Abstract
Adeno-associated virus type 2 (AAV-2) capsid proteins have eight sequence motifs that are potential sites for O- or N-linked glycosylation. Three are in prominent surface locations, close to the sites of cellular receptor attachment and to neutralizing epitopes on or near protrusions surrounding the three-fold axes, raising the possibility that AAV-2 might use glycosylation as a means of immune escape or for preventing reattachment on release of progeny virus. Peptide mapping and structural analysis by Fourier transform ion cyclotron resonance mass spectrometry demonstrates, however, no glycosylation of the capsid protein for virus prepared in cultured HeLa cells.
Adeno-associated viruses (AAVs) are small single-stranded DNA parvoviruses (36). There may be more than the eight serotypes originally identified, with humans or other primates as their primary hosts (13, 16). Although ∼ 80% of the population is seropositive due to natural exposure to AAV (11, 34), AAV has not been associated with disease (3). This is one of the reasons that it is being developed as a vector for in vivo gene therapy (5, 33).
Parvoviruses are small (250 Å) unenveloped viruses in which a single-stranded DNA (ssDNA) genome is surrounded by a T=1 icosahedrally symmetric capsid containing 60 copies of the capsid protein. Parvoviruses contain three or four variants of the capsid protein, differing in length at the N terminus (36). In AAV type 2 (AAV-2), viral protein 3 (VP3; 533 amino acids) constitutes 80% (by mass) of the capsid (60). Alternative mRNA splicing gives variants VP2 and VP1, which are extended at the N terminus by 65 and 202 residues, respectively. Although present in the crystals, the VP1 and VP2 unique regions were not seen in the atomic structure (58)—it appears that the 533 residues common to VP1, VP2, and VP3 occupy symmetry-equivalent positions in the capsid. The VP1 unique addition encodes nuclear localization signals (22, 56) and a phospholipase A2 domain (15) that is likely needed for the virus to escape into the cytoplasm from the endosome (12a). The location of the VP1 unique region has been debated, but it looks as if can move from the inner to outer surfaces (12, 26, 59, 60).
The primary cellular receptor for AAV serotypes 1 to 3 is heparan sulfate (HS) proteoglycan (50). Coreceptors are likely also involved, possibly fibroblast growth factor receptor and integrin αVβ5 (39, 40, 42, 49). AAV enters cells through endosomes (1) and is transported quickly into the nucleus (1, 44). Newly synthesized capsid proteins must be transported to the nucleolus for assembly of the DNA-containing virions (41), and specific binding of a nucleolus-targeting protein to VP2, and of nucleolin to AAV-2 virions has been reported (22, 41). Thus, the natural life cycle requires interactions with a number of other macromolecules in which glycosylation of the capsid proteins, if present, might be important.
Indeed, glycoproteins figure prominently in a number of other viruses, especially enveloped viruses, where they are often exposed on the outer surface. Mutations at the glycosylation sites often interfere with viral entry, infectivity, tissue specificity, or host range, implying roles in cell recognition, membrane fusion, and cell entry (8, 27, 43, 46, 53). Among nonenveloped viruses, glycosylation of structural proteins is less common. However, the fiber proteins of adenovirus, specifically types 2 and 5, are O glycosylated (6). The presence of the carbohydrate modulates the antigenicity in adenovirus (6). In rotavirus, it has been possible to select monoclonal antibody neutralization escape mutants (29) that have new sites of glycosylation in the epitope. Indeed, it has been suggested for human immunodeficiency virus type 1 (57) that glycosylation sites provide variability that allows the virus to escape immune detection of nearby conserved amino acids at the cellular-receptor binding site.
Our characterization of potential AAV-2 glycosylation sites started with a search through the sequence for likely motifs. It is possible to search for putative sites for N-linked glycosylation, for which there are several motifs (2), but for O-linked glycosylation, searching for motifs is more challenging (18, 24). The sequences of the capsid proteins were screened by use of MOTIFS of the Wisconsin Package (Accelrys, Inc.) and the PROSITE database (23). Within the common region of VP1 and VP2, there were two SGXG motifs at positions 157 (SGTG) and 195 (SGLG) (VP1 numbering) (4). These motifs are associated with glycosaminoglycan modification, but only the first at position 157 is preceded by two acidic amino acids in the −5 to −3 positions (EPDSSSGTG) closely resembling the consensus that requires two acidic residues between −4 and −2 of the SGXG conserved sequence (4). Searches for NXS/T motifs that can be associated with N-linked glycosylation (14) led to six AAV-2 sequences all within the part common to VP1, VP2, and VP3. Two (VP1223-225 and VP1408-410) are near the inner surface of the capsid (Fig. 1), but the four on the outer surface are easier to imagine having relevance to virus-host interactions. NPS656-658 (VP1 numbering) can be eliminated from consideration because the presence of a proline strongly reduces the likelihood of glycosylation (14). NGS382-384, NNS496-498, and NKS705-707 remain as candidate sites.
