Abstract
Mutations in CCHH, the putative zinc finger motif, apparently do not play an important role in virus replication in MDCK cells in culture (E. K.-W. Hui, K. Ralston, A. K. Judd, and D. P. Nayak, J. Gen. Virol. 84:3105-3113, 2003). In this report, however, we demonstrate that the CCHH motif plays a critical role in virulence in mice and that some CCHH mutants are highly attenuated in BALB/c mice. Some of the mutant viruses replicated the least in mice lungs, induced little or no lung lesions, and caused highly reduced morbidity and mortality. Furthermore, growth patterns of mutant viruses in different cell lines (MDCK, MLE12, 3LL, A549, and 293T) varied. Mutant viruses that were attenuated in mice also grew poorly in mouse and human cells in culture. However, wild-type (WT) and all mutant viruses replicated to the same titer in MDCK (canine) cells or embryonated chicken eggs. Attenuation in mice correlated with reduced growth in mouse cells in culture, suggesting that potential attenuation in a given host can be predicted from the growth characteristics of the virus in cultured cells (preferably lung cells) from the same species. In challenge experiments, mice immunized by infection with attenuated mutant viruses were fully protected from lethal challenge with WT virus. In summary, the replication and attenuating properties of these mutants suggest that the CCHH motif provides a critical determinant for virulence in mouse and that mutations in the CCHH motif yield potential vaccine candidates for the development of live species-specific attenuated influenza virus vaccines.
Influenza is a global infectious viral disease of great public health concern that is caused by influenza viruses, members of the Orthomyxoviridae family (34). Influenza viruses are enveloped, negative-stranded, segmented RNA viruses. The viral genome consists of eight (influenza A and B viruses) or seven (influenza C virus) separate RNA segments, each encoding one or more proteins (63). Virulence of influenza virus is multigenic and complex in nature (10). Influenza virus virulence is largely controlled by the cleavability of the hemagglutinin (HA) precursor (HA0) into two disulfide-linked subunits, HA1 and HA2 (HA0→HA1 + HA2), which is absolutely required for infection (31, 62, 66). However, cleavage of HA alone is not sufficient for pantropism and neurovirulence of influenza viruses as was shown by gene reassortment experiments (40, 55, 65). Other genes, including M and NS genes, also contribute to viral neurovirulence (40) and pantropism (7, 14). However, the specific function of M and NS genes in influenza virus neurovirulence and pantropism is not known. Similarly, although the cleavability of the HA molecule plays a major role in the virulence of highly pathogenic H5 and H7 subtypes in birds, other genes have been shown to contribute to virulence (3, 30). For example, a mutation in the PB2 protein was shown to influence the outcome of H5N1/97 infection in mice (17). In addition, genes other than PB2 and HA were also shown to contribute to virulence in mice (18).
In several experimental systems, the M gene was shown to have a significant effect on virus replication and virulence (4, 16, 60, 61, 64, 72, 74). For example, single-gene reassortants made with the cold-adapted M gene from influenza A/Ann Arbor/6/60 (H2N2) and other viruses revealed that the M gene alone may be the attenuating factor in cold-adapted viruses (64). In addition, the M gene also contributed to the neurovirulence of the WSN strain (65). Moreover, M1 was found to be an important determinant in the spread of virus in the alveolar environment, which may be controlled by the ability of the virus to replicate in lung cells, and M1 also functions as a stimulator of the immune system through macrophage recruitment (61), suggesting the involvement of M1 in viral pathogenesis. Recently, a double mutation in the nuclear localization signal in M1 was shown to cause strong attenuation in mice (35). In addition, a single amino acid change in M1 was shown to be responsible for the higher yield in mouse lungs (4, 16, 38, 60, 61).
In our previous study (25), we observed that the mutations in the conserved CCHH motif (the putative zinc finger motif) present in helix nine (H9) and the adjacent region of M1 did not affect virus replication in Madin-Darby canine kidney (MDCK) cells in culture (25). However, the significance of the CCHH motif in influenza virus biology is an enigma, since the zinc finger peptide (peptide 6 [CATCEQIADSQHRSHRQMV]) had been shown to be effective against type A influenza virus infection in BALB/c mice (28). Therefore, we have examined the role of the M1 protein in virulence in mice. In this report, we show that CCHH, the putative zinc finger motif, which does not play an important role in virus growth in MDCK cells in culture (25), plays a critical role in virulence in mice. We found that some CCHH mutant viruses were highly attenuated in mice. However, vaccination with these highly attenuated mutant viruses provided strong protection against lethal wild-type (WT) virus challenge. Furthermore, growth of these mutant viruses in cultured cells from different species suggested that the mutations in the CCHH motif yielded species-specific attenuation and that the growth and replication of viruses in cultured cells may predict the attenuation of influenza virus virulence in vivo in a given animal species. These results suggest that mutations of CCHH may generate attenuated influenza A viruses, which may be useful in the development of new master strains for live influenza virus vaccine.
MATERIALS AND METHODS
Cell lines and viruses.
MDCK cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Rockville, Md.) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals, Norcross, Ga.) and antibiotics (100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml Fungizone). Human embryonic 293T kidney cells (a derivative of the 293 line into which the gene for simian virus 40 T antigen was inserted) were maintained in OPTI-MEM-I medium (Invitrogen) supplemented with 5% FBS and antibiotics. Human type II alveolar epithelial A549 cells (ATCC CCL-185) were maintained in F-12K medium (Mediatech Inc., Herndon, Va.) supplemented with 10% FBS and antibiotics. Mouse lung epithelium cells (3LL from Saroji Basak, David Geffen School of Medicine, UCLA) were maintained in RPMI 1640 medium (Mediatech, Inc., Herndon, VA) supplemented with 10% FBS and antibiotics. Mouse lung epithelium MLE 12 cells (ATCC CRL-2100) were maintained in Dulbecco's modified Eagle's medium/F-12 medium (Cambrex Bio Science Walkersville, Walkersville, Md.) supplemented with 2% FBS and antibiotics. All cells were maintained at 37°C in 5% CO2. WT influenza A virus strain A/WSN/33 (H1N1) was used in these experiments, and the stock viruses were prepared as reported previously (23, 24). Plaque assays were done in MDCK cells in the presence of trypsin (0.5 μg/ml) (25).
Generation of transfectant viruses using reverse genetics and preparation of mutant stock viruses.
Mutagenesis reactions were performed by using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Transfectant viruses were generated by transfecting 293T cells with eight plasmids (1 μg each; seven polymerase [Pol I]-Pol II constructs of WT HA, NA, NP, NS, PA, PB1, PB2, and either the WT or mutated M gene) as previously reported (21, 22, 25). Individual plaques were isolated, resuspended in virus dilution buffer (23, 24), and inoculated into MDCK cells at a multiplicity of infection (MOI) of 0.001. Infected cells were incubated in virus growth medium (VGM) (25) with TPCK-treated trypsin (0.5 mg/ml), and the supernatants were harvested at 48 h postinfection (p.i.) and analyzed by plaque assay (22). Mutations in each transfectant virus were checked by reverse transcription-PCR and DNA sequencing as described previously (25).
Labeling and immunoprecipitation of M1 mutant proteins.
At 18 h posttransfection, transfected 293T cells were starved and pulse-labeled with 100 μCi 35S-protein labeling mix (Perkin-Elmer Life & Analytical Sciences, Boston, Mass.) for 2 h (22, 26). Cells were then lysed in 1 ml radioimmunoprecipitation assay buffer, immunoprecipitated with anti-M1 antibody (Biodesign International, Saco, Maine), and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%).
Mice.
BALB/c mice are an established model for influenza virus infection (37, 44, 57, 58, 68). Four-week-old female (10 to 12 g) specific-pathogen-free BALB/c mice (Charles River Labs, Wilmington, Mass.) were intranasally inoculated. Mice were quarantined 48 h prior to use and maintained in a bioclean condition in the facility for animal experiments of the Laboratory Animal Research Center of Utah State University. They were fed standard mouse chow and tap water ad libitum.
Infection of mice.
To determine the pathogenicity of the mutant viruses in the infected mice, mice were anesthetized with ketamine (100 μg/kg) by intraperitoneal injection. They were then infected intranasally with various doses of virus using a 50-μl inoculum. Infected mice in each group (five to six mice per group) were held for observation. Morbidity of the mice (measured by weight loss) and the mortality rate (indicated by mean death time) were determined.
