Abstract
Seventy-two full genomes corresponding to nine mammalian (67 strains) and two avian (5 strains) polyomavirus species were analyzed using maximum likelihood and Bayesian methods of phylogenetic inference. Our fully resolved and well-supported (bootstrap proportions > 90%; posterior probabilities = 1.0) trees separate the bird polyomaviruses (avian polyomavirus and goose hemorrhagic polyomavirus) from the mammalian polyomaviruses, which supports the idea of spitting the genus into two subgenera. Such a split is also consistent with the different viral life strategies of each group. Simian (simian virus 40, simian agent 12 [Sa12], and lymphotropic polyomavirus) and rodent (hamster polyomavirus, mouse polyomavirus, and murine pneumotropic polyomavirus [MPtV]) polyomaviruses did not form monophyletic groups. Using our best hypothesis of polyomavirus evolutionary relationships and established host phylogenies, we performed a cophylogenetic reconciliation analysis of codivergence. Our analyses generated six optimal cophylogenetic scenarios of coevolution, including 12 codivergence events (P < 0.01), suggesting that Polyomaviridae coevolved with their avian and mammal hosts. As individual lineages, our analyses showed evidence of host switching in four terminal branches leading to MPtV, bovine polyomavirus, Sa12, and BK virus, suggesting a combination of vertical and horizontal transfer in the evolutionary history of the polyomaviruses.
Members of the family Polyomaviridae (polyomaviruses) are small, nonenveloped, double-stranded DNA viruses, which are widely distributed among vertebrates. They share a common genome structure consisting of a 4.8- to 5.5-kbp circular double-stranded DNA that encodes five main proteins: two multifunctional regulatory proteins referred to as the large and small T antigens and three structural proteins (VP1, VP2, and VP3), which form the icosahedral viral capsid. At the initiation of this study, at least 16 polyomavirus species were described, but a full-length genomic sequence was available for only 11 (Table 1). Those were nine mammalian and two avian polyomaviruses. Among the mammalian polyomaviruses, there were three full-length genomes available from simian hosts. Lymphotropic polyomavirus (LPV) or African Green monkey polyomavirus was first isolated from a B-lymphoblastoid cell line derived from an African green monkey (43, 75). Serological surveys have revealed that many monkeys and apes, as well as humans, show evidence of infection by viruses antigenically related to LPV (67). However, the natural host of the original isolate and the pathogenicity of LPV are unknown at present. Simian virus 40 (SV40) was identified as a contaminant of monkey kidney cultures used to prepare the first poliovirus vaccines during the late 1950s (54, 65). The natural host of SV40 is the rhesus macaque (56). Simian agent 12 (Sa12, baboon polyomavirus type 1) was first isolated from kidney cells of a vervet monkey (69), but the chacma baboon is considered the natural host because only this species has shown high titers of Sa12 antibodies (3). We recently sequenced the full-length genome of this virus and included these data in the present study. Subsequent to our effort, the complete genome of a slightly different variant of Sa12 was reported by Cantalupo and colleagues (5). Our sequence and the published genome only differ in the viral noncoding regions. An antigenically distinct polyomavirus, referred to as the baboon polyomavirus type 2, was isolated from cultures of baboon kidney cells (17). A complete genome sequence is not available for this virus, so it will not be included in this study. Several polyomaviruses with available full-length genomic sequences have been obtained from rodents. Hamster polyomavirus (HaPV) was originally described as a virus associated with skin epitheliomas of laboratory colony-bred Syrian hamsters (18). A search for the virus reservoir in weanling hamsters demonstrated virus in the spleen and thymus but, distinct from other mammalian polyomaviruses, not in the kidney (48). Mouse polyomavirus (MPV) was first identified as the etiological agent of a wide range of solid tumors in newborn mice injected with cell extracts of leukemic tissues (22, 59). The virus has been studied extensively as a model agent of cell transformation and virus-host interactions leading to the development of tumors (1). The Kilham strain of mouse polyomavirus or murine pneumotropic polyomavirus (MPtV) is a second murine member of the polyomavirus family (35). MPtV, in contrast to other mammalian polyomaviruses, can cause severe disease. Infection of newborn mice causes interstitial pneumonia with high mortality (19). Unique to this species, the virus replicates in vascular endothelial cells of the lung, liver, and spleen (21). However, in immunocompetent mice, MPtV leads to a persistent and unapparent infection (19) and the virus localizes mainly to the kidney following primary infection (21), as seen in other mammalian polyomavirus infections. Full-length genomic sequence data are available from one polyomavirus species infecting bovids (49). Bovine polyomavirus is a frequent contaminant of commercial bovine serum (50); it has no known clinical significance for bovids. Two mammalian polyomaviruses are known to infect humans. In 1971, JC virus (JCV) was isolated from the brain of a patient with progressive multifocal leukoencephalopathy (40), and in the same year, BK virus (BKV) was cultivated from the urine of a renal transplant recipient (16). The viruses are ubiquitous, with BKV seroprevalence reaching nearly 100% within the first 5 years of life and JCV seroprevalence approaching 70% by early adulthood (62). The route of transmission is uncertain but most likely either fecal, oral, or respiratory (for a review, see reference 55). Primary infections with JCV and