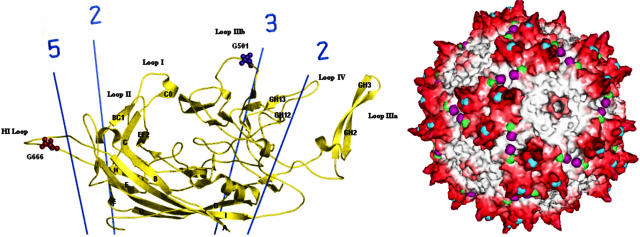
FIG. 1.
(Left) Sequence motifs characteristic of glycosaminoglycan modification are shown as spheres on the part of the capsid subunit structure that is common to VP1, VP2, and VP3 (58). Asn496 to Ser498 and Asn519 to Ser521 are on outer loops near the three- and five-fold axes respectively. (Right) In the assembled capsid, these sequences are in prominent positions on the outer surface. Asn496 to Ser498 are shown in blue near the tips of protrusions that surround each three-fold axis. Asn519 to Ser521 are shown in green near the five-fold axes.
Examination of the location of the candidate sites on the atomic structure of AAV-2 (58) shows that all are in prominent surface locations (Fig. 1). Of particular interest was the location of NNS496-498 at the tip of the protrusions that surround each three-fold axis. It is in a location where glycosylation could impact both cell receptor binding and antigenicity. Heparan sulfates are usually bound by basic residues (35). Electrostatic calculations from the atomic structure showed that residues RGNR585-588 were at the center of a positively charged surface patch that was predicted to be the core of the receptor binding site (58), a prediction that was subsequently confirmed through mutagenesis (25, 38). Although far apart in the structure of an individual subunit, NNS496-498 and RGNR585-588 come together in the assembled capsid, as adjacent loops from neighboring three-fold related subunits (58). The candidate glycosylation sequence is at the tip of the protrusion whereas the receptor-binding site is on the inner side of the protrusion (58), but separated by only 3.1 Å, i.e., in immediate van der Waals contact and well within a likely footprint of the heparan sulfate receptor. Furthermore, NNS496-498 is within the major linear epitope of neutralizing antibody C37B, a monoclonal antibody that inhibits receptor attachment (55). The remaining two candidate glycosylation sites (NGS382-384, and NKS705-707) are well separated within a subunit but come together on surface when contributed by adjacent subunits. They are on the opposite side of the RGNR585-588 receptor-site, but only 21 to 25 Å away, still conceivably within a possible receptor footprint. Neither of these candidate glycosylation sites is within a neutralizing epitope yet mapped, but they are close. NGS382-384, is immediately adjacent in primary structure to epitope 2 of monoclonal antibody A20, and His134, which is a surface neighbor of both NGS382-384 and NKS705-707, is at the start of A20 epitope 1 (55, 58). With these candidate glycosylation sites having potentially significant impact upon both antigenicity and receptor binding, it became imperative to characterize glycosylation experimentally.
For experimental characterization, AAV-2 was prepared according to Xie et al. (60). The methods involve propagation of AAV-2 in cultured human (HeLa) cells, so it is not unreasonable to expect that glycosylation patterns should resemble those in a human host. There might be some differences based on cell type and with the higher in vitro propagation levels that could saturate posttranslational modification pathways. To reduce the potential for such heterogeneity, the virus was harvested 48 h postinfection (cf. 72 h). It is reasonable to expect that at least some of the virus produced would have glycosylation mimicking that occurring naturally. Virus was purified by cesium chloride density gradient ultracentrifugation repeated three times.
