Abstract
The RNA replication complexes of mammalian positive-stranded RNA viruses are generally associated with (modified) intracellular membranes, a feature thought to be important for creating an environment suitable for viral RNA synthesis, recruitment of host components, and possibly evasion of host defense mechanisms. Here, using a panel of replicase-specific antisera, we have analyzed the earlier stages of severe acute respiratory syndrome coronavirus (SARS-CoV) infection in Vero E6 cells, in particular focusing on the subcellular localization of the replicase and the ultrastructure of the associated membranes. Confocal immunofluorescence microscopy demonstrated the colocalization, throughout infection, of replicase cleavage products containing different key enzymes for SARS-CoV replication. Electron microscopy revealed the early formation and accumulation of typical double-membrane vesicles, which probably carry the viral replication complex. The vesicles appear to be fragile, and their preservation was significantly improved by using cryofixation protocols and freeze substitution methods. In immunoelectron microscopy, the virus-induced vesicles could be labeled with replicase-specific antibodies. Opposite to what was described for mouse hepatitis virus, we did not observe the late relocalization of specific replicase subunits to the presumed site of virus assembly, which was labeled using an antiserum against the viral membrane protein. This conclusion was further supported using organelle-specific marker proteins and electron microscopy. Similar morphological studies and labeling experiments argued against the previously proposed involvement of the autophagic pathway as the source for the vesicles with which the replicase is associated and instead suggested the endoplasmic reticulum to be the most likely donor of the membranes that carry the SARS-CoV replication complex.
In the spring of 2003, a novel respiratory disease in humans emerged in Southeast Asia and suddenly gripped the world. This atypical and life-threatening form of pneumonia was termed severe acute respiratory syndrome (SARS) (for a review, see reference 41), and a novel coronavirus (SARS coronavirus [SARS-CoV]) was identified as the etiological agent (9, 11, 26, 42). Coronaviruses are enveloped, positive-stranded RNA viruses with an ∼27- to 31-kb genome, of which about two-thirds is occupied by a gene encoding the viral nonstructural proteins, or replicase. By analogy with other members of the order Nidovirales (for reviews, see references 27, 57, 59, and 72), to which Coronaviridae belong, the replicase gene of SARS-CoV is comprised of open reading frame 1a (ORF1a) and ORF1b, with expression of the latter involving a ribosomal frameshift near the 3′ end of ORF1a. Consequently, genome translation produces two polyproteins (pp1a and pp1ab) of unprecedented complexity and size (4,382 and 7,073 amino acids, respectively). The pp1a and pp1ab primary translation products are subject to extensive proteolytic processing and, based on comparisons with other coronaviruses and recent experimental studies, are predicted to give rise to a total of 16 mature nonstructural proteins (Fig. 1) (31, 47, 58, 65, 73). These replicase cleavage products engage in minus-strand RNA synthesis, genome replication, and the production of subgenomic RNAs (for reviews, see references 39 and 52). The latter are used to express the genes in the 3′-proximal third of the genome, which encode structural and accessory proteins.
FIG. 1.
SARS-CoV replicase polyprotein organization, depicted in the form of the 7,071-amino-acid pp1ab. The border of amino acids encoded in ORF1a and ORF1b is indicated as RFS (ribosomal frameshift), and arrowheads represent sites that are cleaved by the nsp3 PLpro (gray) or the nsp5 Mpro (black). The 16 proteolytic cleavage products (nonstructural proteins) are numbered, and within the cleavage products key replicase domains have been highlighted (see text also). These include putative transmembrane domains (TM) and the four ORF1b-encoded domains (RdRp, Z, Hel, and NendoU) that are conserved in all nidoviruses. Abbreviations, from the N terminus to the C terminus: aa, amino acids; ADRP, ADP-ribose-1′′-monophosphatase; RBD, RNA-binding domains; Z, (putative) zinc-binding domain; Hel, helicase domain; Exo, (putative) exonuclease; MT, (putative) ribose-2′-O-methyltransferase.
Although many coronavirus nonstructural proteins remain to be studied in detail, a number of key enzymatic functions have been identified and characterized through a combination of theoretical and experimental approaches. Examples of such domains (Fig. 1) are the ORF1a-encoded papain-like accessory proteinase (PLpro) and 3C-like main proteinase (Mpro), residing in nonstructural protein 3 (nsp3) and nsp5, respectively, and the ORF1b-encoded RNA-dependent RNA polymerase (RdRp) (in nsp12), RNA helicase (in nsp13), and recently described nidovirus uridylate-specific endoribonuclease (NendoU) (in nsp15) (2, 3, 13, 15, 16, 18, 55, 58, 65, 73). Additional enzymatic domains include ADP-ribose-1′′-monophosphatase (in nsp3), a putative exoribonuclease (in nsp14), and a predicted ribose-2′-O-methyltransferase (in nsp16) (46, 49, 58, 69). Furthermore, a number of small proteins encoded in the 3′-proximal part of ORF1a (nsp7, nsp8, and nsp9) have been shown to possess RNA-binding properties and may operate as accessory factors for the RdRp complex (10, 43, 63, 70). In view of the potential for reemergence of SARS-CoV and the limited means currently available to combat such an outbreak, the replicative enzymes are prime targets for the development of antiviral drugs. Consequently, it is essential both to characterize individual nonstructural proteins in vitro and to understand the intracellular working environment in which nonstructural proteins interact and cooperate during viral RNA synthesis, the SARS-CoV RNA replication complex (RC).
Over the past decade, the RCs of a wide variety of eukaryotic positive-strand RNA viruses have been found to be associated with (modified) intracellular membranes (for recent reviews, see references 1, 30, 38, and 51). Membrane association of the RC is thought to be important for creating a suitable (micro)environment for viral RNA synthesis and may also aid in preventing the activation of certain host defense mechanisms that can be triggered by double-stranded RNA intermediates of RNA virus replication. In the case of nidoviruses, the subcellular localization of the RC and ultrastructure of the associate membranes have been studied in some detail only for the arterivirus equine arteritis virus (EAV) (40, 67) and the coronavirus mouse hepatitis virus (MHV) (5, 14, 44, 56, 66). For both viruses, when infected cells are analyzed by confocal immunofluorescence (IF) microscopy, the majority of replicase subunits (including RdRp and helicase) colocalize, presumably on virus-induced double-membrane vesicles (DMVs). Specific ORF1a-encoded transmembrane domains (residing in nsp3, nsp4, and nsp6 in the case of SARS-CoV [Fig. 1]) have been implicated in DMV formation and membrane association of the nidovirus RC. In the case of the arterivirus EAV, in the absence of other viral components, the expression of two specific transmembrane domain-containing nonstructural proteins (nsp2 and nsp3) induced DMV formation, presumably from endoplasmic reticulum (ER) membranes (60). For the coronavirus MHV, studies of the origin of the RC-carrying membranes have remained equivocal thus far, which may be partially due to the fact that these studies used different cell lines, different cellular marker proteins, and antisera recognizing different nonstructural proteins. Several intracellular compartments (including the Golgi complex, endosomal membranes, ER, and autophagosomes) were previously implicated in MHV RC formation (14, 44, 66).