BKV are generally asymptomatic. The viruses persist indefinitely in the infected individual, primarily but perhaps not exclusively in the kidney and are reactivated in times of immunologic impairment. Disease occurs largely in immunocompromised individuals. Progressive multifocal leukoencephalopathy is a rare demyelinating fatal disorder of the central nervous system caused by JCV, which was a frequent complication of human immunodeficiency virus infection prior to the advent of effective antiviral therapy. BKV infection has been associated with hemorrhagic cystitis, ureteral stenosis, nephritis/nephropathy, and less commonly, pneumonitis. Mammalian polyomaviruses for which none or only partial sequence data are available include rabbit polyomavirus (25), rat polyomavirus (72), baboon polyomavirus type 2 (17), cynomolgus polyomavirus (70), and the recently described chimpanzee polyomavirus (32). Those are not included in this study. Avian polyomaviruses (APV) were first identified in psittacine species (2) and latter in a wide range of bird species (33, 61). The prototype APV is the budgerigar fledging disease virus, which is responsible for a fulminating disease in neonate budgerigars (37). A closely related (34) and also lethal (23) polyomavirus is the goose hemorrhagic polyomavirus (GHPV), which causes the hemorrhagic nephritis enteritis of geese. The biology of the avian polyomaviruses appears to be markedly different from that of the mammalian polyomaviruses. APV exhibits a broad host range compared to the highly specific host range of most mammalian polyomaviruses. Compared to asymptomatic infection, which is characteristic of mammalian polyomaviruses in immunocompetent hosts, the avian polyomaviruses are frequently associated with acute fatal disease. However, because surveys of polyomavirus infection in avian species have not been conducted, the existence of asymptomatic infection cannot be excluded.
TABLE 1.
Biological characteristics of the polyomaviruses included in this study
| Virus name | Abbreviation | Natural host | Species specificity | Tissue specificity | Pathogenicity in normal host |
|---|---|---|---|---|---|
| Lymphotropic polyomavirus or African Green monkey polyomavirus | LPV | Human and nonhuman primates | High | B lymphocyte | ?Inapparent/mild |
| Simian virus 40 | SV40 | Rhesus macaque | High | Kidney | Inapparent/mild |
| SA12 or Baboon polyomavirus type 1 | Sa12 | Baboon | High | Kidney | Inapparent/mild |
| Hamster papovavirus | HaPV | Hamster | High | Kidney/skin | Inapparent/mild or skin epithelioma |
| Mouse polyomavirus | MPV | Mouse | High | Kidney | Inapparent/mild |
| Murine pneumotropic virus or Kilham mouse polyomavirus | MPtV | Mouse | High | Lung/kidney | Inapparent/mild or pneumonitis |
| Bovine polyomavirus | BPV | High | Kidney | Inapparent/mild | |
| JC virus | JCV | Human | High | Kidney | Inapparent/mild |
| BK virus | BKV | Human | High | Kidney | Inapparent/mild |
| Avian polyomavirus | APV | Birds (no goose) | Low | Kidney/small intestine | Lethal/multiorgan involvement |
| Goose hemorrhagic polyomavirus | GHPV | Goose | High | Kidney | Lethal nephritis/enteritis |
Mammalian polyomaviruses adhere to a persistent life strategy (71). They cause unapparent or mild primary infections in young animals, followed by lifelong, nonpathogenic, persistent maintenance of nonintegrated, nondefective, episomal viral DNA (10). They replicate preferentially in the kidneys of a single species or a group of closely related species (4), so they are often described as having cospeciated with their hosts (52, 53, 71). APV, on the contrary, adheres to an acute viral life strategy (71) because it infects a broad array of bird species and is pathogenic (60). GHPV, MPtV, and HaPV do not completely fit these two patterns (Table 1). GHPV, like APV, is the causative agent of a fatal disease in the natural host, but it appears to have a restricted host range like the mammalian polyomaviruses, as it is only known to propagate in goose kidney cells (23). MPtV and HaPV exhibit a unique tissue tropism and the ability to occasionally cause disease. MPtV is unique in its ability to replicate in vascular endothelial cells (20) and cause a fatal lung infection (19). HaPV has the ability to infect hair follicle keratinocytes and cause skin epitheliomas (48), yet each virus exhibits restricted host specificity.
Present assumptions of polyomavirus-host cospeciation are largely the result of the seminal phylogenetic study carried out by Shadan and Villareal (52), although similar observations have been reported previously (58). Their phylogenetic analysis of seven polyomaviruses and their natural hosts (52) showed visual congruence between five mammalian polyomaviruses (HaPV, MPV, BKV, JCV, and SV40) and their host trees, which was taken as an indication of virus-host cospeciation. However, no apparent relationship to the host phylogeny was observed when APV and MPtV were introduced into the analysis, which was attributed to either dislinkage of the viruses from host evolution or phylogenetic independence of the viruses related to a different tissue specificity (52). Another factor that could also cause the observed phylogenetic incongruence is phylogenetic uncertainty, although this problem was not considered at that point. Subsequent studies and reviews of this topic (53, 71) reaffirm the general view that mammalian polyomaviruses, with the exception of MPtV, cospeciated with their hosts but APV did not and used this assessment to argue about different polyomavirus life strategies. Because of failure to account for phylogenetic uncertainty coupled with lack of a formal statistical hypothesis testing framework for codivergence (for an example, see reference 28), we believe the hypothesis of codivergence between polyomaviruses and their natural vertebrate hosts remains untested.