Immunization and challenge of mice.
Mice were first infected by intranasal administration of mutant viruses (105 PFU/50 μl or 104 PFU/50 μl). Four weeks after the vaccination, immunized mice were challenged intranasally, under anesthesia, with 100 50% lethal doses (LD50) of the WT WSN virus. For determination of virus titers in lungs after WT virus challenge, lungs were harvested at day 2 postchallenge, homogenized, and titrated for virus yield by PFU on MDCK cells. The remaining animals were observed for clinical signs and symptoms of infection for 14 days after challenge.
Gross and histopathological examination.
Mice lungs on day 6 p.i. were examined by gross (lung consolidation scores and lung weights) and histopathological examination, and the expression of cytokines from lungs and blood was also determined. Lung consolidation was scored from 0 (normal) to 4 based upon plum discoloration, as previously reported (59). For histopathology, the lung blocks were removed and fixed in 10% phosphate-buffered formalin, embedded in paraffin, sectioned (5 μm), and stained with standard hematoxylin and eosin for routine histology and with periodic acid-Schiff stain to assess mucus. The following compartments of the lungs were examined by histopathology: alveolar spaces, airways at all levels, interstitium, and vessels (both arteries and veins). Inflammatory infiltrates were evaluated for location, severity, and composition (cell types included small mononuclear cells, transformed lymphocytes, neutrophils, and eosinophils). The degrees of inflammation were graded as lung consolidation scores as follows: 0 (no infiltrate/no cells in alveoli), 1+ (most vessels have an infiltrate up to four cells thick); 2+ (most vessels have an infiltrate five to seven cells thick), 3+ (most vessels have an infiltrate greater than seven cells thick), 4+ (total consolidation). These scores also include blood or edema fluid in the tissue space and alveoli.
Lung virus determination.
To determine whether attenuated mutant viruses replicated less in the lungs, we analyzed the virus titer in lungs. Virus titers are usually at the maximum on day 2 or 3 p.i. Therefore, on day 2 p.i., five mice from each group were sacrificed. Lungs were removed, scored, weighed, and frozen at −80°C. Later, lungs were thawed and homogenized in 1 ml cell culture medium using stomacher bags. The tissues were effectively homogenized by rolling a pipette over them in the stomacher bag. Prior to titrating, the homogenized lung samples were centrifuged at 3,200 × g for 5 min to pellet cell debris, and the supernatant was adjusted to 1 g/10 ml medium. Samples were serially diluted in 10-fold increments and titrated by PFU assay in MDCK cells.
Measurement of IL-6 and IFN-α.
The levels of interleukin-6 (IL-6) and alpha interferon (IFN-α) in lungs and blood were measured using a commercially available mouse IL-6 immunoassay kit (BioSource, Camarillo, Calif.) and a mouse IFN-α enzyme-linked immunosorbent assay (ELISA) kit (PBL Biomedical Laboratories, Piscataway, NJ), respectively.
Production of viruses in embryonated chicken eggs.
Ten-day-old specific-pathogen-free embryonated chicken eggs (Charles River Laboratories, North Franklin, CT) were inoculated with viruses. After 48 h, allantoic fluids were collected for plaque assay.
Growth properties of CCHH mutant viruses.
MDCK cells (4.2 × 106 cells), MLE 12 cells (4 × 106 cells), 3LL cells (6 × 106 cells), A549 cells (3.2 × 106 cells), and 293T cells (4.5 × 106 cells) were cultured in 25-cm2 flasks for a monolayer. Cells were then infected with each mutant virus at an MOI of 0.001 and maintained in VGM in the presence of 0.5 μg/ml trypsin. At different time points, supernatants were collected, and the virus titers were determined by plaque assay.
RESULTS
Mutational analysis of the zinc finger motif and the adjacent region of H9.
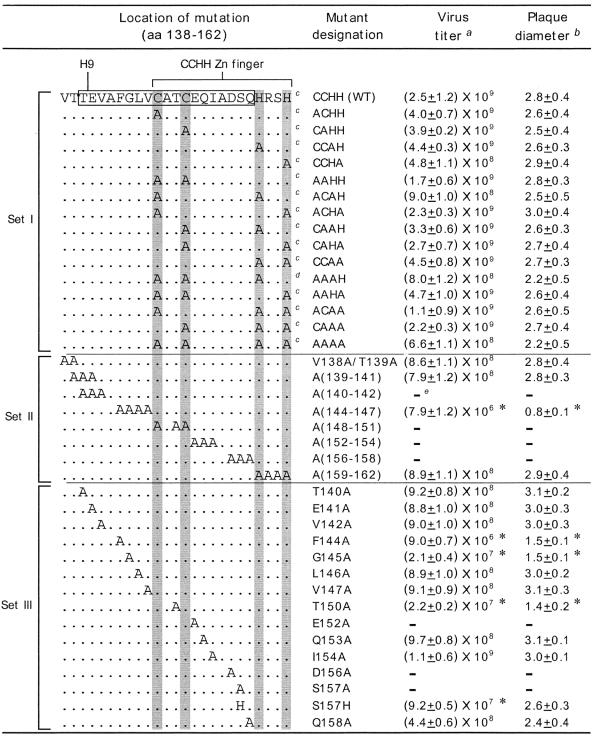
The CCHH motif in helix 9 (H9) of the M1 protein has been presumed to be the zinc finger motif (71). To determine the role of the CCHH motif and the adjacent region of H9, we made single, double, triple, and quadruple mutations in the CCHH residues by alanine replacement as report previously (25). A new triple mutant (AAAH), not reported previously, was also created (Table 1, set I). In addition, we also made single and multiple mutations of the entire H9 region by alanine replacement. We then used a Pol I-Pol II transfection system to rescue mutant influenza virus from each of the mutated cDNAs in human embryonic kidney 293T cells (22, 25). By using reverse genetics, we could rescue mutant viruses from each of mutated CCHH cDNAs. Each of these CCHH mutant viruses, including AAAA, exhibited the WT phenotype in growth and plaque morphology in MDCK cells, as was observed previously (25). The new triple mutant (AAAH), which was not reported previously, also exhibited the WT phenotype in MDCK cells (Table 1, set I). Also, none of these mutant viruses exhibited a temperature-sensitive (ts) phenotype (data not shown). Previous studies showed that the single mutation of a cysteine residue (C148S) had no effect on virus replication (36, 75).
TABLE 1.
Mutations on H9 and the CCHH zinc finger motif
Recombinant viruses were rescued by eight-plasmid transfection of 293T cells. MDCK cells were then infected with transfectant virus at an MOI of 0.001 and maintained in VGM containing 0.5 μg of trypsin/ml as described in Materials and Methods. Supernatants were collected at 48 h p.i. and assayed for PFU/milliliter. Data represent mean values (n = 3). Asterisk, P < 0.05 (versus WT).
Plaque sizes for different mutant viruses on MDCK cells. Plaques were visualized at day 3, and diameters of 10 plaques were measured (millimiters). Data represent mean values (n = 10). Asterisk, P < 0.05 (versus WT).
Results from these mutants in set I agree with our previous result (25).
A new triple mutant in the CCHH motif.
Recombinant virus could not be rescued.
Since the CCHH zinc finger motif is overlapping with H9 (Table 1), we wanted to determine if the neighboring sequences of the CCHH motif played any role in viral replication. Initially, we mutated the residues of H9 in two to four residues (Table 1, set II): V138A/T139A, A(139-141), A(140-142), A(144-147), A(148-151), A(152-154), and A (156-158). All mutant proteins were expressed at levels similar to those of the WT protein (Fig. 1A). Some mutant viruses, such as V138A/T139A, A(139-141), and A(159-162), exhibited WT growth and plaque morphology. However, one mutant, A(144-147), grew to a lower titer and exhibited a small plaque size. This mutant exhibited a partially ts phenotype (Fig. 1B). In addition, four other mutants, A(140-142), A(148-151), A(152-154), and A(156-158), were lethal, and infectious virus could not be rescued even after repeated attempts (Table 1, set II).
FIG. 1.