For SARS-CoV, in a preliminary IF study using an antiserum recognizing the helicase protein (nsp13), we described an initially punctate and subsequently patched staining of the perinuclear region of infected Vero E6 cells and partial colocalization with ER markers (19). Subsequently, Prentice et al. (45) described a large panel of rabbit antisera and identified 12 of the predicted 16 replicase cleavage products in immunoprecipitation analyses of SARS-CoV-infected cell lysates. Similar IF labelings patterns were described for six ORF1a-encoded subunits (nsp1 to nsp4 plus nsp8 and nsp9) and nsp13. In a dual-labeling experiment, the nsp8 staining was found to overlap largely with that of nsp2 and nsp3 and was also claimed to partially colocalize with LC3, a key marker protein of the autophagic pathway (see below). In a recent study of the N-terminal domain of the SARS-CoV replicase (nsp1 to nsp3) and its processing by the nsp3 PLpro, Harcourt et al. (16) showed that, late in infection, most or all of nsp3 localizes to the site where de novo-synthesized viral RNA accumulates, as visualized by metabolic labeling with bromo-UTP. nsp1 and nsp2 only partially colocalized with this presumed site of viral RNA synthesis.
At the ultrastructural level, the early phase of SARS-CoV replication and RC formation has not been studied in great detail. Published electron microscopy (EM) studies (12, 37, 71) focused mainly on nucleocapsid formation and virus production, did not use replicase-specific antisera, and were partially performed at late(r) stages of infection, when cytopathic effects begin to disrupt the cellular infrastructure. Goldsmith et al. (12) showed immunolabeling of cytoplasmic vesicles for (unspecified) viral proteins and RNA. Larger vesicle-containing membrane sacs and DMVs, partially resembling those previously described for MHV, were also observed; however, given the multistep infection and late fixation (3 to 5 days postinfection [p.i.]) used in these experiments, the timing of their appearance in the context of the viral life cycle could not be properly assessed.
In this paper, by using a panel of SARS-CoV replicase-specific antisera and a combination of IF microscopy and EM, we have analyzed the earlier stages of SARS-CoV infection and in particular the formation of membrane structures that are likely involved in viral RNA synthesis. With infected Vero E6 cells, we observed the early formation (4 to 6 h p.i.) and accumulation of typical DMVs, although their preservation strongly depended on the procedure used for fixation of the cells. In immunoelectron microscopy (IEM) labeling studies, vesicular structures could be labeled with SARS-CoV replicase-specific antibodies. Key viral enzymes colocalized throughout infection, and, opposite to what has been described for MHV, the presumed RC appeared to remain fully separated from the site of virus assembly. Studies involving marker proteins point to the ER as the most likely source of the membranes with which the SARS-CoV RC is associated.
MATERIALS AND METHODS
Virus and cells.
SARS-CoV strain Frankfurt 1 (kindly provided by H. F. Rabenau and H. W. Doerr, Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany) was used to infect Vero E6 cells, which release maximum progeny titers by approximately 12 h p.i. (19, 45). Multiplicities of infection of 1 to 10 were used, and cells were fixed between 3 and 18 h p.i. All work with live SARS-CoV was performed inside biosafety cabinets in the biosafety level 3 facility at Leiden University Medical Center.
SARS-CoV antisera.
SARS-CoV-specific antisera (Table 1) were raised in New Zealand White rabbits as described previously (61), using as the antigen either bovine serum albumin-coupled synthetic peptides or His-tagged expression products purified from Escherichia coli. For all sera, the specificity of the immune response was confirmed by a combination of Western blot analysis and/or immunoprecipitation studies (data not shown) and IF microscopy (see below). Mock-infected cells and preimmmune sera were included as negative controls. To allow dual-labeling experiments with two rabbit antisera recognizing different SARS-CoV nonstructural proteins, the immunoglobulin (Ig) fraction was isolated from 1.5 ml of anti-nsp3 (αnsp3) serum by use of a protein A antibody purification kit (Sigma) and directly coupled to Alexa Fluor 488 (AF488) dye by use of an AF488 protein labeling kit (Molecular Probes).
TABLE 1.
SARS-CoV antigens and antisera used in this study
| SARS-CoV antigen | Location in pp1ab (aa)a | Predicted functionb | Region (aa)a used as antigen | Antigen type | Antiserum |
|---|---|---|---|---|---|
| nsp3 | 819-2740 | ADRP | 834-943 | His-tagged protein | αnsp3 |
| SUD | 1208-1557 | His-tagged protein | |||
| PLpro | 1558-2040 | His-tagged protein | |||
| nsp5 | 3241-3546 | Mpro | 3241-3546 | His-tagged protein | αnsp5 |
| nsp8 | 3920-4117 | RBD | 3920-4117 | His-tagged protein | αnsp8 |
| nsp9 | 4118-4230 | RBD | 4209-4230 | Synthetic peptide | αnsp9 |
| nsp12 | 4370-5301 | RdRp | 5283-5301 | Synthetic peptide | αnsp12 |
| nsp13 | 5302-5902 | Helicase | 5885-5902 | Synthetic peptide | αnsp13 |
| nsp15 | 6430-6775 | NendoU | 6762-6775 | Synthetic peptide | αnsp15 |
| M | NAc | Membrane protein | 204-221 | Synthetic peptide | αM |
aa, amino acids.
ADRP, ADP-ribose-1′′-monophosphatase; SUD, SARS-CoV unique domain; Mpro, main proteinase; RBD, RNA-binding domain. See also reference 58.
NA, not applicable.
Marker proteins and antibodies.
To visualize a variety of cellular compartments, a combination of antibodies and green fluorescent protein (GFP)-tagged proteins was used. A cytomegalovirus (CMV) promoter-driven expression vector (pEGFP-N1; Clontech) for human lysosome-associated membrane protein 1 (LAMP1) fused to the N terminus of GFP was kindly provided by Stephane Méresse, University of Marseille (6). A similar vector (pEGFP-C3; Clontech) expressing a fusion of GFP and human LC3B was generously donated by Karla Kirkegaard, Stanford University (20). To construct similar expression vectors expressing fusions of GFP to the A and C isoforms of LC3, the LC3B gene was excised from pEGFP-LC3B by using restriction enzymes EcoRI (which cuts between the GFP and LC3B moieties of the gene) and PstI (downstream of the gene). Subsequently, a PCR product containing the coding sequence for LC3A or LC3C was inserted and the sequence of the insert was verified. The LC3A and LC3C coding sequences were amplified from cDNA clones obtained from the German Resource Center for Genome Research (RZPD). The LC3A PCR product was amplified from cDNA clone DKFZp761L0515Q (GenBank accession number AL833855), and the sequence used was identical to nucleotides 66 to 431 of this entry. The major part of the LC3C PCR product was amplified from cDNA clone HU3_p983B07336D2 (GenBank accession number AA424268), but since the LC3C coding sequence in this clone was found to be incomplete, the 3′-terminal 119 nucleotides were added by three subsequent PCR steps. The final product contained the full-length gene, which was identical to nucleotides 65 to 505 of the LC3C mRNA sequence, as documented for GenBank accession number NM_001004343. Transfection of CMV expression vectors into Vero E6 cells and subsequent SARS-CoV infection were described previously (18).