In this study, we have compared and analyzed all of the 72 complete polyomavirus genomes available (as of October 2005) corresponding to nine mammalian and two avian species using phylogenetic methods that take into account phylogenetic uncertainty. Then, within a statistical framework, we used our best hypothesis of polyomavirus evolutionary relationships and established phylogenies of their hosts to test polyomaviruses-host codivergence using cophylogenetic reconciliation analysis. Our results show how different viral life strategies can be accommodated within a robust hypothesis of polyomavirus-host coevolution.
MATERIALS AND METHODS
Sequence data, alignment, and model selection.
Our data set consisted of one new Sa12 complete genome (accession no. DQ435829) and 71 polyomavirus complete genomes from GenBank (Fig. 1). The coding gene sequences were parsed out using CDSparser v1.2 (http://inbio.byu.edu/faculty/dam83/CDSParser/group.asp). The main five genes of the virus genome (VP1, VP2, VP3, large T antigen, and small T antigen) were used for the phylogenetic analyses. Each gene was aligned first at the amino acid level using MUSCLE 3.3 (11), and then the amino acid alignments were converted into nucleotide alignments. AlignmentHelper 1.0 (http://inbio.byu.edu/faculty/dam83/cdm) was used to translate the nucleotide FASTA files to the amino acid level first and then to create nucleotide alignments based on the amino acid alignments produced by MUSCLE 3.3. Gblocks 0.91b (6) was used on each gene nucleotide alignment to assess character uncertainty (i.e., questionable homology statements of aligned ambiguous regions). Ambiguous characters were deleted, rendering a concatenated aligned data set of 5,310 characters. The following Gblocks parameter values were used: p1 (minimum number of sequences for a conserved position), 37; p2 (minimum number of sequences for a flank position), 37; p3 (maximum number of contiguous nonconserved positions), 21; p4 (minimum length of a block), 9; p5 (allowed gap positions), half, per individual gene nucleotide alignment. The best model of DNA substitution based on the Akaike criterion implemented in Modeltest 3.06 (44) was chosen for each resulting Gblock from each gene and all of them concatenated. The following evolutionary models were selected: HKY (26) plus I (invariable sites) plus Γ (gamma distribution) for VP1, TVM (68) plus I plus Γ for VP2 and VP3, GTR (68) plus I plus Γ for large T antigen, small T antigen, and all the concatenated genes.
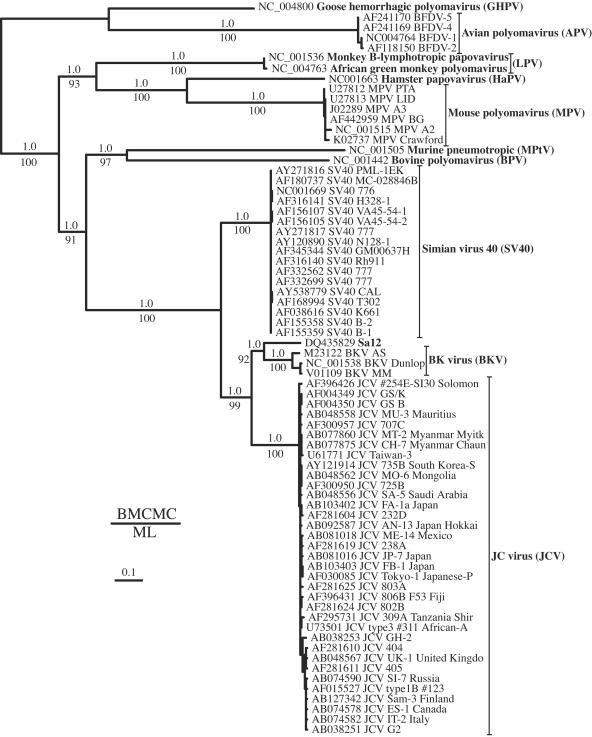
FIG. 1.
Midpoint-rooted Bayesian estimation of polyomavirus phylogeny. The same topology was obtained from a maximum likelihood analysis. The branch lengths are drawn proportional to the amount of change along that branch based on the mean branch length distribution (scale shown).
Phylogenetic analyses.
Polyomavirus evolutionary relationships were estimated using maximum likelihood (ML) and Bayesian coupled with Markov chain Monte Carlo (BMCMC) methods of phylogenetic inference. ML searches (13) of the concatenated gene data set were performed in PAUP* v4.0b10 (66) under the GTR plus I plus Γ model with the following model parameters: π (base frequency)A, 0.317; πC, 0.198; πG, 0.223; πT, 0.262; r (substitution rate)CT, 1.793; rCG, 3.095; rAT, 1.517; rAG, 1.918; rAC, 3.889; I, 0.092; α (shape parameter of the gamma distribution), 1.394. We conducted ML heuristic searches with 10 random addition replicates and tree bisection and reconnection branch swapping. Uncertainty in the resulting ML relationships was assessed using the nonparametric bootstrap procedure (12) with 100 bootstrap replicates, tree bisection and reconnection branch swapping, one random addition replicate, and starting trees obtained by neighbor joining (using model-corrected distances). BMCMC searches (30) of the unlinked five virus genes were performed in MrBayes v3.0 (47) under the models above. This approach offers several practical advantages over more traditional hill climbing heuristic searches, including simultaneous assessment of both tree and clade support and the ability to accommodate phylogeny and model uncertainty (28). Model parameters were treated as unknown variables with uniform priors and were estimated as part of the analysis. We ran four Markov chains (4.0 × 106 cycles) simultaneously, which were started from random trees and sampled every 1,000th cycle. To check that stationarity had been reached, we monitored the fluctuating value of the likelihood and all the phylogenetic parameters graphically using Tracer v1.2 (46) and repeated each simulation four times starting from different random trees. All sample points prior to reaching stationarity were discarded as “burn in.” The posterior probabilities for individual clades obtained from separate analyses were compared for congruence (27, 29, 39) and then combined and summarized on a majority rule consensus tree (29).