Expression of M1 mutant proteins and ts phenotype of mutant viruses. (A) cDNA-transfected 293T cells were pulse-labeled at 18 h posttransfection for 2 h. Cells were then lysed in radioimmunoprecipitation assay buffer, immunoprecipitated with anti-M1 antibody, and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (B) MDCK cells were infected with A(144-147), F144A, G145A, or T150A virus at an MOI of 0.001 and maintained in VGM containing 0.5 μg/ml trypsin at either 33°C or 39.5°C. Supernatants were collected at 48 h p.i. and assayed for virus titer by plaque assay. Data represent mean titers (n = 3). *, P < 0.001 (versus 33°C).
Next, we replaced single amino acid residues with alanine (Table 1, set III). We rescued mutant viruses from all single alanine replacement mutations except E152A, D156A, and S157A. Again, all mutant proteins were expressed at similar levels in cDNA-transfected 293T cells (Fig. 1A). Among the 14 rescued mutants, 4 (F144A, G145A, T150A, and S157H) exhibited reduced titers, and 10 virus mutants exhibited the WT phenotype (Table 1, set III). G145A exhibited a ts phenotype (Fig. 1B).
Reduced virulence of CCHH mutants in mice.
Although the mutational analysis suggested that the CCHH motif did not have any role in virus replication and growth in cultured MDCK cells, it has been previously reported that a peptide containing the CCHH motif affected virus pathogenesis in mice (28, 41). We therefore wanted to determine if the CCHH motif has any role in virus pathogenicity in a mouse model. The virulence of mutant viruses was tested by measuring mortality in mice infected with different dosages. For this experiment, two single, six double, four triple, and the quadruple alanine mutants of the CCHH motif were used. All of these mutant viruses exhibited the WT phenotype with regard to virus growth and plaque morphology in MDCK cells (Table 1). Accordingly, groups of BALB/c mice (five to six mice per group) were infected intranasally with WT or mutant viruses at different dosage (106, 105, 104, and 103 PFU/mouse). Results (Table 2) showed that the pathogenicities of mutant viruses in mice varied and can be grouped into three classes. All mice infected with WT WSN virus using 106, 105, and 104 PFU/mouse died. Two single mutants (CCAH and CAHH) and two double mutants (CAHA and ACHA) exhibited pathogenicity similar that of the WT. Some double mutants (CCAA, CAAH, and AAHH) and triple mutants (CAAA and AAHA) were 10 times less virulent than the WT virus, as they required 10 times more virus than the WT virus to cause similar mouse mortality. Four mutants (ACAH, ACAA, AAAH, and AAAA), on the other hand, did not kill mice even with an inoculum of 105 PFU and were therefore 100 times less virulent than WT virus (Table 2). The two least virulent viruses (AAAH and AAAA) were also tested at 106 PFU/mouse and still did not cause death. These results show that for the CCHH motif, the first C and H were critical for virulence, as ACAH, ACAA, AAAH, and AAAA viruses were highly avirulent in mice. Also, the average mean day of death correlated with mortality, i.e., mice exhibiting a higher percentage of mortality died earlier compared to mice exhibiting a lower mortality rate (Table 2).
TABLE 2.
Survival data for BALB/c mice after WT and mutant virus infection
| Virus | Expt | No. of survivors/total no. (MDD)a for infectious dose (PFU/mouse) of:
|
Virulence relative to WTb | |||
|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | |||
| WT | ||||||
| CCHH | 1 | 0/5 (6.6 ± 1.1) | 0/5 (8.0 ± 1.7) | 0/5 (9.2 ± 1.1) | 5/5 (>21) | |
| 2 | 0/5 (6.2 ± 1.4) | 0/5 (7.6 ± 1.5) | 0/5 (9.0 ± 1.2) | 3/5 (10.0 ± 0.0) | ||
| 3 | 0/6 (6.7 ± 1.8) | 0/5 (7.4 ± 0.5) | 0/5 (8.6 ± 0.5) | 1/5 (12.8 ± 4.2) | ||
| Single mutations | ||||||
| CCAH | 1 | 0/5 (5.4 ± 0.5) | 0/5 (6.8 ± 0.4) | 0/5 (8.0 ± 0.7) | 1/5 (9.0 ± 1.2) | Same |
| 2 | 0/5 (5.5 ± 0.8) | 1/5 (7.5 ± 0.5) | 0/5 (9.2 ± 1.1) | 5/5 (>21) | ||
| CAHH | 1 | 0/5 (8.4 ± 0.9) | 1/5 (11.5 ± 6.4) | 1/5 (12.8 ± 1.7) | 5/5 (>21) | Same |
| Double mutations | ||||||
| CCAA | 1 | 0/5 (6.7 ± 1.0) | 0/5 (8.6 ± 1.5) | 4/5 (12.0) | 5/5 (>21) | 10× less |
| CAHA | 1 | 0/5 (6.2 ± 0.8) | 0/5 (7.0 ± 0.7) | 1/5 (9.0 ± 0.0) | 4/5 (10.0) | Same |
| CAAH | 1 | 0/5 (7.0 ± 1.4) | 0/5 (8.4 ± 1.1) | 4/5 (13.0) | 5/5 (>21) | 10× less |
| ACHA | 1 | 0/5 (7.0 ± 1.0) | 0/5 (8.0 ± 0.0) | 1/5 (9.5 ± 1.7) | 5/5 (>21) | Same |
| ACAH | 1 | 5/5 (>21) | 5/5 (>21) | 5/5 (>21) | 5/5 (>21) | >100× less |
| 2 | 6/6 (>21) | 5/5 (>21) | 5/5 (>21) | 4/5 (12.0) | ||
| 3 | 2/5 (9.3 ± 0.6) | 3/5 (10.5 ± 3.5) | 5/5 (>21) | 5/5 (>21) | ||
| AAHH | 1 | 0/5 (8.2 ± 1.4) | 0/5 (9.2 ± 1.8) | 4/5 (9.0) | 5/5 (>21) | 10× less |
| Triple mutations | ||||||
| CAAA | 1 | 0/5 (7.8 ± 0.4) | 0/5 (10.6 ± 1.1) | 4/5 (11.0) | 4/5 (11.0) | |
| 2 | 0/5 (7.8 ± 1.5) | 0/5 (13.4 ± 3.2) | 5/5 (>21) | 5/5 (>21) | 10× less | |
| ACAA | 1 | 0/5 (7.6 ± 0.5) | 3/5 (12.5 ± 0.7) | 5/5 (>21) | 5/5 (>21) | 100× less |
| 2 | 2/5 (13.0 ± 2.0) | 5/5 (>21) | 5/5 (>21) | 5/5 (>21) | ||
| AAHA | 1 | 0/5 (7.6 ± 0.5) | 0/5 (9.2 ± 1.6) | 5/5 (>21) | 5/5 (>21) | 10× less |
| 2 | 0/5 (7.6 ± 1.1) | 0/5 (9.0 ± 1.2) | 4/5 (8.0) | 5/5 (>21) | ||
| AAAH | 1 | 6/6 (>21) | 6/6 (>21) | 5/5 (>21) | 5/5 (>21) | 100× less |
| Quadruple mutation | ||||||
| AAAA | 1 | 0/5 (8.0 ± 1.0) | 5/5 (>21) | 5/5 (>21) | 5/5 (>21) | 100× less |
| 2 | 5/6 (11.0) | 6/6 (>21) | 6/6 (>21) | 6/6 (>21) | ||
Number of surviving mice/number of mice in group. MDD, mean day of death of mice that died within 21 days of infection.
Compared to WT virus.
Body weight changes and lung infection parameters during infection.