Mouse monoclonal antibodies (MAbs) recognizing protein disulfide isomerase (PDI) (MAb 1D3, marker for the ER [68]) and human ERGIC-53 (MAb G1/93, ER-Golgi intermediate compartment [ERGIC] marker; Alexis Biochemicals) were used. Rabbit antisera against rat LC3, raised using either a synthetic peptide (serum no. 2-3) or recombinant rat LC3 (serum SK2-6), were kindly provided by Tamotsu Yoshimori and Takahiro Kamimoto, National Institute of Genetics, Japan (21). A bovine serum albumin-coupled synthetic peptide (NH2-MPSEKTFKQRRTFEQRVEDKK-COOH) representing the human LC3B N-terminal domain was used to raise an additional rabbit antiserum (αLC3B), as described above. The reactivity of this antiserum was confirmed using the GFP-LC3B fusion protein described above (see Fig. 7A; also data not shown).
FIG. 7.
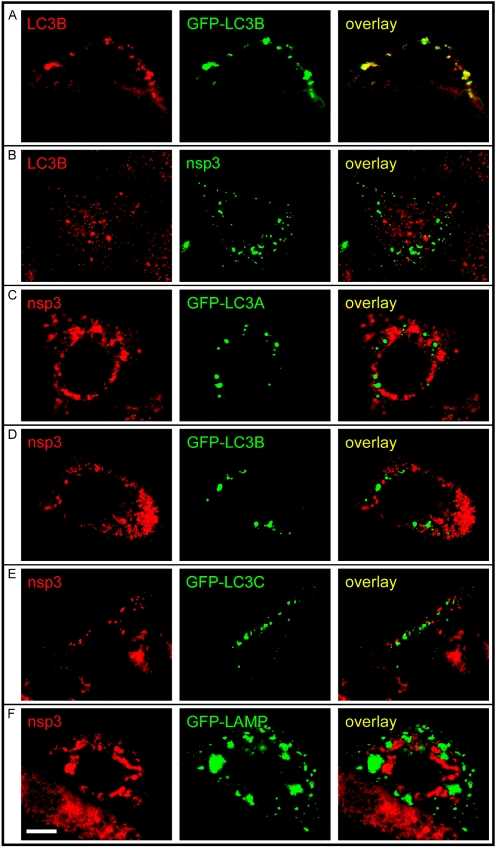
IF microscopy analysis of the overlap between autophagosomes (visualized by means of the LC3 marker protein) and the SARS-CoV RC in infected Vero E6 cells (9 h p.i.). (A) Staining of GFP-LC3B-expressing, transfected Vero E6 cells with the αLC3B rabbit antiserum that was raised using an N-terminal synthetic peptide (see Materials and Methods). (B) IF double-labeling analysis showing a (relatively rare) example of a SARS-CoV-infected cell with a convincing LC3B labeling pattern which is clearly distinct from the staining for the viral replication complex (nsp3). (C to E) Staining of pGFP-LC3A, -LC3B, -LC3C-transfected and SARS-CoV-infected Vero E6 cells, showing complete separation of compartments positive for GFP-LC3A, GFP-LC3B, or GFP-LC3C and structures carrying the viral replication complex (stained with αnsp3). (F) Staining of pLAMP-GFP-transfected and SARS-CoV-infected Vero E6 cells, showing complete separation of compartments positive for LAMP1-GFP and structures carrying the viral replication complex (stained with αnsp3). Bar, 10 μm.
Immunofluorescence microscopy.
SARS-CoV-infected Vero E6 cells on glass coverslips were fixed with 3% paraformaldehyde (for at least 16 h) at various time points after infection and were processed for immunofluorescence microscopy essentially as described by van der Meer et al. (67). Following permeabilization, single- or dual-labeling IF assays were carried out with rabbit antisera and/or mouse monoclonal antibodies, which were detected using indocarbocyanine (Cy3)-conjugated donkey anti-rabbit Ig and Alexa Fluor 488-conjugated goat anti-mouse Ig secondary antibodies, respectively (Molecular Probes/Invitrogen). Samples were examined with a Zeiss Axioskop 2 fluorescence microscope (equipped with the appropriate filter sets, a digital Axiocam HRc camera, and Zeiss Axiovision 4.2 software) or with a Zeiss LSM510 confocal laser scanning microscope, constantly using the same pinhole setting for both channels to give optical sections with a standard thickness of 0.8 μm. Images were optimized with Adobe Photoshop 6.0.
Electron microscopy.
For ultrastructural morphological investigation, SARS-CoV-infected Vero E6 cells were initially fixed at various time points after infection with 1.5% glutaraldehyde in 100 mM cacodylate buffer for 60 min at room temperature (RT), postfixed with 1% osmium tetroxide in phosphate buffer for 60 min at 4°C, dehydrated in a graded ethanol series up to 100%, and embedded in epoxy LX-112 resin. In follow-up experiments, infected cells were prefixed overnight at RT with 3% paraformaldehyde and then cryofixed by high-speed plunge freezing in liquid ethane. This step was followed by freeze substitution with 1% osmium tetroxide and 0.5% uranyl acetate in acetone, dehydration in acetone, and subsequent embedment in epoxy LX-112 resin. Ultrathin sections were contrasted with uranyl acetate and lead hydroxide.
For immunoelectron microscopy, cells were fixed with 3% paraformaldehyde and 0.2% glutaraldehyde in PHEM buffer {60 mM PIPES [piperazine-1,4-bis(2-ethanesulfonic acid)], 25 mM HEPES, 2 mM MgCl2, 10 mM EGTA} for 2 h at RT. After being washed in phosphate-buffered saline, the cells were scraped from the dish, pelleted, and embedded in 12% gelatin. The pellet was cut into ∼1-mm3 cubes, which were cryoprotected in 2.3 M sucrose and subsequently snap-frozen in liquid nitrogen. Ultrathin cryosections were labeled with SARS-CoV-specific rabbit serum αnsp3 (1:500), αnsp13 (1:300), or αM (1:200) or with anti-PDI MAb 1D3 (1:30).
Rabbit antibodies were detected with protein A-gold particles, whereas mouse MAbs were indirectly labeled with protein A-gold particles via a rabbit anti-mouse IgG bridging antibody (1:200) (Dako Cytomation, Denmark). For single-labeling experiments, 15-nm colloidal gold particles were used, and for double-labeling experiments, both 10- and 15-nm colloidal gold particles were used. The sections were contrasted with uranyl acetate and embedded in methyl cellulose.
All specimens were viewed with a Philips CM-10 transmission electron microscope (Eindhoven, The Netherlands) at 80 kV.
RESULTS AND DISCUSSION
Colocalization of key SARS-CoV replicative proteins in infected cells.
We first studied the subcellular localization of a variety of SARS-CoV nonstructural proteins by using our panel of rabbit antisera (Table 1) with IF assays. Five proteins that contain key enzymatic functions for coronavirus replication were included in this analysis: the two proteinases (nsp3 and nsp5), RdRp (nsp12), helicase (nsp13), and NendoU (nsp15). In addition, we successfully raised antisera recognizing two small ORF1a-encoded subunits with RNA-binding properties, nsp8 and nsp9 (10, 63, 70; also data not shown).