Alternative phylogenetic hypotheses were compared using the Shimodaira and Hasegawa (S-H) test (57). Ten thousand replicates were performed for each S-H topology test, resampling the partial likelihoods for each site (RELL model) using PAUP*. All trees were rooted using midpoint rooting.
Cophylogenetic reconciliation analysis.
A tanglegram (41) composed of a pruned version of our best phylogenetic hypothesis of polyomavirus relationships, including all of the main polyomavirus lineages and published phylogenetic hypotheses of their hosts, was tested using cophylogenetic reconciliation analysis (Fig. 2). The relationships among the main mammal hosts were depicted from previously published BMCMC and ML trees (38) based on a 16.4-kbp data set. Extensive morphological (for examples, see references 14, 15) and molecular (for examples, see references 24, 42, 45) evidence support the relationships among the four simian polyomavirus hosts, as depicted in Fig. 2 (see the TOL website [http://tolweb.org/tree/phylogeny.html] for further information and references).
FIG. 2.
Tanglegram of polyomaviruses and their hosts with associated taxa connected by dotted lines.
Cophylogenetic mapping.
Reconciliation analysis of the associations established in Fig. 2 was explored through cophylogeny mapping (31) using TreeMap v.2.02 (7). Accordingly, event costs for codivergence, duplication, loss, and switching were set to 0, 1, 1, and 1, respectively, under the assumption that congruence via codivergence is the null hypothesis. Bounds for the reconciliation analysis were set at the maximum possible: 77 maximum noncodivergence events, a minimum of 0 lineage codivergences, 20 lineage duplications, 10 host switches, 77 lineage losses, and a maximum parasite load of 11. The significance of each reconciliation (jungle) was evaluated in TreeMap via Markov randomization of the virus tree 100 times, bounded by the specific properties of each solution (i.e., number of noncodivergence events, codivergence events, lineage duplications, host switches, and parasite load).
RESULTS
Phylogenetic relationships.
ML and BMCMC analyses that account for phylogenetic uncertainty resulted in the same backbone topology (Fig. 1). Evolutionary relationships among different polyomaviruses were strongly supported by high bootstrap (>90%) and posterior probability (pP) (1.0) values. All of the trees showed a major grouping of each of the well-characterized polyomaviruses. BKV and Sa12 formed a clade sister to the JCV and, altogether, to the SV40 clade. MPV and HaPV were sister taxa to a clade of virus infecting monkeys (LPV). More distantly related are the GHPV and APV. Interestingly, the MPtV is clustered with the bovine polyomavirus (BPV), and both are sister taxa to the simian polyomaviruses, excluding the LPV. An alternative rodent monophyletic clade (MPtV, MPV, and HaPV) was significantly rejected by the S-H test (P < 0.001) and has a pP value of <0.001. Similarly, a simian polyomavirus monophyletic cluster (BKV, Sa12, JCV, SV40, and LPV) was also rejected (P and pP < 0.001). As for the JCV evolutionary relationships, our trees agree with previous phylogenetic hypotheses (63, 64, 73, 74), although our trees showed less resolution among and within types and subtypes and lower clade support. This is not surprising considering that we applied an alignment strategy adequate for assessing deep taxonomic relationships; hence, it is too conservative (i.e., less informative) for resolving JCV relationships.
Cophylogenetic mapping and reconciliation analyses.
The source phylogenies for hosts and viruses and their associations are depicted in the tanglegram (Fig. 2). Six optimal reconstructions showing a significant degree of congruence (P < 0.01) with their host phylogenies were recovered (Table 2). All of them involved 12 codivergence events and different combinations of host switches and losses per node (Table 2). It is ill advised to discriminate between solutions using the number of events because noncodivergence events are not necessarily comparable and not all the events are observable (31); hence, a consensus diagram showing the origin of associated lineages across these optimal reconstructions is presented instead (Fig. 3). This diagram summarizes the number of times each lineage arose from each kind of event. Out of 20 lineages, 9 lineages arose from codivergent events and 11 from noncodivergent events, 4 of which were dominated by host switches, 4 by duplications, and 3 by losses.
TABLE 2.
