. [DOI] [PubMed] [Google Scholar]
- 20.Ding, Y. H., B. M. Baker, D. N. Garboczi, W. E. Biddison, and D. C. Wiley. 1999. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 11:45-56. [DOI] [PubMed] [Google Scholar]
- 21.Esnouf, R. M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 22.Fontenot, J. D., J. M. Gatewood, S. V. Mariappan, C. P. Pau, B. S. Parekh, J. R. George, and G. Gupta. 1995. Human immunodeficiency virus (HIV) antigens: structure and serology of multivalent human mucin MUC1-HIV V3 chimeric proteins. Proc. Natl. Acad. Sci. USA 92:315-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galanakis, P. A., G. A. Spyroulias, A. Rizos, P. Samolis, and E. Krambovitis. 2005. Conformational properties of HIV-1 gp120/V3 immunogenic domains. Curr. Med. Chem. 12:1551-1568. [DOI] [PubMed] [Google Scholar]
- 24.Gelin, B. R., and M. Karplus. 1979. Side-chain torsional potentials: effect of dipeptide, protein, and solvent environment. Biochemistry 18:1256-1268. [DOI] [PubMed] [Google Scholar]
- 25.Ghiara, J. B., D. C. Ferguson, A. C. Satterthwait, H. J. Dyson, and I. A. Wilson. 1997. Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site. J. Mol. Biol. 266:31-39. [DOI] [PubMed] [Google Scholar]
- 26.Ghiara, J. B., E. A. Stura, R. L. Stanfield, A. T. Profy, and I. A. Wilson. 1994. Crystal structure of the principal neutralization site of HIV-1. Science 264:82-85. [DOI] [PubMed] [Google Scholar]
- 27.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler II, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 159:5114-5122. [PubMed] [Google Scholar]
- 30.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, X.-H. Wang, S. Buda, T. Kimura, F. A. J. Konings, A. Nadas, C. A. Anyangwe, P. Nyambi, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. Submitted for publication.
- 32.Gorny, M. K., J. Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635-643. [PubMed] [Google Scholar]
- 33.Gupta, G., G. M. Anantharamaiah, D. R. Scott, J. H. Eldridge, and G. Myers. 1993. Solution structure of the V3 loop of a Thailand HIV isolate. J. Biomol. Struct. Dyn. 11:345-366. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman, T. L., and R. W. Doms. 1999. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 16:57-65. [DOI] [PubMed] [Google Scholar]
- 35.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, X., J. J. Barchi, Jr., F. D. Lung, P. P. Roller, P. L. Nara, J. Muschik, and R. R. Garrity. 1997. Glycosylation affects both the three-dimensional structure and antibody binding properties of the HIV-1 IIIB GP120 peptide RP135. Biochemistry 36:10846-10856. [DOI] [PubMed] [Google Scholar]
- 37.Huang, X., M. C. Smith, J. A. Berzofsky, and J. J. Barchi, Jr. 1996. Structural comparison of a 15 residue peptide from the V3 loop of HIV-1 IIIb and an O-glycosylated analogue. FEBS Lett. 393:280-286. [DOI] [PubMed] [Google Scholar]
- 38.Huisman, J. G., A. Carotenuto, A. F. Labrijn, C. H. Papavoine, J. D. Laman, M. M. Schellekens, M. H. Koppelman, and C. W. Hilbers. 2000. Recognition properties of V3-specific antibodies to V3 loop peptides derived from HIV-1 gp120 presented in multiple conformations. Biochemistry 39:10866-10876. [DOI] [PubMed] [Google Scholar]
- 39.Jelinek, R., T. D. Terry, J. J. Gesell, P. Malik, R. N. Perham, and S. J. Opella. 1997. NMR structure of the principal neutralizing determinant of HIV-1 displayed in filamentous bacteriophage coat protein. J. Mol. Biol. 266:649-655. [DOI] [PubMed] [Google Scholar]
- 40.Jelinek, R., A. P. Valente, K. G. Valentine, and S. J. Opella. 1997. Two-dimensional NMR spectroscopy of peptides on beads. J. Magn. Reson. 125:185-187. [DOI] [PubMed] [Google Scholar]
- 41.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krachmarov, C. P., W. J. Honnen, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. Submitted for publication.