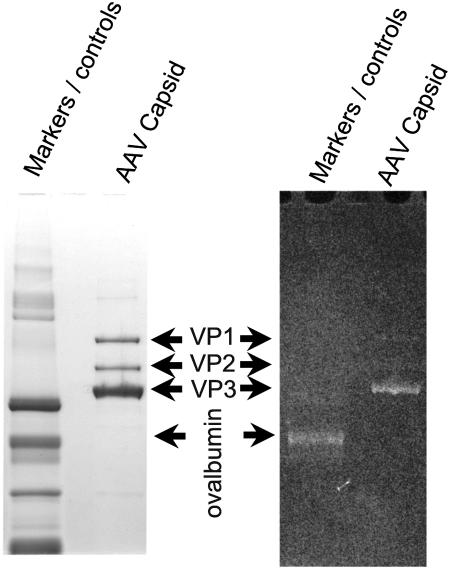
Various enzymatic, chemical, and immunochemical analyses can reveal the presence of glycans (21, 28, 51). For preliminary screening, methods were chosen that had broad specificity—chemical oxidations detected with either fluorescence or immunoassay. For both assays, capsid proteins from denatured AAV-2 particles were electrophoretically separated on a 4 to 12% Bis-Tris gel (NuPage, Inc.). The Glycoprofile III fluorescent glycoprotein detection kit (Sigma, Inc.) was applied to a fixed gel according to the manufacturer's instructions. Briefly, potential glycoconjugates were oxidized with a periodic acid solution, stained with dansyl hydrazide, destained, and imaged using a UV transilluminator with a yellow filter. EZBlue (Sigma, Inc) was then used to stain all proteins. The band for VP3 fluoresced with an intensity similar to that of ovalbumin (Fig. 2). This band could correspond to approximately an 8-mer attached at a single site or smaller modifications at several sites. The weak fluorescent staining of VP1 and VP2 is not inconsistent with the 10-fold-lower levels of VP1 and VP2, and with modification of a site or sites common to VP1, VP2, and VP3.
FIG. 2.
Glycosylation detection through fluorescence labeling. The left panel shows an EZBlue-stained gel with denatured AAV-2 capsids run alongside the ProteoProfile PTM markers (Sigma-Aldrich, Inc.). The right panel shows the gel stained with Glycoprofile III (Sigma-Aldrich, Inc.) with fluorescent imaging. VP3 fluorescent staining suggests glycosylation commensurate with ovalbumin. The weak fluorescent staining of VP1 and VP2 is not inconsistent with their ∼10-fold-lower concentration and glycosylation of a region common to VP1, VP2, and VP3.
The second assay used the digoxigenin (DIG) glycan detection kit (Roche, Inc.) according to the manufacturer's instructions. Potential carbohydrates were oxidized on the intact capsid with periodate caproic acid hydrazide and then labeled with digoxigenin. The particles were then denatured and separated electrophoretically. The proteins were transferred to a nitrocellulose membrane, then hybridized with an antidigoxigenin antibody conjugated to alkaline phosphatase so that activity could be measured colorimetrically. VP3 stained positively, but so did several unglycosylated proteins tested as negative controls. Thus, both chemical assays were giving inconclusive results. These types of tests are known to be difficult, with the real possibility of false positives. A more definitive method was sought, and the positive, if equivocal, chemical detection results motivated a mass spectrometric analysis.
Our choice would be liquid chromatography (LC)-Fourier transform ion cyclotron mass spectrometry (FT-ICR MS), taking advantage of the resources of the National High Magnetic Field Laboratory (NHMFL). There are important differences relative to the more familiar matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). The preseparation of the samples by LC circumvents the problem of selective desorption/ionization, while the high resolution and mass accuracy provided by ICR permit the analysis of large glycopeptides (48). Also, the analysis of intact glycopeptides permits the assignment of specific sugar structures to a specific amino acid residue (19). Thus, it is preferable with FT-ICR MS to characterize directly the glycopeptides, rather than cleave off the carbohydrate as might be needed with MALDI-TOF MS, a particular challenge with O-linked glycosylation due to the lack of suitable enzymes.
One disadvantage of FT-ICR MS is the relatively large amount of sample needed. For the liquid chromatography mass spectrometry-mass spectrometry (LCMS-MS) methods used to confirm the identity of potentially glycosylated peptides, larger quantities (up to 200 μg) would be needed than for MALDI-TOF MS. Such quantities are orders of magnitude greater than those normally prepared in culture for virological research. Replication of AAV-2 in HeLa cells is high yield relative to other cell types (37), and we took advantage of extensive past efforts to maximize production for crystallographic structure determination (60)—no further development of AAV-2 production in HeLa cells was required for the FT-ICR MS characterization.