Reconciliation analysisa
| OR | No. of:
|
z | ||||
|---|---|---|---|---|---|---|
| CE | HS | D | L | NCE | ||
| A | 12 | 2 | 8 | 6 | 16 | 16 |
| B | 12 | 2 | 8 | 6 | 16 | 16 |
| C | 12 | 4 | 8 | 2 | 14 | 14 |
| D | 12 | 2 | 8 | 6 | 16 | 16 |
| E | 12 | 1 | 8 | 9 | 18 | 18 |
| F | 12 | 0 | 8 | 13 | 21 | 21 |
OR, optimal reconstruction (P < 0.01); CE, codivergences; HS, host switches; D, duplications; L = losses; NCE, noncodivergence events; z, total event cost (based on assigned event costs).
FIG. 3.
Consensus diagram showing the origin of associate lineages across six optimal reconstructions resulting from reconciliation analysis. Evolutionary events are separated by periods and presented as follows: codivergences, host switches, duplications, and losses.
DISCUSSION
Phylogenetic implications.
Our phylogenetic trees based on 72 taxa and 5 genes concatenated (ML) and unlinked (BMCMC) did not agree with previous phylogenetic hypotheses based on the analysis of fewer genes and taxa (23). Our phylogenetic estimate resulted in two reciprocally monophyletic clades separating the polyomaviruses of birds (APV and GHPV) from the mammalian polyomaviruses, which supports the idea of placing these two clades into distinct subgenera (34, 61). This phylogenetic association is also supported by the unique biological characteristic that separates APV and GHPV from the other polyomaviruses: both are pathogens with a lethal progression (acute life strategy). Rodent and simian viruses do not seem to form monophyletic assemblages, as previously reported (9, 34). Moreover, all of the alternative topologies grouping these two polyomavirus groups together were significantly worse than our best estimate of phylogeny (Fig. 1). The different evolutionary paths followed by the HaPV, MPV, and MPtV lineages could be driven by their different tissue preferences and/or pathogenicity (Table 1). All simian polyomaviruses but LPV form a robust clade separated by a long branch from their immediate mammal relatives, which fall in an intermediate position in the tree, as indicated by the midpoint rooting. This addresses the fact that, although suspected to be simian, the actual natural host of the LPV is unknown and leaves room for a nonsimian origin of the LPV. If the origin were proven to be murine, host and virus evolutionary paths would be more congruent than reported here.
Codivergence analysis.
Previous studies (for example, see reference 52) based on a limited number of taxa and genes did not phylogenetically separate APV and MPtV from their mammalian relatives and concluded that only HaPV, MPV, BKV, JCV, and SV40 cospeciated with their hosts (53, 71). Our more extensive and sophisticated phylogenetic and cophylogenetic analyses showed within a statistical framework that polyomaviruses, as a family, codiverged with their vertebrate hosts (Table 2 and Fig. 3). A basal spilt on the polyomavirus tree separates both avian and mammalian viruses (Fig. 1). Congruence near the base of the reconciled tree (Fig. 2) is an effective indicator that codivergence is the characteristic dynamic, since historically, the signal has not been obliterated by subsequent evolutionary events (31). Such dynamics have been identified in other DNA viruses such as Herpesvirus (36). Acute (bird polyomaviruses) and persistent (mammalian polyomavirues) viral life strategies can now be naturally accommodated within this cophylogentic framework.
Mammalian polyomaviruses, as most DNA viruses, adhere to a persistent life strategy (71). Among other biological characteristics (Table 1) they are highly species specific and cospeciate with their hosts. Five lineages in the consensus tree (Fig. 3) leading to MPV, HaPV, MPtV plus BPV, and JCV plus Sa12 plus BKV clades support this view; since they are mostly dominated by codivergence events. However, all the other branches in the tree are dominated by noncodivergence events, and four of those, leading to MPtV, BPV, Sa12, and BKV, are dominated by host switch events. An equal number of switches were observed between MPtV and BPV (2 each way), but four switches were observed from BKV to Sa12 (baboon) and only two were observed from Sa12 to the human BKV and JCV (one each way). Host switching has never been demonstrated in polyomaviruses, so it is assumed that codivergence is the norm. Our analyses suggest that host switching could be more common than expected, at least in recent times. Hantavirus of mice, Spumavirus of humans (31), and primate lentiviruses (8) indicate that substantial biological boundaries can be overcome under certain circumstances. Nevertheless, our cospeciation analysis does not take into account uncertainty in host-virus associations. Mammalian polyomaviruses were initially isolated from laboratory animals or tissue culture, and little is known about infection in the natural host. For example, BPV was recovered from tissue cultures and is believed to be a contaminant with the calf serum, but no study we are aware of reported its natural host. Some doubt is cast on the bovid origin of BPV by the lack of correlation between detection of viral DNA by PCR and the presence of virus-specific antibodies in serum (51). Similarly, the natural host of the LPV is not known for certain and serological studies have not been able to address this uncertainty. Without such information, it is difficult to judge whether an apparent host-switching is real or the result of isolation from or replication in a transient host. It is thus possible that these apparent cases of host switching might really be examples of misidentification of naturally codiverging hosts. Our phylogenetic analyses may then provide some insight into the origin of these viruses; if, as suggested by our trees, these natural hosts were to be murine, a better fitted coevolutionary pattern would be observed. As for the human-baboon polyomaviruses, all evidence indicates that these are their natural hosts. Hence, if we are to accept a model of codivergence for mammalian polyomaviruses these latter presumptive noncodivergence events will require future mechanistic investigation.