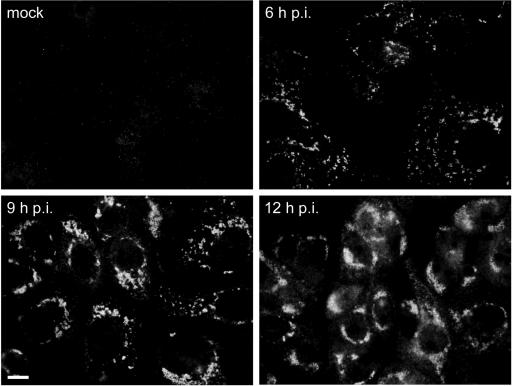
The replication cycle of SARS-CoV in Vero E6 cells takes about 12 h (16, 19, 45). The first signal for each of the nonstructural proteins mentioned above, including that for the previously uncharacterized Mpro (nsp5), RdRp (nsp12), and NendoU (nsp15) subunits, could be detected between 4 and 6 h p.i. In each case, as exemplified for nsp3 in Fig. 2, the early labeling pattern is a punctate cytoplasmic staining, which develops into a number of densely labeled areas close to the nucleus later in infection. In single-labeling experiments, the staining patterns for all nonstructural proteins studied were essentially similar (data not shown).
FIG. 2.
Time course IF labeling experiment showing the development of SARS-CoV replicase signal in infected Vero E6 cells, as exemplified by labeling for nsp3. The initially punctate cytoplasmic staining (6 h p.i.) develops into a number of densely labeled areas close to the nucleus later in infection (9 and 12 h p.i.). Bar, 10 μm.
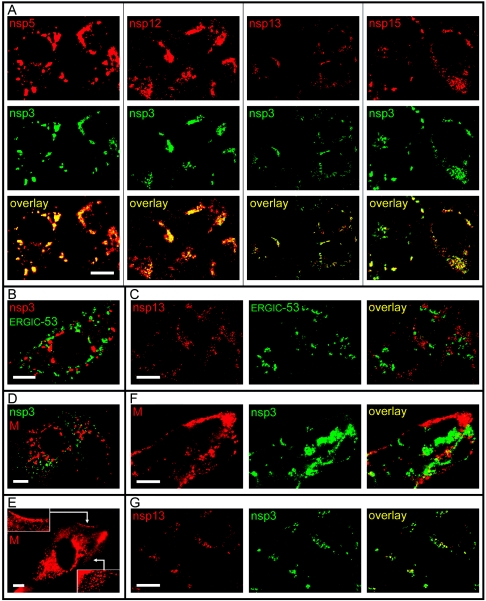
To analyze the extent of colocalization between different nonstructural proteins, the Ig fraction from the αnsp3 serum was purified and coupled directly to the fluorescent AF488 dye. This allowed us to perform dual-labeling experiments by incubating the fixed cells first with a regular αnsp rabbit serum, then with a Cy3-labeled anti-rabbit Ig conjugate, and finally, after extensive washing (four to five buffer changes in 30 min), with the AF488-labeled αnsp3 Ig. Experiments with antisera recognizing proteins that do not colocalize with nsp3 (e.g., the SARS-CoV M protein [Fig. 3D ], described below) demonstrated that this protocol prevents cross-reaction of the Cy3-labeled anti-rabbit Ig conjugate and the AF488-labeled αnsp3 Ig fraction. Using this approach, we assessed the colocalization of nsp3 with the other nonstructural proteins at 6, 9, and 12 h p.i. Representative dual-labeling images, recorded with a confocal microscope, are shown in Fig. 3A for the 9-h p.i. time point. Generally, throughout infection, the extent of colocalization between the nsp3 staining and that for nsp5, nsp12, nsp13, and nsp15 was large to very large, although some variation between cells was observed and small numbers of single-labeled spots were also visible. Similar observations were made for nsp8 and nsp9 (data not shown). Our IF data confirm and extend results published by others (16, 45) and are in line with the expected formation of a membrane-bound RC containing most of the replicase cleavage products in SARS-CoV-infected cells.
FIG. 3.
Confocal IF microscopy analysis of the intracellular distribution of various SARS-CoV replicase subunits in infected Vero E6 cells. (A) Double-labeling experiments (9 h p.i.) using an AF488-coupled IgG fraction purified from an anti-nsp3 serum and antisera recognizing nsp5, nsp12, nsp13, and nsp15. Extensive colocalization of these five nonstructural proteins was observed throughout infection. (B) Double-labeling experiment (9 h p.i.) for SARS-CoV nsp3 and the ERGIC-53 cellular marker protein. (C) Double-labeling experiment (18 h p.i.) for the SARS-CoV nsp13 helicase and the ERGIC-53 cellular marker protein, illustrating the complete separation of the nsp13 and the ERGIC at late time points in infection. (D) Double-labeling experiment (6 h p.i.) for SARS-CoV nsp3 and the viral M protein, which localizes to the Golgi complex at this time point. (E) Labeling for the SARS-CoV M protein at 9 h p.i., showing the spread of the protein throughout the cytoplasm, presumably due to the traffic of progeny virions towards the plasma membrane. Insets illustrate the strong labeling of the region just beneath the plasma membrane. (F) Double-labeling experiment (18 h p.i.) for SARS-CoV nsp3 and M protein, confirming the almost-complete separation of the two proteins also at late time points in infection. (G) Double-labeling experiment (18 h p.i.) using an AF488-coupled IgG fraction purified from an anti-nsp3 serum and an antiserum recognizing nsp13, illustrating the colocalization of the two proteins also at late stages of infection. In general, late in infection, the nsp13 signal was found to decline more rapidly than that of nsp3, suggesting differences in turnover of these two proteins. Bar, 10 μm.
Separation of membranes involved in SARS-CoV RC formation and virus assembly.
For MHV, it was previously reported that, at late time points after infection, a subset of nonstructural proteins (including nsp1 and the nsp13 helicase) relocalize to the presumed site of virus assembly, which was identified using an antiserum against the triple-spanning membrane protein (M protein), a major component of virus particles. Based on this observation, Bost et al. (5) and Brockway et al. (7) proposed a link between these components of the RC and the regulation of RNA packaging and/or virus assembly. On the other hand, it should be noted that the analysis of MHV-infected cells late in infection is complicated by strong syncytium formation, which induces major changes in the cellular infrastructure. In our studies, the fact that SARS-CoV infection only rarely induces syncytia in Vero E6 cells was a clear advantage and prompted us to assess the extent of overlap between RC-containing regions and the site of virus assembly.
The coronavirus M protein is a major determinant of virion budding (for a recent review, see reference 8 and references therein) which has been reported to occur in the ERGIC (24). However, the documented site of M accumulation in coronavirus-infected cells is not the ERGIC but the Golgi complex (23), although the exact localization within this organelle can range from the cis to the trans side (25, 29). In the Golgi complex, the M protein may be present either incorporated in maturing virions or inserted in the membranes of the organelle itself. In an expression system, the SARS-CoV M protein was also targeted to the Golgi complex (36); however, to our knowledge, the localization of the protein in SARS-CoV-infected cells has not yet been described. We have previously reported the complete separation of the SARS-CoV nsp13 helicase staining and the Golgi complex, which was labeled using a Golgi-GFP marker protein (19). In follow-up experiments, the staining with a MAb recognizing an established marker protein for the ERGIC (ERGIC-53 [54]) was also found to be separated from the nsp3/nsp13 labeling throughout infection (Fig. 3B and C; also data not shown).