Other parameters of infection besides mortality were also investigated to assess the morbidity of mice infected with mutant viruses. Mice receiving single doses of these viruses (105 PFU/mouse) were compared with those infected with WT virus (Table 3). Severe weight loss occurred following infection with the WT as well as with the CAAA and ACAA viruses, and mild weight loss or minimal weight gain was seen after infection with the ACAH, AAHH, and AAHA viruses. On the other hand, appreciable weight gain was observed after infection with the AAAH and AAAA viruses. Weight loss correlated with mice mortality, as depicted in Table 2. Lung infection parameters of animals sacrificed on day 6 were also determined, and lungs were examined for gross (weight, pneumonia, and consolidation) as well as histopathologic (cellular infiltration, hemorrhage, etc.) lung lesions. The triple mutants CAAA, ACAA, and AAHA induced lung consolidation that resulted in scores that were not significantly different from those of WT virus infection. Lower lung lesion scores were seen in mice infected with the ACAH and AAHH double mutant viruses. No lung consolidation was observed in animals infected with the AAAH and AAAA viruses. An increase in lung weight was observed after infection with most viruses, except for AAAH and AAAA viruses. The lungs were sent to a separate laboratory for histopathological examination. Severity of microscopic lung lesions and hemorrhage varied somewhat from animal to animal, and overall patterns seen in each group of mice are depicted in Table 3. In spite of the limitations that greater histopathological changes occurred near death and that the data represent just one segment during the time of infection, the two groups of animals exhibiting the lowest lung pathology scores were those infected with the AAAH and AAAA viruses. Overall, a decrease in body weight and increase in lung weight and higher lung pathology scores correlated with the virulence (mortality) of mutant and WT viruses.
TABLE 3.
Body weight changes and lung infection parameters during infection with WT and mutant virusesf
| Virusa | Mean % body wt change (g)b | Lung virus titer (day 2 p.i.) (% of WT PFU) | Lung parameters (day 6)
|
|||
|---|---|---|---|---|---|---|
| Consolidation scorec | Histopathologyd
|
|||||
| Lung wt (g) | Lesions | Hemorrhage | ||||
| WT | ||||||
| CCHH | −15.8 | (5.3 ± 0.9) × 108 (100)e | 2.4 ± 0.2 | 226 ± 28 | +/++ | +/++ |
| Double mutants | ||||||
| ACAH | +0.8 | (9.6 ± 0.7) × 106* (1.8) | 0.5 ± 0.4** | 143 ± 30** | − | +/++ |
| AAHH | −2.5 | (4.1 ± 0.3) × 107* (7.7) | 0.9 ± 0.3** | 185 ± 31 | +/+++ | −/+ |
| Triple mutants | ||||||
| CAAA | −19.0 | (4.9 ± 0.5) × 107* (9.2) | 2.1 ± 0.3 | 195 ± 6 | +/++ | −/+ |
| ACAA | −15.6 | (1.1 ± 0.8) × 107* (2.1) | 1.8 ± 0.8 | 210 ± 52 | + | −/+ |
| AAHA | −3.6 | (7.3 ± 0.7) × 107* (13.8) | 1.9 ± 0.8 | 203 ± 26 | ++ | −/+ |
| AAAH | +11.3 | (1.1 ± 0.1) × 102** (<0.001) | 0.0 ± 0.0** | 120 ± 21** | − | + |
| Quadruple mutant | ||||||
| AAAA | +11.9 | (6.8 ± 0.1) × 101** (<0.001) | 0.0 ± 0.0** | 123 ± 13** | − | + |
The infecting virus dose was 105 PFU/mouse.
Between day 0 and day 6 of infection (groups of five mice).
Consolidation score is based upon the overall appearance of lungs on day 6. A higher score (range, 0 to 4) indicates more consolidation.
Severity scores: −, no lesions or hemorrhage; +, mild; ++, moderate; +++, severe. The results represent the average from groups of five mice.
Percentage of WT (CCHH virus) PFU.
*P < 0.05; **P < 0.01 compared to WT (CCHH virus).
Next, to determine if the virus growth in lungs corresponded to the severity of infection and mortality, we examined virus growth in the lungs of infected mice. Since virus titers reached the maximum at around day 2 or 3 p.i. in lungs, we determined the virus titers in the lungs on day 2 p.i. Accordingly, groups of four mice infected with different viruses were sacrificed on day 2 p.i., and the pooled lungs were used for determining virus titers by plaque (PFU) assay (Table 3). The lungs of mice infected with WT virus had the highest titer (5.3 × 108 PFU/ml), and the majority of the double or triple mutant viruses exhibited titers around 1.5 logs less in mouse lungs. However, two mutants (AAAH and AAAA) had highly reduced virus titers, about 6 to 7 logs less, compared to the WT virus. These results indicate that CCHH was a critical determinant for virulence in mice and that mutations in the CCHH motif led to decreased virus growth in lungs and reduced virulence.
Chemokine response in infected mice.
Chemokines are small secretory molecules involved in recruiting inflammatory cells to the site of infection (73). Cytokines activate host antiviral defense systems upon infection. IFNs are considered the body's first line of innate antiviral defense (8, 13, 15). Type I IFNs (IFN-α/β) are the key cytokines produced by influenza A virus-infected epithelial cells, monocytes/macrophages, and spleen cells (5, 29, 39, 52, 54, 70) and function as antiviral factors. Several studies have demonstrated that an early rise in the levels of IFN-α, tumor necrosis factor (TNF), IL-1α, IL-1β, and IL-6 in bronchoalveolar lavage fluids or lung homogenates is associated with symptom development and lung pathology in influenza virus-infected mice (19, 33, 47, 69). Therefore, the levels of IL-6 and IFN-α in serum samples and lungs of the mutant virus-infected mice were determined. Accordingly, the lungs (extracts of homogenized lung in cell culture medium) and serum samples from the mice infected with WT and mutant viruses were assayed for IL-6 and IFN-α levels by ELISA. Results show that mutant viruses AAAH and AAAA, which exhibited the least lung pathology and virus growth, also induced the lowest IL-6 and IFN-α levels in lungs and blood (Fig. 2).
FIG. 2.
Chemokine production by virus-infected mice. Mice were inoculated intranasally at 105 PFU/mouse. On day 2, groups of five mice were sacrificed. The lungs (open bars) and serum samples (gray bars) were collected. Samples were then assayed for IL-6 (A) and IFN-α (B) levels. Data represent mean titers (n = 5). *, P < 0.05 (versus CCHH); **, P < 0.001 (versus CCHH). IFN-α levels for AAAH and AAAA are the background.
Growth of mutant viruses in different cultured cell lines.
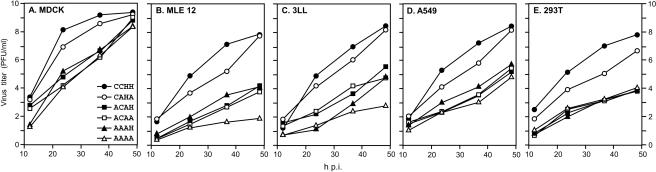
The above-described results demonstrated two antagonistic observations with CCHH mutants: (i) in MDCK cells, alanine replacement of the CCHH motif had little or no effect on virus replication, and mutant viruses essentially grew to the same titer as the WT virus (25); (ii) on the other hand, in infected mice, some CCHH mutants were highly attenuated and grew to a very low titer in mouse lungs, implying that the CCHH motif played a critical role in virus growth and virulence in mice, as observed here and as has been implicated previously (28). Therefore, we wanted to determine if reduced virulence in mice could be at least partly due to species-specific attenuation in virus growth caused by a mutation of CCHH. Therefore, we investigated the growth behavior and virus yield of CCHH mutant viruses in a number of cell lines from different species. Accordingly, MDCK, 293T, MLE 12, A549, and 3LL cells were infected with WT virus (CCHH) and five mutant viruses (CAHA, ACAH, ACAA, AAAH, and AAAA) at a very low MOI (0.001 PFU/ml) and propagated in the presence of trypsin (0.5 μg/ml) for 48 h (Fig. 3). Supernatants were collected at different times postinfection, and virus titers were determined by the PFU assay of MDCK cells. MDCK and 293T cells are kidney cell lines from canines (Canis familiaris) and humans (Homo sapiens), respectively, which are established model cells for influenza virus replication. MLE 12 and 3LL cell lines are mouse (Mus musculus) lung epithelium cells and may therefore represent the cells infected at physiological sites of influenza virus infection in the mouse model. A549 cells, a human type II alveolar epithelial cell line, may represent the lung cells involved in influenza virus infection in human beings.
FIG. 3.
Growth properties of CCHH mutant viruses. MDCK, MLE 12, 3LL, A549, and 293T cells were infected with each virus at an MOI of 0.001. At the indicated time points, virus titers (PFU) in the supernatant were determined. The values are the means of results from triple experiments. The standard deviation is less than 0.5 for each sample.