Acknowledgments
We thank the Brigham Young University Office of Research and Creative Activities and the Brigham Young University Cancer Research Center for financial support for this study (R.G.C., D.A.M., and K.A.C.). This work was also supported by NIH R01 AI50217 (R.P.V. and K.A.C.), GM66276 (K.A.C.), and the PhRMA Foundation.
REFERENCES
- 1.Benjamin, T. L. 2001. Polyoma virus: old findings and new challenges. Virology 289:167-173. [DOI] [PubMed] [Google Scholar]
- 2.Bozeman, L. H., R. B. Davis, D. Gaudry, P. D. Lukert, O. J. Fletcher, and M. J. Dykstra. 1981. Characterization of a papovavirus isolated from fledgling budgerigars. Avian Dis. 25:972-980. [PubMed] [Google Scholar]
- 3.Braun, L., S. S. Kalter, L. A. Yakovleva, V. R. Kaschula, and K. V. Shah. 1980. Neutralizing antibodies to simian papovavirus SA12 in Old World primates in laboratory colonies: high prevalence in baboons. J. Med. Primatol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, B. A., and L. P. Villarreal. 1985. Host species specificity of polyomavirus DNA replication is not altered by simian virus 40 72-base-pair repeats. Mol. Cell. Biol. 5:1534-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantalupo, P., A. Doering, C. S. Sullivan, A. Pal, K. W. Peden, A. M. Lewis, and J. M. Pipas. 2005. Complete nucleotide sequence of polyomavirus SA12. J. Virol. 79:13094-13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 7.Charleston, M. A., and R. D. M. Page. 2002. TREEMAP v2.0, application for Apple Macintosh.
- 8.Charleston, M. A., and D. L. Robertson. 2002. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst. Biol. 51:528-535. [DOI] [PubMed] [Google Scholar]
- 9.Crandall, K., M. Perez-Losada, R. G. Christensen, D. A. McClellan, and R. P. Viscidi. 2005. Phylogenomics and molecular evolution of polyomaviruses, p. 1-14. In N. Ahsan (ed.), Polyomaviruses and human diseases. Landes Biosciences, Georgetown, Tex. [DOI] [PubMed]
- 10.Dubensky, T. W., and L. P. Villarreal. 1984. The primary site of replication alters the eventual site of persistent infection by polyomavirus in mice. J. Virol. 50:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 14.Fleagle, J. G., and W. S. McGraw. 2002. Skeletal and dental morphology of African papionins: unmasking a cryptic clade. J. Hum. Evol. 42:267-292. [DOI] [PubMed] [Google Scholar]
- 15.Fleagle, J. G., and W. S. McGraw. 1999. Skeletal and dental morphology supports diphyletic origin of baboons and mandrills. Proc. Natl. Acad. Sci. USA 96:1157-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner, S. D., A. M. Field, D. V. Coleman, and B. Hulme. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253-1257. [DOI] [PubMed] [Google Scholar]
- 17.Gardner, S. D., W. A. Knowles, J. F. Hand, and A. A. Porter. 1989. Characterization of a new polyomavirus (polyomavirus papionis-2) isolated from baboon kidney cell cultures. Arch. Virol. 105:223-233. [DOI] [PubMed] [Google Scholar]
- 18.Graffi, A., T. Schramm, I. Graffi, D. Bierwolf, and E. Bender. 1968. Virus-associated skin tumors of the Syrian hamster: preliminary note. J. Natl. Cancer Inst. 40:867-873. [PubMed] [Google Scholar]
- 19.Greenlee, J. E. 1981. Effect of host age on experimental K virus infection in mice. Infect. Immun. 33:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenlee, J. E., S. H. Clawson, R. C. Phelps, and W. G. Stroop. 1994. Distribution of K-papovavirus in infected newborn mice. J. Comp. Pathol. 111:259-268. [DOI] [PubMed] [Google Scholar]
- 21.Greenlee, J. E., R. C. Phelps, and W. G. Stroop. 1991. The major site of murine K papovavirus persistence and reactivation is the renal tubular epithelium. Microb. Pathog. 11:237-247. [DOI] [PubMed] [Google Scholar]
- 22.Gross, L. 1953. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc. Soc. Exp. Biol. Med. 83:414-421. [DOI] [PubMed] [Google Scholar]
- 23.Guerin, J. L., J. Gelfi, L. Dubois, A. Vuillaume, C. Boucraut-Baralon, and J. L. Pingret. 2000. A novel polyomavirus (goose hemorrhagic polyomavirus) is the agent of hemorrhagic nephritis enteritis of geese. J. Virol. 74:4523-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, E. E., and T. R. Disotell. 1998. Nuclear gene trees and the phylogenetic relationships of the mangabeys (Primates: Papionini). Mol. Biol. Evol. 15:892-900. [DOI] [PubMed] [Google Scholar]
- 25.Hartley, J. W., and W. P. Rowe. 1964. New papovavirus contaminating shope papillomata. Science 143:258-260. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa, M., K. Kishino, and T. Yano. 1985. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 27.Huelsenbeck, J. P., and N. S. Imennov. 2002. Geographic origin of human mitochondrial DNA: accommodating phylogenetic uncertainty and model comparison. Syst. Biol. 51:155-165. [DOI] [PubMed] [Google Scholar]