Subsequently, an anti-SARS-CoV M rabbit serum (Table 1) was used to visualize compartments involved in virion assembly and maturation. As expected, the αM labeling was found to change dramatically during the course of infection (Fig. 3D, E, and F). Whereas the early staining was restricted to the Golgi complex (Fig. 3D; also data not shown), the protein was seen in spots throughout the cytoplasm when the cells entered the productive stage of infection (Fig. 3E), presumably due to the traffic of progeny virions towards the plasma membrane. This notion was supported by strong labeling of the area just beneath the plasma membrane (Fig. 3E) and material outside the cells.
Finally, infected cells were double labeled using the AF488-labeled αnsp3 Ig fraction and the αM serum and analyzed by confocal microscopy. It was found that throughout infection, despite the considerable changes in M protein staining, the two signals remained almost completely separated (Fig. 3F). Given the large overlap between the staining for nsp3 and that for all other nonstructural proteins studied here, including the nsp13 helicase (Fig. 3A and G), this result makes it unlikely that these SARS-CoV nonstructural proteins undergo the late relocalization to compartments involved in virus assembly that was described for MHV. To establish whether this is due to the absence of syncytium formation in the case of SARS-CoV, to the use of different cell lines in studies with MHV, or to a specific difference between these two coronaviruses, more detailed comparative studies are required, and these are in progress.
SARS-CoV infection induces DMV formation.
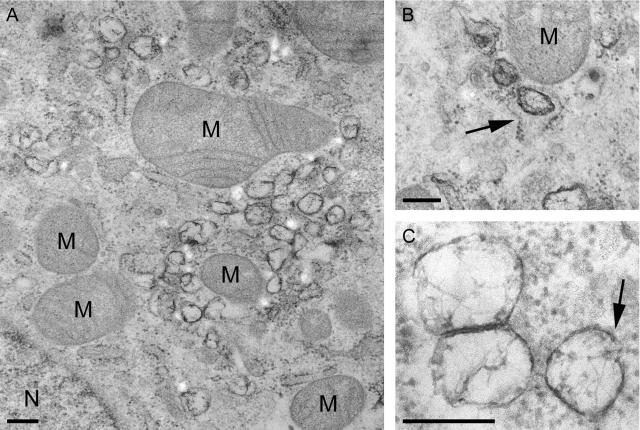
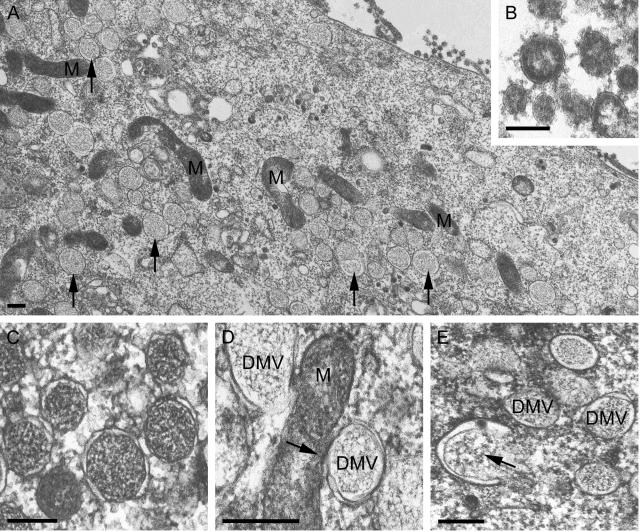
At various time points after infection, the ultrastructural changes in SARS-CoV-infected Vero E6 cells were investigated by preparing samples for transmission electron microscopy using conventional chemical fixation followed by embedment in an epoxy resin. At 6 h p.i., conspicuous vesicular structures, which were absent in mock-infected cells, were readily observed. These structures appeared to have a single membrane (although small pieces of double membrane could occasionally be discerned [Fig. 4C ]). The vesicles were irregular in shape and often displayed a spider web-like content (Fig. 4A and C). They were clearly associated with the (dilated) rough ER and were regularly found located within this organelle (Fig. 4B). Furthermore, they frequently clustered in the perinuclear region, in areas with many mitochondria, which showed normal morphology (Fig. 4A). These vesicle clusters had increased in size and number by 9 h p.i.
FIG. 4.
EM analysis of SARS-CoV-infected Vero E6 cells (panels A and B, 6 h p.i.; panel C, 9 h p.i.) fixed using conventional chemical fixation and embedded in epoxy LX-112 resin. (A) Low-magnification overview of a cluster of virus-induced vesicles in the perinuclear region of the cell (N, nucleus), which is also rich in mitochondria (M). Whereas other membranes, like those of mitochondria, were generally well preserved, the virus-induced vesicles were quite electron lucent and the surrounding membranes were poorly visible. (B) Virus-induced vesicles were often observed to occur in association with the ER or inside the lumen of the (dilated) ER (arrow). (C) Close-up of virus-induced vesicles, showing their electron-lucent interior with a spider web-like content. Only occasionally, a part of a surrounding double membrane was observed (arrow). The images presented in this figure illustrate the poor conservation of the virus-induced vesicles when standard procedures for fixation and embedment were used. Bar, 250 nm.
The morphological characteristics of the SARS-CoV-induced vesicles differed in several respects from those described for MHV-infected cells. Although the size (200 to 350 nm) and irregular shape resembled those of MHV DMVs (14), the double membrane (reported to be often fused into a trilayer in the case of MHV) was lacking and the interior of the vesicles was not as electron lucent as that of the empty vesicles described for MHV. Taken together, these observations suggested that these membranous structures associated with the early phase of SARS-CoV infection had not been well preserved by the routine chemical fixation used in the initial experiments.
To test this hypothesis, we subsequently applied cryofixation by high-speed plunge freezing in liquid ethane, followed by freeze substitution with 1% osmium tetroxide and 0.5% uranyl acetate in acetone. This method resulted in a strikingly different morphology of the membranous structures (Fig. 5). They were now spherical, with an electron density similar to that of the cytoplasm, and limited by a clear double membrane (Fig. 5A and C). With the exception of a significant size difference (average diameters of 200 to 300 versus 80 to 100 nm), the characteristics of these DMVs corresponded with those described for cells infected with the distantly related arterivirus EAV (40, 67). Furthermore, in addition to the previously observed association with the rough ER (Fig. 4B), the outer membrane of the SARS-CoV DMVs was occasionally seen to be continuous with the outer membrane of a mitochondrion (Fig. 5D). Some of the observed profiles were reminiscent of the “protrusion and detachment” model, described by Pedersen et al. (40) as one of the possibilities for the formation of EAV DMVs (Fig. 5E).
FIG. 5.
EM analysis of SARS-CoV-infected Vero E6 cells (panels A, B, D, and E, 9 h p.i.; panel C, 6 h p.i.) cryofixed by high-speed plunge freezing in liquid ethane, a step followed by freeze substitution with 1% osmium tetroxide and 0.5% uranyl acetate in acetone and embedment in epoxy LX-112 resin. (A) Low-magnification overview of a region rich in virus-induced DMVs (arrows) and mitochondria (M). The interior of the virus-induced vesicles was strikingly different from that in the images presented in Fig. 4, and clear double membranes were now found to surround the structures. (B) Close-up of virions outside of the cell, with the spikes on the virion surface illustrating the general high quality of samples prepared using cryofixation. (C) Close-up of virus-induced DMVs, showing the double membrane of the structure and the high electron density of the interior compared to those shown in Fig. 4C. (D) Example of apparent continuity (arrow) between the outer membrane of a DMV and a mitochondrion (M), as was occasionally observed. (E) Example of a possible intermediate (arrow) in DMV formation, reminiscent of the previously proposed “protrusion and detachment” model (40). Bars, 250 nm (A, C, D, and E) and 100 nm (B).