Results (Fig. 3A and Table 4) show that in a canine cell line (MDCK), as expected, each of the mutant viruses, whether possessing double (CAHA and ACAH), triple (ACAA and AAAH), or even quadruple (AAAA) mutations in the CCHH motif, exhibited the WT (CCHH) titer at 48 h p.i. (1 × 108 to 1 × 109 PFU/ml), as was reported previously (25). However, differences in virus yield between the WT and several mutant viruses were observed at 24 and 36 h p.i.; these variations are likely to be due to variations in the multiplicity of infecting viruses at a very low MOI.
TABLE 4.
Growth characteristics of WT and mutant influenza viruses in different cell linesa
| Virus | Titer (PFU/ml) for cell type:
|
||||
|---|---|---|---|---|---|
| Canine kidney (MDCK) | Mouse lung
|
Human
|
|||
| MLE 12 | 3LL | Lung (A549) | Kidney (293T) | ||
| WT | |||||
| CCHH | (2.4 ± 1.2) × 109 | (6.7 ± 0.8) × 107 | (8.0 ± 1.0) × 108 | (4.3 ± 0.5) × 108 | (9.8 ± 0.7) × 107 |
| Double mutations | |||||
| CAHA | (1.9 ± 0.6) × 109 | (5.3 ± 0.6) × 107 | (5.3 ± 0.7) × 108 | (1.7 ± 0.7) × 108 | (1.0 ± 0.7) × 107 |
| ACAH | (7.0 ± 0.3) × 108 | (2.3 ± 0.2) × 104 | (8.0 ± 1.0) × 105 | (3.0 ± 0.3) × 105 | (3.3 ± 0.4) × 104 |
| Triplet mutations | |||||
| ACAA | (1.1 ± 0.5) × 109 | (1.0 ± 0.1) × 104 | (1.3 ± 0.3) × 105 | (6.7 ± 0.7) × 105 | (6.0 ± 0.2) × 104 |
| AAAH | (2.5 ± 1.0) × 108 | (1.6 ± 0.1) × 104 | (1.0 ± 0.2) × 105 | (1.8 ± 0.3) × 106 | (4.6 ± 0.1) × 104 |
| Quadruple mutation | |||||
| AAAA | (2.4 ± 0.5) × 108 | (2.3 ± 0.1) × 102 | (1.7 ± 0.1) × 103 | (1.9 ± 0.2) × 105 | (5.8 ± 0.1) × 104 |
Different cells (MDCK, MLE 12, 3LL, A549, and 293T) were infected with WT and mutant influenza viruses at an MOI of 0.001. Culture supernatants were harvested at 48 h p.i. Virus titers are given as PFU/milliliter. Mean results from three experiments are shown.
In mouse cell lines (MLE 12 and 3LL), results (Fig. 3B and C) show that mutant viruses could be divided into three groups. The first group (WT and CAHA) grew normally to a final titer of about 5 × 107 to 8 × 108 PFU/ml at 48 h p.i. The second group (ACAH, ACAA, and AAAH) produced virus titers of 1 × 104 to 2.3 × 104 PFU/ml. The third group (AAAA) had the lowest titers (2 × 102 to 2 × 103 PFU/ml) at 48 h p.i. (Table 4).
In human cell lines (A549 and 293T), viruses could be divided into two major groups (Fig. 3D and E). The first group (WT and CAHA) grew normally to a final titer of about 9 × 107 to 4.5 × 108 PFU/ml at 48 h p.i. The second group (ACAH, ACAA, AAAH, and AAAA) produced virus titers about 2 to 3 log10 lower at 48 h p.i. (approximately 3.3 × 104 to 6.7 × 105 PFU/ml) (Table 4). The growth behavior of these mutant viruses in human cells (A549 and 293T) was essentially same as that in mouse cells. Some grew to same titer as the WT virus, while others yielded 1,000-fold-less virus. These results show that growth in mouse cells essentially corresponded to the behavior of these viruses in infected mice; i.e., viruses that grew to a higher titer were more pathogenic in mice than the viruses that grew to a lower titer. Furthermore, the growth of viruses in mouse and human cell lines followed a similar pattern.
Protective effect of mutant viruses in mice.
Since some of the mutant viruses were highly attenuated in mice, we wanted to investigate whether these mutant viruses could protect mice against a lethal challenge of WT virus. Accordingly, mice were first infected by intranasal administration of mutant viruses (105 PFU/50 μl or 104 PFU/50 μl). Four weeks after the vaccination, immunized mice were challenged intranasally, under anesthesia, with 100 LD50 of the WT WSN virus. For determination of virus titers in lungs after WT virus challenge, lungs were harvested at day 2 p.i., homogenized, and titrated on MDCK cells for PFU. The remaining animals were observed for clinical signs and symptoms of infection for 14 days after challenge. Results (Table 5) show that some mortality occurred during the vaccination phase of the experiment in the AAHA and CAAA groups at 105 PFU/mouse but not at 104 PFU/animal, and one death occurred with the double mutant ACAH vaccination. All mice in the ACAA-, AAAH-, and AAAA-immunized groups survived. Body weight gains after immunization were seen in these groups as well, indicating low virulence of these vaccinating viruses. All of the mice that survived the vaccinations were protected after WT virus challenge, demonstrating high protective efficacy against the subsequent WT virus challenge. After challenge, all immunized mice gained weight, showed no sign of illness or infection, and did not have any detectable virus or either IL-6 or IFN-α in their lungs (Table 5). These results show that highly attenuated viruses, including AAAH and AAAA viruses, were highly protective against lethal WT virus challenge.
TABLE 5.
Protection against lethal virus challenge in immunized BALB/c mice
| Virus | Vaccine dose (PFU/mouse) | After immunizationa
|
After WT virus challengeb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of survivors/ total no. | MDDc | % Wt changed | No. of survivors/ total no. | MDDc | % Wt changed | Virus titere | IL-6f (pg/ml) | IFN-αf (pg/ml) | ||
| Mock | 20/20 | >30 | +9.6 | 0/10 | 6.5 ± 1.1 | −31.8 | 2.8 ± 0.1 × 107 | 3,033 ± 13 | 584 ± 50 | |
| Double mutant | ||||||||||
| ACAH | 105 | 19/20 | 14 | −8.0 | 10/10 | >21 | +2.2 | <33 | NM | NM |
| Triple mutants | ||||||||||
| CAAA | 104 | 20/20 | >30 | +8.1 | 10/10 | >21 | +2.3 | <33 | NM | NM |
| 105 | 13/20 | 9.4 ± 1.2 | −14.0 | 10/10 | >21 | +3.4 | <33 | NM | NM | |
| ACAA | 104 | 20/20 | >30 | +8.8 | 10/10 | >21 | +1.7 | <33 | NM | NM |
| 105 | 20/20 | >30 | +1.5 | 10/10 | >21 | +2.8 | <33 | NM | NM | |
| AAHA | 104 | 20/20 | >30 | +9.6 | 10/10 | >21 | +4.0 | <33 | NM | NM |
| 105 | 0/20 | 9.6 ± 1.8 | −19.7 | NAg | NA | NA | NA | NA | NA | |
| AAAH | 104 | 20/20 | >30 | +13.2 | 10/10 | >21 | +0.6 | <33 | NM | NM |
| 105 | 20/20 | >30 | +14.0 | 10/10 | >21 | +0.6 | <33 | NM | NM | |
| 106 | 20/20 | >30 | +8.8 | 10/10 | >21 | +1.7 | <33 | NM | NM | |
| Quadruple mutant | ||||||||||
| AAAA | 104 | 20/20 | >30 | +8.1 | 10/10 | >21 | +2.9 | <33 | NM | NM |
| 105 | 20/20 | >30 | +10.3 | 10/10 | >21 | +2.9 | <33 | NM | NM | |
| 106 | 20/20 | >30 | +2.2 | 10/10 | >21 | +1.7 | <33 | NM | NM | |
Intranasally immunized with a 50-μl volume of mutant virus at the indicated dose (PFU/mouse).
Mice were challenged intranasally with 100 LD50 (106 PFU/animal) of the WT (CCHH) WSN virus 30 days after vaccination.
MDD, mean day of death of mice that died after immunization or after WT virus challenge.