- 28.Huelsenbeck, J. P., B. Larget, and M. E. Alfaro. 2004. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 21:1123-1133. [DOI] [PubMed] [Google Scholar]
- 29.Huelsenbeck, J. P., B. Larget, R. E. Miller, and F. Ronquist. 2002. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 51:673-688. [DOI] [PubMed] [Google Scholar]
- 30.Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollback. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310-2314. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, A. P., and M. A. Charleston. 2004. A cophylogenetic perspective of RNA-virus evolution. Mol. Biol. Evol. 21:45-57. [DOI] [PubMed] [Google Scholar]
- 32.Johne, R., D. Enderlein, H. Nieper, and H. Muller. 2005. Novel polyomavirus detected in the feces of a chimpanzee by nested broad-spectrum PCR. J. Virol. 79:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johne, R., and H. Muller. 1998. Avian polymavirus in wild birds: genome analysis of isolates from Falconiformes and Psittaciformes. Arch. Virol. 143:1501-1512. [DOI] [PubMed] [Google Scholar]
- 34.Johne, R., and H. Muller. 2003. The genome of goose hemorrhagic polyomavirus, a new member of the proposed subgenus Avipolyomavirus. Virology 308:291-302. [DOI] [PubMed] [Google Scholar]
- 35.Kilham, L., and H. W. Murphy. 1953. A pneumotropic virus isolated from C3H mice carrying the Bittner milk agent. Proc. Soc. Exp. Biol. Med. 82:133-137. [DOI] [PubMed] [Google Scholar]
- 36.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 37.Müller, H., and R. Nitschke. 1986. A polyoma-like virus associated with an acute disease of fledgling budgerigars (Melopsittacus undulatus). Med. Microbiol. Immunol. (Berlin) 175:1-13. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, W. J., E. Eizirik, S. J. O'Brien, O. Madsen, M. Scally, C. J. Douady, E. Teeling, O. A. Ryder, M. J. Stanhope, W. W. de Jong, and M. S. Springer. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348-2351. [DOI] [PubMed] [Google Scholar]
- 39.Nylander, J. A., F. Ronquist, J. P. Huelsenbeck, and J. L. Nieves-Aldrey. 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53:47-67. [DOI] [PubMed] [Google Scholar]
- 40.Padgett, B. L., D. L. Walker, G. M. ZuRhein, R. J. Eckroade, and B. H. Dessel. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257-1260. [DOI] [PubMed] [Google Scholar]
- 41.Page, R. D. M. 1995. Parallel phylogenies: reconstructing the history of host-parasite assemblages. Cladistics 10:155-173. [Google Scholar]
- 42.Page, S. L., and M. Goodman. 2001. Catarrhine phylogeny: noncoding DNA evidence for a diphyletic origin of the mangabeys and for a human-chimpanzee clade. Mol. Phylogenet. Evol. 18:14-25. [DOI] [PubMed] [Google Scholar]
- 43.Pawlita, M., A. Clad, and H. zur Hausen. 1985. Complete DNA sequence of lymphotropic papovavirus: prototype of a new species of the polyomavirus genus. Virology 143:196-211. [DOI] [PubMed] [Google Scholar]
- 44.Posada, D., and K. A. Crandall. 1998. Modeltest: Testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 45.Purvis, A. 1995. A composite estimate of primate phylogeny. Philos. Trans. R. Soc. Lond. B 348:405-421. [DOI] [PubMed] [Google Scholar]
- 46.Rambaut, A., and A. J. Drummond. 2003. Tracer: MCMC trace analysis tool, 1.2 ed. University of Oxford, Oxford, United Kingdom. [Online.] http://evolve.zoo.ox.ac.uk.
- 47.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 48.Scherneck, S., R. Ulrich, and J. Feunteun. 2001. The hamster polyomavirus—a brief review of recent knowledge. Virus Genes 22:93-101. [DOI] [PubMed] [Google Scholar]
- 49.Schuurman, R., C. Sol, and J. van der Noordaa. 1990. The complete nucleotide sequence of bovine polyomavirus. J. Gen. Virol. 71(Pt 8):1723-1735. [DOI] [PubMed] [Google Scholar]
- 50.Schuurman, R., B. van Steenis, and C. Sol. 1991. Bovine polyomavirus, a frequent contaminant of calf serum. Biologicals 19:265-270. [DOI] [PubMed] [Google Scholar]
- 51.Schuurman, R., B. van Steenis, A. van Strien, J. van der Noordaa, and C. Sol. 1991. Frequent detection of bovine polyomavirus in commercial batches of calf serum by using the polymerase chain reaction. J. Gen. Virol. 72(Pt 11):2739-2745. [DOI] [PubMed] [Google Scholar]
- 52.Shadan, F. F., and L. P. Villarreal. 1993. Coevolution of persistently infecting small DNA viruses and their hosts linked to host-interactive regulatory domains. Proc. Natl. Acad. Sci. USA 90:4117-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shadan, F. F., and L. P. Villarreal. 1995. The evolution of small DNA viruses of eukaryotes: past and present considerations. Virus Genes 11:239-257. [DOI] [PubMed] [Google Scholar]
- 54.Shah, K., and N. Nathanson. 1976. Human exposure to SV40: review and comment. Am. J. Epidemiol. 103:1-12. [DOI] [PubMed] [Google Scholar]
- 55.Shah, K. V. 1996. Polyomaviruses. Lippencott-Raven Publishers, Philadelphia, Pa.