Taken together, our data suggested that preparative procedures are critical during studies aimed at understanding the intracellular membrane changes that are thought to accompany the formation of the SARS-CoV RC. This was also true for the visualization of virus particles that were secreted from infected cells (Fig. 5A). In the cryofixed samples, these particles displayed strikingly well-preserved features, including clearly visible spikes (Fig. 5B), which were only rarely observed after conventional chemical fixation (reference 50 and references therein; also data not shown).
SARS-CoV nonstructural proteins localize to virus-induced DMVs.
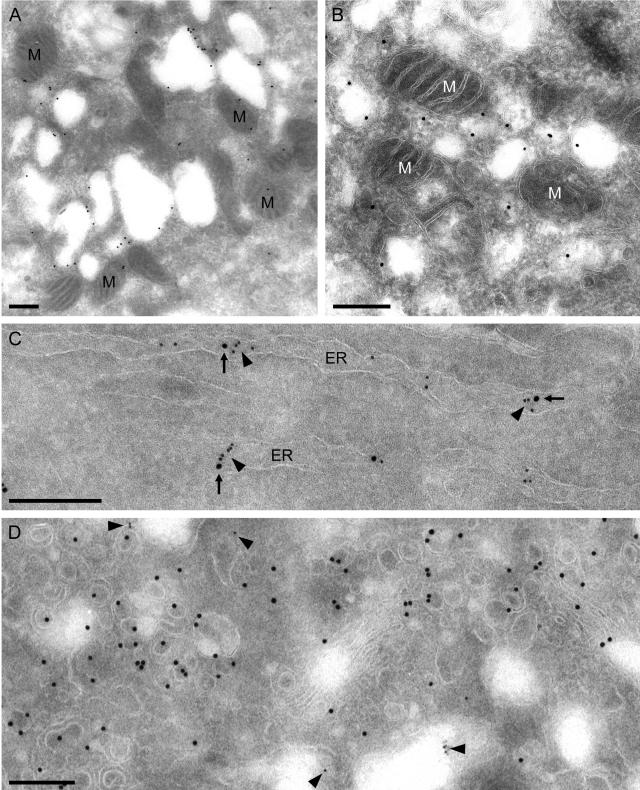
To establish the presence of SARS-CoV nonstructural proteins on the DMV structures, we employed IEM. Ultrathin cryosections of chemically fixed, SARS-CoV-infected Vero E6 cells were used for immunogold-labeling experiments. Unfortunately, this IEM protocol is not compatible with the cryofixation procedures that were employed for our morphology studies. Still, when the chemical fixation required for subsequent IEM was used, infected cells from 6 h p.i. onward showed vesicles similar to those found in the epoxy resin-embedded samples (Fig. 6A and B). Clusters of irregularly shaped vesicles were observed in the perinuclear area, and their size was comparable to those in the epoxy-embedded samples described above. Structures of this kind were not observed in mock-infected control cells (data not shown). However, the interior of the vesicles now appeared to be empty, probably due to the mild fixation procedure required for IEM. At the same time, the membranes and general morphology of other organelles in these specimens, like mitochondria (Fig. 6A and B), were well preserved. This indicated that the poor preservation of the (putative) DMVs is specific for these structures and that they require special processing for visualization at the ultrastructural level, in particular for subsequent immunolabeling studies.
FIG. 6.
IEM analysis of SARS-CoV-infected Vero E6 cells (panels A, B, and D, 9 h p.i.; panel C, 6 h p.i.). Ultrathin cryosections of chemically fixed, SARS-CoV-infected Vero E6 cells were used for immunogold-labeling experiments. Although this protocol was not compatible with the preservation of the interior of DMV-infected cells, many virus-induced vesicles were observed. (A) Cluster of irregularly shaped vesicles in the perinuclear area, which again also contained many mitochondria (M). The boundary of the structures could be labeled specifically using the αnsp3 serum and protein A-gold (15 nm). (B) Higher magnification of structures as shown in panel A but now labeled with the antiserum directed against the viral helicase (αnsp13). (C) Example of ER stacks double positive for nsp13 (visualized using 15-nm gold; arrows) and the cellular protein PDI (visualized using 10-nm gold; arrowheads). (D) Double labeling using the αnsp13 serum (visualized using 10-nm gold; arrowheads) and the αM serum (visualized using 15-nm gold). The αM serum labeled the Golgi area on the infected cell and new virus particles but did not label the vesicles that were positive for nonstructural proteins (and vice versa for the αnsp13 serum). Bar, 250 nm.
Our panel of SARS-CoV-specific rabbit antisera (Table 1) was used for IEM on this material. Positive results were obtained with the antisera recognizing nsp3, nsp8, nsp13, and M. The SARS-CoV-induced vesicles were specifically labeled with αnsp3 (Fig. 6A), αnsp8 (data not shown), and αnsp13 (Fig. 6B). In addition to these vesicles, structures presumed to be ER were positive for these nonstructural proteins. Such membranes could be double labeled for SARS-CoV nonstructural proteins and PDI, an ER marker protein (Fig. 6C), but no colocalization was found on the vesicles themselves. This may be explained either by the apparent loss of their interior contents upon use of the IEM protocol or by the fact that PDI is a luminal protein that may be (largely) excluded from the space between the two tightly apposed membranes that form a DMV.
In line with our IF observations on the separation between the labeling for SARS-CoV nonstructural proteins and that for M protein (Fig. 3), the αM serum did not label the vesicles that were positive for nonstructural proteins. Only Golgi stacks and virions were heavily labeled with this antiserum (Fig. 6D), supporting our conclusion that different membrane populations are involved in RC formation and virus assembly in SARS-CoV-infected cells.
A link between the SARS-CoV RC and the cellular autophagy pathway?
A recent study of MHV suggested a connection between coronavirus replication and autophagy (44), a cellular housekeeping process and stress response that results in the degradation and recycling of cytoplasmic constituents (22, 28). The initial autophagosome contains two distinct membranes, which was the primary basis for the proposed link to the DMVs implicated in MHV RNA synthesis (14). Also, work from the Kirkegaard laboratory has implicated autophagy in the formation of membrane vesicles associated with the poliovirus RC (20, 22, 53, 62), although data from Bienz and coworkers (48) suggested the ER as an alternative membrane source.
Prentice et al. (44) recently described that MHV replication induces autophagy and was seriously affected (>4-log-reduced progeny virus titers) in a knockout cell line for APG5, the gene encoding one of the key proteins in cellular autophagy (Apg5). Upon restoration of Apg5 synthesis by transfection of the knockout cell line with an expression plasmid, MHV replication was restored to normal levels, suggesting that the product of the gene is important during some stage of the virus life cycle. Upon MHV infection, EM studies revealed that the same autophagy-incompetent APG5 knockout cells lacked DMVs and developed hyperswollen membranes, presumably of ER origin. Prentice et al. (44) also reported that, in IF dual-labeling experiments, the staining for MHV nonstructural proteins showed significant overlap with that for two marker proteins of autophagic vacuoles, Apg12 and LC3. Subsequently, the same laboratory reported the colocalization of SARS-CoV nsp8 with LC3 (45). In particular, LC3, the homologue of Saccharomyces cerevisiae Apg8, is considered a marker protein for the autophagic pathway since it is known to be retained in autophagosomal membranes until their maturation is complete (21, 22, 34).