- 43.Krachmarov, C., A. Pinter, W. J. Honnen, M. K. Gorny, P. N. Nyambi, S. Zolla-Pazner, and S. C. Kayman. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J. Virol. 79:780-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krachmarov, C. P., S. C. Kayman, W. J. Honnen, O. Trochev, and A. Pinter. 2001. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1737-1748. [DOI] [PubMed] [Google Scholar]
- 45.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 46.Kuiken, C. L., B. Foley, B. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, J. I. Mullins, S. Wolinsky, and B. Korber (ed.). 1999. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 47.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold. Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 48.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence, M. C., and P. M. Colman. 1993. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234:946-950. [DOI] [PubMed] [Google Scholar]
- 50.Levitt, M., and M. F. Perutz. 1988. Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 201:751-754. [DOI] [PubMed] [Google Scholar]
- 51.Li, Y., M. Urrutia, S. J. Smith-Gill, and R. A. Mariuzza. 2003. Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen. Biochemistry 42:11-22. [DOI] [PubMed] [Google Scholar]
- 52.Markert, R. L., H. Ruppach, S. Gehring, U. Dietrich, D. F. Mierke, M. Kock, H. Rubsamen-Waigmann, and C. Griesinger. 1996. Secondary structural elements as a basis for antibody recognition in the immunodominant region of human immunodeficiency viruses 1 and 2. Eur. J. Biochem. 237:188-204. [DOI] [PubMed] [Google Scholar]
- 53.Martin, A. C., and J. M. Thornton. 1996. Structural families in loops of homologous proteins: automatic classification, modelling and application to antibodies. J. Mol. Biol. 263:800-815. [DOI] [PubMed] [Google Scholar]
- 54.Mbah, H. A., S. Burda, M. K. Gorny, C. Williams, K. Revesz, S. Zolla-Pazner, and P. N. Nyambi. 2001. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J. Virol. 75:7785-7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDonald, I. K., and J. M. Thornton. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238:777-793. [DOI] [PubMed] [Google Scholar]
- 56.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 57.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 58.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R. S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 3:57-69. [DOI] [PubMed] [Google Scholar]
- 59.Rini, J. M., R. L. Stanfield, E. A. Stura, P. A. Salinas, A. T. Profy, and I. A. Wilson. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc. Natl. Acad. Sci. USA 90:6325-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 61.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 62.Rosen, O., J. Chill, M. Sharon, N. Kessler, B. Mester, S. Zolla-Pazner, and J. Anglister. 2005. Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization. Biochemistry 44:7250-7258. [DOI] [PubMed] [Google Scholar]
- 63.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 64.Sarma, A. V., T. V. Raju, and A. C. Kunwar. 1997. NMR study of the peptide present in the principal neutralizing determinant (PND) of HIV-1 envelope glycoprotein gp120. J. Biochem. Biophys. Methods 34:83-98. [DOI] [PubMed] [Google Scholar]
- 65.Sharon, M., N. Kessler, R. Levy, S. Zolla-Pazner, M. Gorlach, and J. Anglister. 2003. Alternative conformations of HIV-1 V3 loops mimic β hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure 11:225-236. [DOI] [PubMed] [Google Scholar]
- 66.Sharpe, S., N. Kessler, J. A. Anglister, W. M. Yau, and R. Tycko. 2004. Solid-state NMR yields structural constraints on the V3 loop from HIV-1 gp120 bound to the 447-52D antibody Fv fragment. J. Am. Chem. Soc. 126:4979-4990. [DOI] [PubMed] [Google Scholar]
- 67.Sheriff, S., W. A. Hendrickson, and J. L. Smith. 1987. Structure of myohemerythrin in the azidomet state at 1.7/1.3 Å resolution. J. Mol. Biol. 197:273-296. [DOI] [PubMed] [Google Scholar]
- 68.Sheriff, S., E. W. Silverton, E. A. Padlan, G. H. Cohen, S. J. Smith-Gill, B. C. Finzel, and D. R. Davies. 1987. Three-dimensional structure of an antibody-antigen complex. Proc. Natl. Acad. Sci. USA 84:8075-8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shirai, H., A. Kidera, and H. Nakamura. 1996. Structural classification of CDR-H3 in antibodies. FEBS Lett. 399:1-8. [DOI] [PubMed] [Google Scholar]