Samples for mass spectrometry were extracted from denatured gels and proteolytically fragmented. Stained VP3 and VP1 bands from eight lanes loaded with 8 μg denatured virus were excised from the gel and destained in 50% acetonitrile-25 mM ammonium bicarbonate until no color remained. Gel pieces were vacuum dried and then incubated overnight at 37°C with 15 μl of 10 μg/μl with trypsin, Asp-N, or Glu-C (Sigma-Aldrich, Inc.). Peptides were eluted from the gel pieces with 75% acetonitrile and 0.25% formic acid. VP1 required additional deionization with a ZipTip (Millipore, Inc). Samples were stored at −20°C. VP3, available in larger amounts, was analyzed first, followed by VP1.
Protein digests were dissolved in acetonitrile-0.5% formic acid (aqueous) at a concentration of approximately 10 μM and ionized by microelectrospray (9, 10). In order to obtain improved signal/noise ratio, some digests were purified with a miniaturized system (ZipTips, Millipore, Bedford, MA) according to the manufacturer's instructions. Accurate mass analysis was performed with data obtained with a custom-built 7T or 9.4T Fourier transform ion cyclotron mass spectrometer (31, 45, 54). A one-dimensional nano-liquid chromatography (nano-LC) system (Eksigent, Livermore, CA) was interfaced with microelectrospray to the 7T FT-ICR MS and used for separation of protein digests over a 125-nl C18 cartridge (Opti-Pak, Optimize Technologies, Inc., Oregon City, OR). The nano-LC setup permitted efficient “peak parking” of proteolytic peptide ions. Peak-parked ions were stored waveform inverse Fourier transform (SWIFT) (17, 32) isolated in the ICR cell and fragmented by infrared multiphoton dissociation (IRMPD) (30).
In total, 402 out of 533 amino acid residues, or 75.4% of the VP3 sequence, were mapped as unmodified or methionine oxidized (Fig. 3; and see Table S2 in the supplemental material). Oxidation of methionine residues frequently occurs during electrophoresis. The N-terminal peptide was not detected, although it was searched for against the mass data considering the possibility of N-terminal acetylation and/or terminal methionine truncation. The N terminus was also blocked when subjected to Edman sequencing and thus is probably posttranslationally modified. If modified by lipidation, it is unlikely that the modified peptide would be extracted from the gel and detected by mass spectrometry. It is notable that all of the unmapped regions of the sequence of VP3 contain at least one cysteine residue. Because cysteine is a reactive amino acid, it is likely that these residues became covalently modified during protein separation with undetermined ligands. That modification would lead to an unknown change in mass, which would preclude the assignment of the modified peptides.
FIG. 3.
Coverage of the VP1 (clear) and VP3 (shaded) sequences by peptide mapping. Residues mapped are marked in boldface. For VP1, 599/733 (82%) of the amino acids in the sequence were covered. For VP3, 402/533 (75%) of the amino acids in the sequence were covered by the VP3 analysis or 415/533 (78%) were covered in either the VP3 or VP1 analysis (underlined).
For VP1, 599 out of a total of 733 amino acid residues, or 82% of the amino acid sequence, were mapped as unmodified or methionine-oxidized peptides (see Fig. S4 and Tables S5 to S7 in the supplemental material). The N terminus was examined for the possibility of acetylation and/or truncation, but no such modification was observed. The missing peptide segments for VP1 all contained at least one cysteine residue, except for three short segments of 2 to 5 amino acids.
Mass spectrometry fragment masses from VP3 and VP1 that did not match unmodified peptide masses from these proteins were analyzed with the program GlycoMod (7). That software searches for matches with all predicted proteolytic fragments and possible N- and O-linked oligosaccharide additions comprising all possible underivatized, acetylated, and methylated monosaccharide units. We have previously employed the GlycoMod tool in studies of plant and human glycoproteins (19, 20, 47, 48), in which we identified possible glycopeptides with bioinformatic assistance and confirmed the structures by IRMPD. The use of high-mass-accuracy data in searches with GlycoMod greatly reduces the number of possible glycopeptide matches because a narrow-mass search window is allowed. However, confirmation of the structure of modifying glycans by tandem mass spectrometry (MS/MS) is necessary because of the risk of false-positive identifications. IRMPD is an important tool because it dissociates glycosidic bonds efficiently but preserves peptide bonds, thereby greatly simplifying the determination of the modifiying glycan structure (19).