Unfortunately, in our hands, the use of the same αLC3 rabbit antisera used in the MHV studies resulted in IF images with a high background signal. A newly produced anti-human LC3B rabbit serum (Fig. 7A), raised using a synthetic peptide, performed slightly better, but as described by others (32), immunodetection of LC3 was often problematic. However, in the small number of SARS-CoV-infected cells that did show convincing LC3B labeling, the signal was completely separated from that of nsp3 (Fig. 7B), although it should be noted that the SARS-CoV nsp3 signal in general seemed to be less well developed in such cells.
The technical problems described above made us turn to the use of GFP-tagged LC3 produced from a CMV promoter-driven vector for transient expression. This approach to visualize autophagosomal structures has been well established (21, 32, 35, 64) and was recently used to show colocalization of GFP-LC3 and poliovirus replicase proteins (20). Also, a GFP fusion to LAMP1, a protein acquired by autophagosomes during the later stages of their maturation (see reference 22 and references therein), was used in these studies and found to localize to poliovirus-induced vesicles. He et al. (17) recently described the existence of three isoforms of human LC3, which were all concluded to localize to autophagic membranes and which displayed similar, punctate labeling patterns. Expression vectors for fusions of GFP to human LC3A, LC3B, and LC3C were generated and used for colocalization studies (Fig. 7C to E).
We transfected Vero E6 cells with expression plasmids for either GFP-LC3A, -LC3B, or -LC3C or LAMP1-GFP and, 5 h later, infected them with SARS-CoV. Expression of all four fusion proteins was observed, and the marker proteins localized to distinct cytoplasmic structures (Fig. 7C to F, GFP panels). Upon double labeling with antisera recognizing SARS-CoV nonstructural proteins, the separation of the marker proteins and viral replicase subunits was complete (Fig. 7C to F, overlay panels). For LAMP1, this result was confirmed independently by double labeling with a LAMP1-specific monoclonal antibody (data not shown).
Concluding remarks.
All mammalian positive-strand RNA viruses rely on membrane surfaces as the scaffold for their RC, but the structures induced and the compartment used as the membrane donor can apparently be quite different (1, 4, 30, 38, 51). How specific the interaction between a viral RC and such a membrane compartment really is remains to be established. In fact, it was recently reported that retargeting of the flock house virus RC from mitochondria to the ER was not only tolerated but surprisingly resulted in a sixfold increase of RNA replication efficiency (33). In this light, it clearly remains to be established to which extent the mechanisms for membrane association of the RC have been conserved between members of the highly diverged nidovirus group. In all nidoviruses (67), the ORF1a-encoded part of the replicase contains three conspicuous hydrophobic domains (located in nsp2, nsp3, and nsp5 in arteriviruses and in nsp3, nsp4, and nsp6 in coronaviruses), of which the last two flank the viral main proteinase. Due to ORF1a/ORF1b ribosomal frameshifting, these subunits are overexpressed relative to the key enzymatic domains encoded in ORF1b, which seems compatible with a “structural protein” role during the formation of structures that carry the RC. For the arterivirus EAV, coexpression of nsp2 and nsp3 suffices to induce DMVs, which are probably derived from the ER and strikingly resemble those induced in infected cells (60).
In comparative studies using Vero E6 cells, which can be infected with both viruses, we noticed a striking resemblance between the localization of EAV and SARS-CoV nonstructural proteins at the level of IF microscopy. In a double infection experiment, using dual labeling for their respective nsp3 proteins, both viruses were found to induce similar but distinct punctate labeling patterns, suggesting that each virus generated its own specific structures in the same region of the cell, while excluding the proteins of the other virus (data not shown). In our experience, the MHV RC staining pattern in mouse cell lines is somewhat different, but a direct comparison between EAV, SARS-CoV, and MHV has not yet been possible due to the lack of a cell line that can be infected with all three viruses. Consequently, also given the limited information available for other coronaviruses, the existence of significant differences between MHV and SARS-CoV can certainly not be excluded at present.
Also, another issue addressed in this paper, the separation between the RC and the site of virus assembly, points to possible differences between SARS-CoV and MHV. For the latter virus, relocalization of specific nonstructural proteins to the site of virus assembly was reported (5, 7), based on dual-labeling experiments involving the viral transmembrane protein M, a key determinant of virus assembly. In our studies with SARS-CoV (Fig. 3), we did not detect relocalization of nonstructural proteins (including the nsp13 helicase, which was reported to relocalize in the case of MHV) and observed an almost-complete separation of RC and M protein throughout infection. The separation of viral RNA synthesis and virion assembly was further supported by the complete separation of SARS-CoV nonstructural protein labeling and markers for the ERGIC (Fig. 3B) and the Golgi complex (19). Our EM studies also supported the separation of DMVs and assembling viruses and of marker proteins thought to be specific for both processes (Fig. 6).
In the case of MHV, disruption of the autophagic pathway had a pronounced effect on virus replication as a whole. Autophagic membranes were implicated in DMV formation, in particular because viral nonstructural proteins were found to colocalize with the autophagic marker protein LC3 (44). For SARS-CoV, Prentice et al. (45) reported a similar colocalization between LC3 and nsp8, a protein that colocalizes with a variety of other nonstructural proteins, like nsp2 and nsp3 (45) and nsp5, nsp9, nsp12, nsp13, and nsp15 (this study). However, using an approach based on recent studies aimed at identifying the origin of poliovirus-induced RC-containing vesicles (20), we did not obtain evidence for colocalization of LC3 or GFP-LC3 with the SARS-CoV RC (Fig. 7). In fact, our observations were in line with our previous conclusion (19), based on IF microscopy studies using ER marker proteins, that the staining for nsp13 shows a significant overlap with the ER. This result has now been confirmed at the EM level (Fig. 6C), and also our morphological observations (Fig. 4B) suggest a link between SARS-CoV DMVs and the ER. The nucleation of autophagosomes and the origin of autophagic membranes are poorly understood (see references 22 and 28 and references therein), and a role for the ER in these aspects of autophagy is considered likely, thus leaving the possibility of a (direct or indirect) link between ER membranes, DMVs, and autophagosomes. Still, in our opinion, the fact that membranes positive for SARS-CoV nonstructural proteins appear to lack a specific autophagic marker, like LC3, but do contain an ER-resident protein, like PDI, argues against the involvement of the autophagic pathway in DMV formation.
Finally, an important technical aspect of this study concerns the fragility of the SARS-CoV DMVs. Although the corresponding structures of EAV and MHV could be readily visualized following conventional fixation and embedment, preservation of SARS-CoV DMVs required high-speed plunge freezing and freeze substitution (Fig. 5). In particular, the inner membrane and DMV contents, which were likely converted into the spider web-like structure upon conventional fixation (Fig. 4C), appear to be very sensitive. Previously, for MHV, the double membrane could be visualized following conventional fixation, but DMVs appeared to be empty (14). The fragility of coronavirus DMVs is reminiscent of studies with poliovirus reporting that special fixation protocols are required to visualize DMVs induced by this virus (53). For SARS-CoV and other nidoviruses, this also leaves the possibility that DMVs are derived from structures and/or intermediates that may only be detected using advanced cryofixation procedures, a possibility that is currently being investigated in our laboratory. In combination with the ongoing biochemical dissection of nidovirus RCs and their associated membrane structures, these studies may provide more definitive evidence concerning origin, morphogenesis, and composition of these virus-induced structures. Moreover, they may reveal why these nidovirus RC-carrying vesicles have a double membrane and where on these structures viral RNA synthesis occurs.