Difference between starting weight (either on the day of immunization or on the day of WT virus challenge) and weight 6 days later.
PFU in lungs on day 2 postchallenge.
In lungs on day 2 postchallenge. NM, not measurable (i.e., no significant difference from the WT background).
NA, not applicable.
Virus replication in embryonated chicken eggs.
Since vaccines at present are made by growing viruses in embryonated chicken eggs, we wanted to determine if mutations in CCHH affected virus growth in chicken embryos. Accordingly, 10-day-old embryonated specific-pathogen-free chicken eggs were inoculated with WT and mutant viruses, and virus titers at 48 h p.i. were measured. Results (Table 6) show that WT virus as well as all mutant viruses grew essentially equally in embryonated chicken eggs and MDCK cells, indicating that the CCHH motif was not involved in virus replication in either embryonated chicken eggs or MDCK cells.
TABLE 6.
Titers of WT and mutant viruses in embryonated chicken eggs
| Virusa | Virus titer (PFU/ml)b |
|---|---|
| WT | |
| CCHH | (3.5 ± 0.2) × 108 |
| Double mutant | |
| ACAH | (7.5 ± 0.7) × 107 |
| Triple mutants | |
| CAAA | (3.7 ± 0.5) × 108 |
| AAAH | (6.9 ± 1.1) × 107 |
| Quadruple mutant | |
| AAAA | (9.8 ± 0.8) × 107 |
The inoculating virus dose was 6.5×103 to 1.2×104 PFU/egg.
PFU/milliliter at 48 h postinoculation.
DISCUSSION
In this report, we present the effect of mutations of the CCHH mutant, the putative zinc finger motif, and of the entire H9 of M1, which contains both cysteine residues of the CCHH motif. Our data show that some single or multiple mutations in H9 either were lethal or caused a reduction in virus growth in MDCK cells (Table 1, sets II and III). Although the sequences in H9 are highly conserved, some variations were observed among different influenza A virus strains (Table 7). It should be noted that some naturally occurring avian and human viruses possess Ala157, which was lethal in the WSN M1 background (Table 1, set III). It will be interesting to determine what compensatory sequence(s) in these avian and human strains could reverse the deleterious effect of the S157A mutation in the WSN background.
TABLE 7.
Mutations found in H9 and the CCHH motif of influenza A virus strains
The region (amino acids [aa] 140 to 162) includes the entire H9 amino acid sequence (aa 140 to 158; underlined) and the overlapping zinc finger motif (aa 148 to 162). The positions of the CCHH motif are shown in boldface type. Single mutations on the amino acid residues that either were lethal or exhibited reduced titers (Table 1, set III) are indicated as bullets (•) or thin arrows (↓), respectively.
The arrangement of CCHH, the putative zinc finger motif of influenza virus M1, matches the CX2-4CX2-15HX2-6H (where X is any amino acid residue) motif, which is known to be involved in zinc ion binding (27). However, a typical CCHH-type zinc finger protein contains 12 amino acid residues between the second Cys residue and the first His residue, whereas the M1 protein contains only seven residues between the second Cys and the first His (Table 1). Also, the first two cysteine residues on the M1 CCHH motif (Cys148 and Cys151) are located on the same side of the helix (data not shown), and both cysteines in H9 are accessible from the domain surface (25, 45). However, although the M1 protein actually binds a zinc ion, only a small percentage (∼6 to 9%) of the influenza A virus M1 protein contains zinc, and the zinc content of M1 does not influence the in vitro RNA binding (9). Since CCHH mutant viruses grew efficiently in MDCK cells, it is unlikely that zinc ion binding was involved in M1 function. Furthermore, naturally occurring human and equine viruses (A/Bangkok/1/79 [H3N2], A/environment/Hong Kong/437-6/99 [H5N1], and A/equine/Praque/1/56 [H7N7]) possess a mutation in the CCHH motif (Table 7) (25).
We have previously shown that mutations in the CCHH motif, the putative zinc finger, did not affect virus replication in MDCK cells in culture (25). However, in this report, we provide evidence that the CCHH motif in the influenza A virus M1 protein is an important contributory factor in viral pathogenesis in vivo in mice. Some of the CCHH mutant viruses were highly attenuated, caused much less mortality and morbidity (Table 2), produced lower virus titers in mouse lungs (Table 3), exhibited reduced lung lesions (Tables 3), and produced fewer cytokines after infection (Fig. 2). Similar findings with regard to the reduction of virulence have also been reported for the zinc finger motif of the Sendai virus (SeV) V protein (CX3CX11CXCX2CX3CX2C). The zinc finger mutant SeVs showed viral protein synthesis and growth patterns in cultured LLC-MK2 Rhesus monkey (Macaca mulatta) kidney cells similar to those of WT SeV. However, the SeV mutants were strongly attenuated in ICR/Crj (CD-1 strain) mice (11, 20). Furthermore, in this report, we observed that mice immunized with mutant viruses by intranasal infection were fully protected against lethal challenge with WT virus (Table 5), demonstrating the potential use of these mutant viruses as live, attenuated vaccine. Also, when the growth properties of the mutant viruses in different cell lines were compared (Fig. 3 and Table 4), we observed that mutant viruses grew in a species-specific manner, reflecting their virulence in vivo.
The M1 protein has been implicated in influenza virus growth virulence in a number of studies (4, 16, 60, 61, 64, 72, 74). Our studies reported here suggest that the CCHH motif of M1 may be a contributory factor in virus virulence in a species-specific manner. Our results show that multicycle growth patterns of mutant viruses varied in different cell types when infected at a very low MOI (0.001) (Fig. 3 and Table 4) and that mutant viruses, which were attenuated in mice, had reduced virus titers in 3LL and MLE 12 mouse lung epithelium cells. In addition, viruses that were attenuated in mice also had reduced growth in human kidney (293T) and lung (A549) cells. It has recently been shown that the highly virulent 1918 influenza virus was also virulent in mice without adaptation (32, 67). It will be worthwhile to determine whether the growth pattern of mutant viruses in cultured cells, preferably lung cells, of other species, including ferrets, primates, and different avian species, is also species specific. Results reported here suggest that growth in species-specific cells, preferably lung cells, may correlate with virulence/attenuation in vivo. Therefore, attenuation of virulence of influenza virus for a given species may be predicted from the replication behavior in cultured cells, preferably lung cells of the same species. If this observation, i.e., correlating virus growth in vitro with virus pathogenicity in vivo, is confirmed with further testing, it would provide an easier way to predict attenuation of virulence in a given species of animals.
However, as of yet, it is unclear how CCHH mutations would lead to species-specific attenuation. It is unlikely that the loss of zinc ion binding played a critical role in attenuation of mutant virus, since in that case, any disruption of the CCHH motif by even a single mutation of CCHH would have led to attenuation. The M1 protein plays critically important roles in many aspects of virus replication, including virus entry and uncoating, regulation of transcription and replication of viral RNA, and viral RNP export from the nucleus (34, 42, 43, 56). In addition, it has recently been shown that the M1 protein contains a novel L domain involved in virus budding (21, 22). These processes require the M1 protein to interact with the viral glycoproteins HA and NA (1), viral RNP (2, 6, 46, 50, 51, 53, 75), and other viral and host proteins. M1 associates with a number of cellular proteins, including histones (12, 76), RACK-1 (cellular receptor of activated C kinase 1) (49), nucleosomes (12), VPS28 (vacuolar protein sorting 28), Cdc42 (cell division cycle 42) (21), protein kinase C (49), extracellular signal-regulated kinase (48), and possibly other host proteins. It is likely that mutations in the CCHH motif may affect interactions with one or more host proteins and may thus affect virus growth and virulence in a species-specific manner.
Furthermore, these highly attenuated M1 mutant viruses can provide protection against challenge with a lethal dose of WT influenza virus. These mutant viruses are not temperature sensitive and are therefore unlikely to be cold adapted and are likely to be less thermolabile than the presently used cold-adapted influenza virus vaccine. Data presented here suggest that it is now possible to generate tailor-made virus strains with unique properties that can lead to species-specific attenuation of virulence. Finally, if needed, these mutations can be combined with other mutations affecting virulence to create highly attenuated stable virus strains that can grow to high titers in chicken embryos or cells approved for vaccine production and induce protective immunity in a specific host. These mutants are therefore potential candidates as master strains for producing live, attenuated influenza virus vaccines.