- 70.Stanfield, R., E. Cabezas, A. Satterthwait, E. Stura, A. Profy, and I. Wilson. 1999. Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing Fabs. Struct. Fold. Des. 7:131-142. [DOI] [PubMed] [Google Scholar]
- 71.Stanfield, R. L., J. B. Ghiara, E. O. Saphire, A. T. Profy, and I. A. Wilson. 2003. Recurring conformation of the human immunodeficiency virus type 1 gp120 V3 loop. Virology 315:159-173. [DOI] [PubMed] [Google Scholar]
- 72.Stanfield, R. L., M. K. Gorny, C. Williams, S. Zolla-Pazner, and I. A. Wilson. 2004. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure 12:193-204. [DOI] [PubMed] [Google Scholar]
- 73.Stanfield, R. L., A. Zemla, I. A. Wilson, and B. Rupp. 2006. Antibody elbow angles are influenced by their light chain class. J. Mol. Biol. 357:1566-1574. [DOI] [PubMed] [Google Scholar]
- 74.Tolman, R. L., M. A. Bednarek, B. A. Johnson, W. J. Leanza, S. Marburg, D. J. Underwood, E. A. Emini, and A. J. Conley. 1993. Cyclic V3-loop-related HIV-1 conjugate vaccines. Synthesis, conformation and immunological properties. Int. J. Pept. Protein Res. 41:455-466. [DOI] [PubMed] [Google Scholar]
- 75.Tugarinov, V., A. Zvi, R. Levy, Y. Hayek, S. Matsushita, and J. Anglister. 2000. NMR structure of an anti-gp120 antibody complex with a V3 peptide reveals a surface important for co-receptor binding. Struct. Fold. Des. 8:385-395. [DOI] [PubMed] [Google Scholar]
- 76.Vranken, W. F., M. Budesinsky, F. Fant, K. Boulez, and F. A. Borremans. 1995. The complete consensus V3 loop peptide of the envelope protein gp120 of HIV-1 shows pronounced helical character in solution. FEBS Lett. 374:117-121. [DOI] [PubMed] [Google Scholar]
- 77.Vranken, W. F., M. Budesinsky, J. C. Martins, F. Fant, K. Boulez, H. Gras-Masse, and F. A. Borremans. 1996. Conformational features of a synthetic cyclic peptide corresponding to the complete V3 loop of the RF HIV-1 strain in water and water/trifluoroethanol solutions. Eur. J. Biochem. 236:100-108. [DOI] [PubMed] [Google Scholar]
- 78.Vranken, W. F., F. Fant, M. Budesinsky, and F. A. Borremans. 2001. Conformational model for the consensus V3 loop of the envelope protein gp120 of HIV-1 in a 20% trifluoroethanol/water solution. Eur. J. Biochem. 268:2620-2628. [DOI] [PubMed] [Google Scholar]
- 79.Vu, H. M., R. de Lorimier, M. A. Moody, B. F. Haynes, and L. D. Spicer. 1996. Conformational preferences of a chimeric peptide HIV-1 immunogen from the C4-V3 domains of gp120 envelope protein of HIV-1 CAN0A based on solution NMR: comparison to a related immunogenic peptide from HIV-1 RF. Biochemistry 35:5158-5165. [DOI] [PubMed] [Google Scholar]
- 80.Vu, H. M., D. Myers, R. de Lorimier, T. J. Matthews, M. A. Moody, C. Heinly, J. V. Torres, B. F. Haynes, and L. Spicer. 1999. Nuclear magnetic resonance analysis of solution conformations in C4-V3 hybrid peptides derived from human immunodeficiency virus (HIV) type 1 gp120: relation to specificity of peptide-induced anti-HIV neutralizing antibodies. J. Virol. 73:746-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, G., R. MacKenzie, P. J. Durda, and P. Tsang. 2000. The binding of a glycoprotein 120 V3 loop peptide to HIV-1 neutralizing antibodies. Structural implications. J. Biol. Chem. 275:36645-36652. [DOI] [PubMed] [Google Scholar]
- 82.Zolla-Pazner, S., M. K. Gorny, P. N. Nyambi, T. C. VanCott, and A. Nadas. 1999. Immunotyping of human immunodeficiency virus type 1 (HIV): an approach to immunologic classification of HIV. J. Virol. 73:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zolla-Pazner, S., P. Zhong, K. Revesz, B. Volsky, C. Williams, P. Nyambi, and M. K. Gorny. 2004. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res. Hum. Retrovir. 20:1254-1258. [DOI] [PubMed] [Google Scholar]
- 84.Zvi, A., R. Hiller, and J. Anglister. 1992. Solution conformation of a peptide corresponding to the principal neutralizing determinant of HIV-1 IIIB: a two-dimensional NMR study. Biochemistry 31:6972-6979. [DOI] [PubMed] [Google Scholar]