No N-linked glycopeptide masses could be matched with the GlycoMod tool to any of the experimental masses submitted from either VP3 or VP1. In the case of O glycosylation, more matches are expected because of the large number of Ser and Thr residues in most proteins and because of the large number of possible glycan structures at each site. Five possible glycopeptide matches were found in the LC-MS data set from the tryptic digest of VP3 (see Table S8A in the supplemental material), and 10 possible matches were found in the data from the Asp-N digest of the same protein (see Table S8B in the supplemental material). In neither set was the same glycan composition matched to the same peptide. In order to confirm the structures of the glycopeptides, the digested protein was subjected to LC-MS/MS analysis. Low sample volume and concentration necessitated the use of high-sensitivity nano-LC MS. The Eksigent peak-parking function allowed the flow over the LC column to be greatly reduced, and thus the specific peptide ions of interest could be manipulated in the 7T FT-ICR (SWIFT isolated and IRMPD) for up to 20 min per digest fragment. The increased analysis time produced high-resolution, high-signal/noise spectra with high mass accuracy for definitive MS/MS analysis from low-concentration complex peptide mixtures. In no case did the MS/MS data confirm the presence of modifying sugars. For VP1, several O-linked glycopeptide masses were matched (data not shown), but these were not investigated by LC-MS/MS because of the low level of staining of the protein for glycosylation (Fig. 2).
Thus, in spite of the presence of appropriate sequence motifs at suitably exposed locations on the surface of the capsid, it appears that at least the vast majority of AAV-2 particles are not glycosylated. A conservative estimate of the detection limit of FT-ICR MS is ∼750 fM for a 15-μl sample. Gels loaded with about 10 μg of virus (protein plus DNA) yielded bands of about 5 μg VP3 and 1 μg VP1 and MS samples that were ∼85 and 12 pM, respectively. Although negative mass spectrometry results cannot exclude the possibility that the capsid is ever glycosylated, no glycosylation is detected under conditions where 1% glycosylation of VP3 and 10% of VP1 should be detectable. It is therefore possible to define 1% as the upper bound for glycosylation of intact AAV-2.
Supplementary Material
Acknowledgments
This work was funded by NIH R01 GM66875 (M.S.C.), NSF DMR-00-84173 (A.G.M.), Florida State University, and the National High Magnetic Field Laboratory in Tallahassee, Fla.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blom, N., T. Sicheritz-Ponten, R. Gupta, S. Gammeltoft, and S. Brunak. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633-1649. [DOI] [PubMed] [Google Scholar]
- 3.Bloom, M. E., and N. S. Young. 2001. Parvoviruses, p. 2361-2380. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 4th ed. Lippincott WIlliams & Wilkins, Philadelphia, Pa.
- 4.Bourdon, M. A., T. Krusius, S. Campbell, N. B. Schwartz, and E. Ruoslahti. 1987. Identification and synthesis of a recognition signal for the attachment of glycosaminoglycans to proteins. Proc. Natl. Acad. Sci. USA 84:3194-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, P. J., and R. J. Samulski. 2000. Adeno-associated viral vectors as gene delivery vehicles. Int. J. Mol. Med. 6:17-27. [DOI] [PubMed] [Google Scholar]
- 6.Cauet, G., J. M. Strub, E. Leize, E. Wagner, A. Van Dorsselaer, and M. Lusky. 2005. Identification of the glycosylation site of the adenovirus type 5 fiber protein. Biochemistry 44:5453-5460. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, C. A., E. Gasteiger, and N. H. Packer. 2001. GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1:340-349. [DOI] [PubMed] [Google Scholar]
- 8.Delos, S. E., M. J. Burdick, and J. M. White. 2002. A single glycosylation site within the receptor-binding domain of the avian sarcoma/leukosis virus glycoprotein is critical for receptor binding. Virology 294:354-363. [DOI] [PubMed] [Google Scholar]
- 9.Emmett, M. R., and R. M. Caprioli. 1994. Microelectrospray mass spectrometry: ultra-high-sensitivity analysis of peptides and proteins. J. Am. Soc. Mass Spectrom. 5:605-613. [DOI] [PubMed] [Google Scholar]