Acknowledgments
We thank Ee Ming Yap, Bryan France, Dominic Ho-Ping Tang, Nancy Fong, Tess Lin, and Rilwan Balogun for their help with DNA preparation and ELISA experiments.
This work was supported by USPHS grants (R01 AI16348 and R01 AI41681) to D.P.N. This work was supported in part by contract NO1-AI-15435 from the Virology Branch, NIAID, NIH, to D.F.S.
REFERENCES
- 1.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudin, F., I. Petit, W. Weissenhorn, and R. W. H. Ruigrok. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281:102-108. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., W. Garten, H.-D. Klenk, and R. Rott. 1981. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology 113:725-735. [DOI] [PubMed] [Google Scholar]
- 4.Brown, E., H. Liu, L. Kit, S. Baird, and M. Nesrallah. 2001. Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc. Natl. Acad. Sci. USA 98:6883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brydon, E. W. A., S. J. Morris, and C. Sweet. 2005. Role of apoptosis and cytokines in influenza virus morbidity. FEMS Microbiol. Rev. 29:837-850. [DOI] [PubMed] [Google Scholar]
- 6.Bui, M., G. Whittaker, and A. Helenius. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70:8391-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castrucci, M. R., and Y. Kawaoka. 1993. Biologic importance of influenza virus stalk length in influenza A virus NA. J. Virol. 67:759-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doly, J., A. Civas, S. Navarro, and G. Uze. 1997. Type I interferons: expression and signalization. Cell. Mol. Life Sci. 54:1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elster, C., E. Fourest, F. Baudin, K. Larsen, S. Cusack, and R. W. H. Ruigrok. 1994. A small percentage of influenza virus M1 protein contains zinc but zinc does not influence in vitro M1-RNA interaction. J. Gen. Virol. 75:37-42. [DOI] [PubMed] [Google Scholar]
- 10.Fislová, T., and F. Kostolanský. 2005. The factors of virulence of influenza A virus. Acta Virol. 49:147-157. [PubMed] [Google Scholar]
- 11.Fukuhara, N., C. Huang, K. Klyotani, T. Yoshida, and T. Sakaguchi. 2002. Mutational analysis of the Sendai virus V protein: importance of the conserved residues for Zn binding, virus pathogenesis, and efficient RNA editing. Virology 299:172-181. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Robles, I., H. Akarsu, C. W. Muller, R. W. H. Ruigrok, and F. Baudin. 2005. Interaction of influenza virus proteins with nucleosomes. Virology 332:329-336. [DOI] [PubMed] [Google Scholar]
- 13.García-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 14.García-Sastre, A., R. K. Durbin, H. Zheng, P. Palese, R. Gertner, D. E. Levy, and J. E. Durbin. 1998. The role of interferon in influenza virus tissue tropism. J. Virol. 72:8550-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 16.Govorkova, E. A., A. S. Gambaryan, E. C. Claas, and Y. A. Smirnov. 2000. Amino acid changes in the hemagglutinin and matrix proteins of influenza A (H2) viruses adapted to mice. Acta Virol. 44:241-248. [PubMed] [Google Scholar]
- 17.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 18.Hatta, M., and Y. Kawaoka. 2002. The continued pandemic threat posed by avian influenza viruses in Hong Kong. Trends Microbiol. 10:340-344. [DOI] [PubMed] [Google Scholar]
- 19.Hennet, T., H. J. Ziltener, J. Frei, and E. Peterhans. 1992. A kinetic study of immune mediators in the lungs of mice infected with influenza A virus. J. Immunol. 149:932-939. [PubMed] [Google Scholar]
- 20.Huang, C., K. Kiyotani, Y. Fujii, N. Fukuhara, A. Kato, Y. Nagai, T. Yoshida, and T. Sakaguchi. 2000. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74:7834-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui, E. K.-W., S. Barman, D. H.-P. Tang, B. France, and D. P. Nayak. 2006. YRKL sequence of influenza virus M1 functions as the L domain motif and interacts with VPS28 and Cdc42. J. Virol. 80:2291-2308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Hui, E. K.-W., S. Barman, T. Y. Yang, and D. P. Nayak. 2003. Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motifs. J. Virol. 77:7078-7092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Hui, E. K.-W., and D. P. Nayak. 2001. Role of ATP in influenza virus budding. Virology 290:329-341. [DOI] [PubMed] [Google Scholar]
- 24.Hui, E. K.-W., and D. P. Nayak. 2002. Role of G protein and protein kinase signalling in influenza virus budding in MDCK cells. J. Gen. Virol. 83:3055-3066. [DOI] [PubMed] [Google Scholar]
- 25.Hui, E. K.-W., K. Ralston, A. K. Judd, and D. P. Nayak. 2003. Conserved cysteine and histidine residues in the putative zinc finger motif of the influenza A virus M1 protein are not critical for influenza virus replication. J. Gen. Virol. 84:3105-3113. [DOI] [PubMed] [Google Scholar]
- 26.Hui, E. K.-W., E. M. Yap, D. S. An, I. S. Y. Chen, and D. P. Nayak. 2004. Inhibition of influenza virus matrix (M1) protein expression and virus replication by U6 promoter-driven and lentivirus-mediated delivery of siRNA. J. Gen. Virol. 85:1877-1884. [DOI] [PubMed] [Google Scholar]
- 27.Iuchi, S. 2001. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 58:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Judd, A. K., A. Sanchez, D. J. Bucher, J. H. Huffman, K. Bailey, and R. W. Sidwell. 1997. In vivo anti-influenza virus activity of a zinc finger peptide. Antimicrob. Agents Chemother. 41:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julkunen, I., K. Melén, M. Nyqvist, J. Pirhonen, T. Sareneva, and S. Matikainen. 2001. Inflammatory responses in influenza A virus infection. Vaccine 19:S32-S37. [DOI] [PubMed] [Google Scholar]
- 30.Kawaoka, Y., A. Nestorowicz, D. J. Alexander, and R. G. Webster. 1987. Molecular analyses of the hemagglutinin genes of H5 influenza viruses: origin of a virulent turkey strain. Virology 158:218-227. [DOI] [PubMed] [Google Scholar]
- 31.Klenk, H.-D., and W. Garten. 1994. Activation cleavage of viral spike proteins by host proteases, p. 241-280. In E. Wimmer (ed.), Cellular receptors for animal viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Kobasa, D., A. Takada, K. Shinya, M. Hatta, P. Halfmann, S. Theriault, H. Suzuki, H. Nishimura, K. Mitamura, N. Sugaya, T. Usui, T. Murata, Y. Maeda, S. Watanabe, M. Suresh, T. Suzuki, Y. Suzuki, H. Feldmann, and Y. Kawaoka. 2004. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431:703-707. [DOI] [PubMed] [Google Scholar]
- 33.Kurokawa, M., M. Imakita, C. A. Kumeda, and K. Shiraki. 1996. Cascade of fever production in mice infected with influenza virus. J. Med. Virol. 50:152-158. [DOI] [PubMed] [Google Scholar]
- 34.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 725-769. In D. M. Knipe and P. M. Howley (ed.), Fundamental virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 35.Liu, T., and Z. Ye. 2005. Attenuating mutations of the matrix gene of influenza A/WSN/33 virus. J. Virol. 79:1918-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, T., and Z. Ye. 2002. Restriction of viral replication by mutation of the influenza virus matrix protein. J. Virol. 76:13055-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from human. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCullers, J. A., E. Hoffmann, V. C. Huber, and A. D. Nickerson. 2005. A single amino acid change in the C-terminal domain of the matrix protein M1 of influenza B virus confers mouse adaptation and virulence. Virology 336:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, J. L., and E. M. Anders. 2003. Virus-cell interactions in the induction of type 1 interferon by influenza virus in mouse spleen cells. J. Gen. Virol. 84:193-202. [DOI] [PubMed] [Google Scholar]