- 56.Shah, K. V., and C. H. Southwick. 1965. Prevalence of antibodies to certain viruses in sera of free-living rhesus and of captive monkeys. Indian J. Med. Res. 53:488-500. [PubMed] [Google Scholar]
- 57.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 58.Soeda, E., T. Maruyama, J. R. Arrand, and B. E. Griffin. 1980. Host-dependent evolution of three papova viruses. Nature 285:165-167. [DOI] [PubMed] [Google Scholar]
- 59.Stewart, S. E., B. E. Eddy, and N. Borgese. 1958. Neoplasms in mice inoculated with a tumor agent carried in tissue culture. J. Natl. Cancer Inst. 20:1223-1243. [DOI] [PubMed] [Google Scholar]
- 60.Stoll, R., G. Hobom, and H. Muller. 1994. Host restriction in the productive cycle of avian polyomavirus budgerigar fledgling disease virus type 3 depends on a single amino acid change in the common region of structural proteins VP2/VP3. J Gen. Virol. 75(Pt 9):2261-2269. [DOI] [PubMed] [Google Scholar]
- 61.Stoll, R., D. Luo, B. Kouwenhoven, G. Hobom, and H. Muller. 1993. Molecular and biological characteristics of avian polyomaviruses: isolates from different species of birds indicate that avian polyomaviruses form a distinct subgenus within the polyomavirus genus. J Gen. Virol. 74(Pt 2):229-237. [DOI] [PubMed] [Google Scholar]
- 62.Stolt, A., K. Sasnauskas, P. Koskela, M. Lehtinen, and J. Dillner. 2003. Seroepidemiology of the human polyomaviruses. J. Gen. Virol. 84:1499-1504. [DOI] [PubMed] [Google Scholar]
- 63.Sugimoto, C., M. Hasegawa, A. Kato, H. Y. Zheng, H. Ebihara, F. Taguchi, T. Kitamura, and Y. Yogo. 2002. Evolution of human polyomavirus JC: implications for the population history of humans. J. Mol. Evol. 54:285-297. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki, M., H. Y. Zheng, T. Takasaka, C. Sugimoto, T. Kitamura, E. Beutler, and Y. Yogo. 2002. Asian genotypes of JC virus in Japanese-Americans suggest familial transmission. J. Virol. 76:10074-10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sweet, B. H., and M. R. Hilleman. 1960. The vacuolating virus, S.V. 40. Proc. Soc. Exp. Biol. Med. 105:420-427. [DOI] [PubMed] [Google Scholar]
- 66.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0 beta 10 ed. Sinauer, Sunderland, Mass.
- 67.Takemoto, K. K., and K. Segawa. 1983. A new monkey lymphotropic papovavirus: characterization of the virus and evidence of a related virus in humans. Prog. Clin. Biol. Res. 105:87-96. [PubMed] [Google Scholar]
- 68.Tavaré, S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences, p. 57-86. In R. M. Miura (ed.), Some mathematical questions in biology: DNA sequence analysis. American Mathematical Society, Providence, R.I.
- 69.Valis, J. D., N. Newell, M. Reissig, H. Malherbe, V. R. Kaschula, and K. V. Shah. 1977. Characterization of SA12 as a simian virus 40-related papovavirus of chacma baboons. Infect. Immun. 18:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Gorder, M. A., P. Della Pelle, J. W. Henson, D. H. Sachs, A. B. Cosimi, and R. B. Colvin. 1999. Cynomolgus polyoma virus infection: a new member of the polyoma virus family causes interstitial nephritis, ureteritis, and enteritis in immunosuppressed cynomolgus monkeys. Am. J. Pathol. 154:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Villarreal, L. P., V. R. Defilippis, and K. A. Gottlieb. 2000. Acute and persistent viral life strategies and their relationship to emerging diseases. Virology 272:1-6. [DOI] [PubMed] [Google Scholar]
- 72.Ward, J. M., A. Lock, M. J. Collins, Jr., M. A. Gonda, and C. W. Reynolds. 1984. Papovaviral sialoadenitis in athymic nude rats. Lab. Anim. 18:84-89. [DOI] [PubMed] [Google Scholar]
- 73.Yogo, Y., C. Sugimoto, H. Y. Zheng, H. Ikegaya, T. Takasaka, and T. Kitamura. 2004. JC virus genotyping offers a new paradigm in the study of human populations. Rev. Med. Virol. 14:179-191. [DOI] [PubMed] [Google Scholar]
- 74.Zheng, H. Y., C. Sugimoto, M. Hasegawa, N. Kobayashi, A. Kanayama, A. Rodas, M. Mejia, J. Nakamichi, J. Guo, T. Kitamura, and Y. Yogo. 2003. Phylogenetic relationships among JC virus strains in Japanese/Koreans and Native Americans speaking Amerind or Na-Dene. J. Mol. Evol. 56:18-27. [DOI] [PubMed] [Google Scholar]
- 75.zur Hausen, H., and L. Gissmann. 1979. Lymphotropic papovaviruses isolated from African green monkey and human cells. Med. Microbiol. Immunol. (Berlin) 167:137-153. [DOI] [PubMed] [Google Scholar]