Acknowledgments
We are indebted to Sjoerd van den Worm, Marjolein Kikkert, and the staff of the LUMC animal facility for their assistance with rabbit antiserum production. We thank Christian Cambillau, Bruno Canard, and coworkers (AFMB, CNRS, and University of Marseille, France) and John Ziebuhr and coworkers (Würzburg University, Germany) for providing some of the antigens used in this study. We are grateful to Stephane Méresse (University of Marseille, France), Karla Kirkegaard and William Jackson (Stanford University), and Tamotsu Yoshimori and Takahiro Kamimoto (National Institute of Genetics, Japan) for generously providing expression vectors and antisera used in this study and for helpful discussions. We are grateful to Frans Prins (LUMC Department of Pathology) for assistance with confocal microscopy. We acknowledge the continued support of Peter Bredenbeek, Alexander Gorbalenya, Willy Spaan, and Gijsbert van Willigen for SARS-CoV research at LUMC.
This work was supported (in part) by the European Commission in the context of the activities of the Euro-Asian SARS-DTV Network (SP22-CT-2004-511064).
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CL(pro)) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Bhardwaj, K., L. A. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boucrot, E., C. R. Beuzon, D. W. Holden, J. P. Gorvel, and S. Meresse. 2003. Salmonella typhimurium SifA effector protein requires its membrane- anchoring C-terminal hexapeptide for its biological function. J. Biol. Chem. 278:14196-14202. [DOI] [PubMed] [Google Scholar]
- 7.Brockway, S. M., X. T. Lu, T. R. Peters, T. S. Dermody, and M. R. Denison. 2004. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 78:11551-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, C. A. M., and P. J. M. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64:165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. M. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. M. E. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 10.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouchier, R. A. M., T. Kuiken, M. Schutten, G. van Amerongen, J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. M. E. Osterhaus. 2003. Aetiology—Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarino, L. A., K. Bhardwaj, W. Dong, J. Sun, A. Holzenburg, and C. C. Kao. 2005. Mutational analysis of the SARS virus nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 353:1106-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, H., Y. Dang, F. Dai, Z. Guo, J. Wu, X. She, Y. Pei, Y. Chen, W. Ling, C. Wu, S. Zhao, J. O. Liu, and L. Yu. 2003. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J. Biol. Chem. 278:29278-29287. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, W. T., T. H. Giddings, M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabeya, Y., N. Mizushima, T. Uero, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirkegaard, K., M. P. Taylor, and W. T. Jackson. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2:301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klumperman, J., J. Krijnse Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. M. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krijnse Locker, J., M. Ericsson, P. J. M. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krijnse Locker, J., G. Griffiths, M. C. Horzinek, and P. J. M. Rottier. 1992. O-glycosylation of the coronavirus M protein. Differential localization of sialyltransferases in N-linked and O-linked glycosylation. J. Biol. Chem. 267:14094-14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. X. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. Leduc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 27.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Levine, B., and D. J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6:463-477. [DOI] [PubMed] [Google Scholar]
- 29.Machamer, C. E., S. A. Mentone, J. K. Rose, and M. G. Farquhar. 1990. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc. Natl. Acad. Sci. USA 87:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffin, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. E. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 32.Martinet, W., G. R. De Meyer, L. Andries, A. G. Herman, and M. M. Kockx. 2006. In situ detection of starvation-induced autophagy. J. Histochem. Cytochem. 54:85-96. [DOI] [PubMed] [Google Scholar]
- 33.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 77:12193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima, N. 2004. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 36:2491-2502. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima, N., A. Yamamoto, M. Matsui, T. Yoshimori, and Y. Ohsumi. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nal, B., C. M. Chan, F. Kien, L. Siu, J. Tse, K. Chu, J. Kam, I. Staropoli, B. Crescenzo-Chaigne, I. Escriou, S. van der Werf, K. Y. Yuen, and R. Altmeyer. 2005. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 86:1423-1434. [DOI] [PubMed] [Google Scholar]
- 37.Ng, M. L., S. H. Tan, E. E. See, E. E. Oi, and A. E. Ling. 2003. Proliferative growth of SARS coronavirus in Vero E6 cells. J. Gen. Virol. 84:3291-3303. [DOI] [PubMed] [Google Scholar]
- 38.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasternak, A. O., W. J. M. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [Online.] doi 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peiris, J. S. M., Y. Guan, and K. Y. Yuen. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88-S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peti, W., M. A. Johnson, T. Herrmann, B. W. Neuman, M. J. Buchmeier, M. Nelson, J. Joseph, R. Page, R. C. Stevens, P. Kuhn, and K. Wuthrich. 2005. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 79:12905-12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prentice, E., J. McAuliffe, X. T. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Putics, A., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1′′-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79:12721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. X. Tong, A. Tamin, L. Lowe, M. Frace, J. L. Derisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 48.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. J. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsp3. Structure (Cambridge) 13:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salanueva, I. J., J. L. Carrascosa, and C. Risco. 1999. Structural maturation of the transmissible gastroenteritis coronavirus. J. Virol. 73:7952-7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salonen, A., T. Ahola, and L. Kaariainen. 2004. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective, p. 31-55. In L. Enjuanes (ed.), Coronavirus replication and reverse genetics. Springer, Berlin, Germany.
- 53.Schlegel, A., T. H. J. Giddings, M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweizer, A., J. A. M. Fransen, T. Bachi, L. Ginsel, and H. P. Hauri. 1988. Identification, by a monoclonal antibody, of a 53-Kd protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J. Cell Biol. 107:1643-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siddell, S. G., J. Ziebuhr, and E. J. Snijder. 2005. Coronaviruses, toroviruses, and arteriviruses, p. 823-856. In B. W. Mahy and V. ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections, vol. 1. Virology. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 58.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snijder, E. J., S. G. Siddell, and A. E. Gorbalenya. 2005. The order Nidovirales, p. 390-404. In B. W. Mahy and V. ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections, vol. 1 Virology. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 60.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 61.Snijder, E. J., A. L. M. Wassenaar, and W. J. M. Spaan. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 68:5755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suhy, D. A., T. H. Giddings, and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure (Cambridge) 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanida, I., Y. S. Sou, J. Ezaki, N. Minematsu-Ikeguchi, T. Ueno, and E. Kominami. 2004. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABA(A) receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 279:36268-36276. [DOI] [PubMed] [Google Scholar]
- 65.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 66.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. M. Spaan, and J. Krijnse Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaux, D., J. Tooze, and S. Fuller. 1990. Identification by anti-idiotypic antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature 345:495-502. [DOI] [PubMed] [Google Scholar]
- 69.von Grotthuss, M., L. S. Wyrwicz, and L. Rychlewski. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 16 October 2005, posting date. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, Q. F., J. M. Cui, X. J. Huan, H. Y. Zheng, J. H. Huang, F. Ling, K. P. Li, and J. Q. Zhang. 2004. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 73:332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziebuhr, J. 2004. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 7:412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]