- 40.Nakajima, S., and A. Sugiura. 1980. Neurovirulence of influenza virus in mice. II. Mechanism of virulence as studied in a neuroblastoma cell line. Virology 101:450-457. [DOI] [PubMed] [Google Scholar]
- 41.Nasser, E. H., A. K. Judd, A. Sanchez, D. Anastasiou, and D. J. Bucher. 1996. Antiviral activity of influenza virus M1 zinc finger peptides. J. Virol. 70:8639-8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nayak, D. P., and E. K.-W. Hui. 2002. Assembly and morphogenesis of influenza viruses. Recent Res. Dev. Virol. 4:35-54. [Google Scholar]
- 43.Nayak, D. P., E. K.-W. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106:147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novak, M., Z. Moldoveanu, D. P. Schafer, J. Mestecky, and R. W. Compans. 1993. Murine model for evaluation of protective immunity to influenza virus. Vaccine 11:55-60. [DOI] [PubMed] [Google Scholar]
- 45.Okada, A., T. Miura, and H. Takeuchi. 2003. Zinc- and pH-dependent conformational transition in a putative interdomain linker region of the influenza virus matrix protein M1. Biochemistry 42:1978-1984. [DOI] [PubMed] [Google Scholar]
- 46.Patterson, S., J. Gross, and J. S. Oxford. 1988. The intracellular distribution of influenza virus matrix protein and nucleoprotein in infected cells and their relationship to hemagglutinin in the plasma membrane. J. Gen. Virol. 69:1859-1872. [DOI] [PubMed] [Google Scholar]
- 47.Peper, R. L., and H. Van Campen. 1995. Tumor necrosis factor as a mediator of inflammation in influenza A viral pneumonia. Microb. Pathog. 19:175-183. [DOI] [PubMed] [Google Scholar]
- 48.Pleschka, S., T. Wolff, C. Ehrhardt, G. Hobom, O. Planz, U. R. Rapp, and S. Ludwig. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3:301-305. [DOI] [PubMed] [Google Scholar]
- 49.Reinhardt, J., and T. Wolff. 2000. The influenza A virus M1 protein interacts with the cellular receptor of activated C kinase (RACK) 1 and can be phosphorylated by protein kinase C. Vet. Microbiol. 74:87-100. [DOI] [PubMed] [Google Scholar]
- 50.Ress, P. J., and N. J. Dimmock. 1981. Electrophoretic separation of influenza virus ribonucleoproteins. J. Gen. Virol. 53:125-132. [DOI] [PubMed] [Google Scholar]
- 51.Ress, P. J., and N. J. Dimmock. 1982. Kinetics of synthesis of influenza virus ribonucleoprotein structures. J. Gen. Virol. 59:403-408. [DOI] [PubMed] [Google Scholar]
- 52.Ronni, T., T. Sareneva, J. Pirhonen, and I. Julkunen. 1995. Activation of IFN-α, IFN-γ, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J. Immunol. 154:2764-2774. [PubMed] [Google Scholar]
- 53.Ruigrok, R. W. H., and F. Baudin. 1995. Structure of influenza virus ribonucleoprotein particles. II. Purified RNA-free influenza virus ribonucleoprotein from structures that are indistinguishable from the intact influenza virus ribonucleoprotein particles. J. Gen. Virol. 76:1009-1114. [DOI] [PubMed] [Google Scholar]
- 54.Sareneva, T., S. Matikainen, M. Kurimoto, and I. Julkunen. 1998. Influenza A virus-induced IFN-α/β and IL-18 synergistically enhance IFN-γ gene expression in human T cells. J. Immunol. 160:6032-6038. [PubMed] [Google Scholar]
- 55.Schlesinger, R. W., P. J. Husak, G. L. Bradshaw, and P. P. Panayotov. 1998. Mechanisms involved in natural and experimental neuropathogenicity of influenza viruses: evidence and speculation. Adv. Virus Res. 50:289-379. [DOI] [PubMed] [Google Scholar]
- 56.Schmitt, A. P., and R. A. Lamb. 2005. Influenza virus assembly and budding at the viral budzone. Adv. Virus Res. 64:383-416. [DOI] [PubMed] [Google Scholar]
- 57.Schulman, J. L. 1968. The use of an animal model to study transmission of influenza virus infection. Am. J. Public Health Nations Health 58:2092-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sidwell, R. W., and D. F. Smee. 2001. In vitro and in vivo assay systems for study of influenza virus inhibitors. Antivir. Res. 48:1-16. [DOI] [PubMed] [Google Scholar]
- 59.Sidwell, R. W., D. F. Smee, J. H. Huffman, D. L. Barnard, K. W. Bailey, J. D. Morrey, K. Bush, and Y. S. Babu. 2001. The in vivo influenza-inhibitory effects of the cyclopentane neuraminidase inhibitor RWJ-270201. Antimicrob. Agents Chemother. 45:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smeenk, C. A., and E. G. Brown. 1994. The influenza virus variant A/FM/1/47-MA possesses single amino acid replacements in the hemagglutinin, controlling virulence, and in the matrix protein, controlling virulence as well as growth. J. Virol. 68:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smeenk, C. A., K. E. Wright, B. F. Burns, A. J. Thaker, and E. G. Brown. 1996. Mutations in the hemagglutinin and matrix genes of a virulent influenza virus variant, A/FM/1/47-MA, control different stages in pathogenesis. Virus Res. 44:79-95. [DOI] [PubMed] [Google Scholar]
- 62.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1-20. [DOI] [PubMed] [Google Scholar]
- 63.Steinhauer, D. A., and J. J. Skehel. 2002. Genetics of influenza viruses. Annu. Rev. Genet. 36:305-332. [DOI] [PubMed] [Google Scholar]
- 64.Subbarao, E. K., M. Perkins, J. J. Treanor, and B. R. Murphy. 1992. The attenuation phenotype conferred by the M gene of the influenza A/Ann Arbor/6/60 cold-adapted virus (H2N2) on the A/Korea/82 (H3N2) reassortant virus results from a gene constellation effect. Virus Res. 25:37-50. [DOI] [PubMed] [Google Scholar]
- 65.Sugiura, A., and M. Ueda. 1980. Neurovirulence of influenza virus in mice. I. Neurovirulence of recombinants between virulent and avirulent virus strains. Virology. 101:440-449. [DOI] [PubMed] [Google Scholar]
- 66.Tashiro, M., and R. Rott. 1996. The role of proteolytic cleavage of viral glycoproteins in the pathogenesis of influenza virus infections. Semin. Virol. 7:237-243. [Google Scholar]
- 67.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solórzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. García-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 68.Ulmer, J. B., J. J. Donnelly, S. E. Parker, G. H. Rhodes, P. L. Felgner, V. J. Dwarki, S. H. Gromkowski, R. R. Deck, C. M. DeWitt, A. Friedman, L. A. Hawe, K. R. Leander, D. Martinez, H. C. Perry, J. W. Shiver, D. L. Montgomery, and M. A. Liu. 1993. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259:1745-1749. [DOI] [PubMed] [Google Scholar]
- 69.Vacheron, F., A. Rudent, S. Perin, C. Labarre, A. M. Quero, and M. Guenounou. 1990. Production of interleukin 1 and tumor necrosis factor activities in bronchoalveolar washings following infection of mice by influenza virus. J. Gen. Virol. 71:477-479. [DOI] [PubMed] [Google Scholar]
- 70.Van Reeth, K. 2000. Cytokines in the pathogenesis of influenza. Vet. Microbiol. 74:109-116. [DOI] [PubMed] [Google Scholar]
- 71.Wakefield, L., and G. G. Brownlee. 1989. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 17:8569-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, A. 1997. Virulence of influenza A virus for mouse lung. Virus Genes 14:187-194. [DOI] [PubMed] [Google Scholar]
- 73.Welsh, R. M., and G. C. Sen. 1997. Nonspecific host responses to viral infections, p. 109-141. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa.
- 74.Yasuda, J., D. J. Bucher, and A. Ishihama. 1994. Growth control of influenza A virus by M1 protein: analysis of transfectant viruses carrying the chimeric M gene. J. Virol. 68:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye, Z., T. Liu, D. P. Offringa, J. McInnis, and R. A. Levandowski. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 73:7467-7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhimov, O. P., and H.-D. Klenk. 1997. Histones as a target for influenza virus matrix protein M1. Virology 235:302-310. [DOI] [PubMed] [Google Scholar]