- 10.Emmett, M. R., F. M. White, C. L. Hendrickson, S. D. Shi, and A. G. Marshall. 1998. Application of micro-electrospray liquid chromatography techniques to FT-ICR mass spectrometry to enable high-sensitivity biological analysis. J. Am. Soc. Mass Spectrom. 9:333-340. [DOI] [PubMed] [Google Scholar]
- 11.Erles, K., P. Sebokova, and J. R. Schlehofer. 1999. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV). J. Med. Virol. 59:406-411. [DOI] [PubMed] [Google Scholar]
- 12.Farr, G. A., and P. Tattersall. 2004. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology 323:243-256. [DOI] [PubMed] [Google Scholar]
- 12a.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. USA 102:17148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavel, Y., and G. von Heijne. 1990. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3:433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 16.Grimm, D., and M. A. Kay. 2003. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 3:281-304. [DOI] [PubMed] [Google Scholar]
- 17.Guan, S., and A. G. Marshall. 1996. Stored waveform inverse Fourier transform (SWIFT) ion excitation in trapped-ion mass spectrometry: theory and applications. Int. J. Mass Spectrom. Ion Processes 157/158:5-37. [Google Scholar]
- 18.Gupta, R., H. Birch, K. Rapacki, S. Brunak, and J. E. Hansen. 1999. O-GLYCBASE version 4.0: a revised database of O-glycosylated proteins. Nucleic Acids Res. 27:370-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Håkansson, K., H. J. Cooper, M. R. Emmett, C. E. Costello, A. G. Marshall, and C. L. Nilsson. 2001. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptide yield complementary sequence information. Anal. Chem. 73:4530-4536. [DOI] [PubMed] [Google Scholar]
- 20.Håkansson, K., M. R. Emmett, A. G. Marshall, P. Davidsson, and C. L. Nilsson. 2003. Structural analysis of 2D-gel-separated glycoproteins from human cerebrospinal fluid by tandem high-resolution mass spectrometry. J. Proteome Res. 2:581-588. [DOI] [PubMed] [Google Scholar]
- 21.Harvey, D. J. 2001. Identification of protein-bound carbohydrates by mass spectrometry. Proteomics 1:311-328. [DOI] [PubMed] [Google Scholar]
- 22.Hoque, M., K.-I. Ishizu, A. Matsumoto, S.-I. Han, F. Arisaka, M. Takayama, K. Suzuki, K. Kato, T. Kanda, H. Watanabe, and H. Handa. 1999. Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. J. Virol. 73:7912-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulo, N., C. J. Sigrist, V. Le Saux, P. S. Langendijk-Genevaux, L. Bordoli, A. Gattiker, E. De Castro, P. Bucher, and A. Bairoch. 2004. Recent improvements to the PROSITE database. Nucleic Acids Res. 32:D134-D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung, E., A. L. Veuthey, E. Gasteiger, and A. Bairoch. 2001. Annotation of glycoproteins in the SWISS-PROT database. Proteomics 1:262-268. [DOI] [PubMed] [Google Scholar]
- 25.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kronenberg, S., B. Böttcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn, J. E., B. R. Eing, R. Brossmer, K. Munk, and R. W. Braun. 1988. Removal of N-linked carbohydrates decreases the infectivity of herpes simplex virus type 1. J. Gen. Virol. 69:2847-2858. [DOI] [PubMed] [Google Scholar]
- 28.Kuster, B., T. N. Krogh, E. Mortz, and D. J. Harvey. 2001. Glycosylation analysis of gel-separated proteins. Proteomics 1:350-361. [DOI] [PubMed] [Google Scholar]
- 29.Lazdins, I., B. S. Coulson, C. Kirkwood, M. Dyall-Smith, P. J. Masendycz, S. Sonza, and I. H. Holmes. 1995. Rotavirus antigenicity is affected by the genetic context and glycosylation of VP7. Virology 209:80-89. [DOI] [PubMed] [Google Scholar]
- 30.Little, D. P., J. P. Speir, M. W. Senko, P. B. O'Connor, and F. W. McLafferty. 1994. Infrared multiphoton dissociation of large multiply-charged ions for biomolecule sequencing. Anal. Chem. 66:2809-2815. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, A. G. 2000. Milestones in Fourier transform ion cyclotron resonance mass spectrometry technique development. Int. J. Mass Spectrom. 200:331-356. [Google Scholar]
- 32.Marshall, A. G., T.-C. L. Wang, and T. L. Ricca. 1985. Tailored excitation for Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Chem. Soc. 107:7893-7897. [Google Scholar]
- 33.Monahan, P. E., and R. J. Samulski. 2000. Adeno-associated virus vectors for gene therapy: more pros than cons? Mol. Med. Today 6:433-440. [DOI] [PubMed] [Google Scholar]
- 34.Moskalenko, M., L. Chen, M. van Roey, B. A. Donahue, R. O. Snyder, J. G. McArthur, and S. D. Patel. 2000. Epitope mapping of human anti-adeno-associated virus type 2 neutralizing antibodies: implications for gene therapy and virus structure. J. Virol. 74:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulloy, B., and R. J. Linhardt. 2001. Order out of complexity—protein structures that interact with heparin. Curr. Opin. Struct. Biol. 11:623-628. [DOI] [PubMed] [Google Scholar]
- 36.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2360. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 37.Ogston, P., K. Raj, and P. Beard. 2000. Productive replication of adeno-associated virus can occur in human papillomavirus type 16 (HPV-16) episome-containing keratinocytes and is augmented by the HPV-16 E2 protein. J. Virol. 74:3494-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 40.Qiu, J., H. Mizukami, and N. S. Brown. 1999. Adeno-associated virus 2 co-receptors? Nat. Med. 5:467. [DOI] [PubMed] [Google Scholar]
- 41.Qiu, J., and K. E. Brown. 1999. A 110-kDa nuclear shuttle protein, nucleolin, specifically binds to adeno-associated virus type 2 (AAV-2) capsid. Virology 257:373-382. [DOI] [PubMed] [Google Scholar]
- 42.Qiu, J., A. Handa, M. Kirby, and K. E. Brown. 2000. The interaction of heparin sulfate and adeno-associated virus 2. Virology 269:137-147. [DOI] [PubMed] [Google Scholar]
- 43.Schneider, P. A., C. G. Hatalski, A. J. Lewis, and W. I. Lipkin. 1997. Biochemical and functional analysis of the Borna disease virus G protein. J. Virol. 71:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Brauchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 294:1929-1932. [DOI] [PubMed] [Google Scholar]
- 45.Senko, M. W., C. L. Hendrickson, L. Pasa-Tolic, J. A. Marto, F. M. White, S. Guan, and A. G. Marshall. 1996. Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry at 9.4 Tesla. Rapid Commun. Mass Spectrom. 10:1824-1828. [DOI] [PubMed] [Google Scholar]
- 46.Shirato, K., H. Miyoshi, A. Goto, Y. Ako, T. Ueki, H. Kariwa, and I. Takashima. 2004. Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J. Gen. Virol. 85:3637-3645. [DOI] [PubMed] [Google Scholar]
- 47.Sihlbom, C., P. Davidsson, M. R. Emmett, A. G. Marshall, and C. L. Nilsson. 2004. Glycoproteomics of cerebrospinal fluid in neurodegenerative disease. Int. J. Mass Spectrom. 234:145-152. [Google Scholar]
- 48.Sihlbom, C., P. Davidsson, and C. L. Nilsson. 2005. Enrichment of human glycoproteins prior to structure determination with ion trap dissociation in Fourier transform ion cyclotron resonance mass spectrometry. J. Proteome Res. 4:2294-2301. [DOI] [PubMed] [Google Scholar]
- 49.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 50.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarentino, A. L., R. B. Trimble, and T. H. Plummer, Jr. 1989. Enzymatic approaches for studying the structure, synthesis, and processing of glycoproteins. Methods Cell Biol. 32:111-139. [DOI] [PubMed] [Google Scholar]
- 52.Reference deleted.
- 53.Tuffereau, C., H. Leblois, J. Benejean, P. Coulon, F. Lafay, and A. Flamand. 1989. Arginine or lysine in position 333 of ERA and CVS glycoprotein is necessary for rabies virulence in adult mice. Virology 172:206-212. [DOI] [PubMed] [Google Scholar]
- 54.White, F. M., J. A. Marto, and A. G. Marshall. 1996. An External Source 7 Tesla FT-ICR mass spectrometer with electrostatic ion guide. Rapid Commun. Mass Spectrom 10:1845-1849. [DOI] [PubMed] [Google Scholar]
- 55.Wobus, C. E., B. Hügle-Dörr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 58.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]
- 60.Xie, Q., J. Hare, W. Bu, W. Jackson, J. Turnigan, and M. S. Chapman. 2004. Large-scale preparation, purification and crystallization of wild-type adeno-associated virus 2. J. Virol. Methods 122:17-27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.