Abstract
H5N1 influenza A viruses are widely distributed among poultry in Asia, but until recently, only a limited number of wild birds were affected. During late April through June 2005, an outbreak of H5N1 virus infection occurred among wild birds at Qinghai Lake in China. Here, we describe the features of this outbreak. First identified in bar-headed geese, the disease soon spread to other avian species populating the lake. Sequence analysis of 15 viruses representing six avian species and collected at different times during the outbreak revealed four different H5N1 genotypes. Most of the isolates possessed lysine at position 627 in the PB2 protein, a residue known to be associated with virulence in mice and adaptation to humans. However, neither of the two index viruses possessed this residue. All of the viruses tested were pathogenic in mice, with the exception of one index virus. We also tested the replication of two viruses isolated during the Qinghai Lake outbreak and one unrelated duck H5N1 virus in rhesus macaques. The Qinghai Lake viruses did not replicate efficiently in these animals, producing no evidence of disease other than transient fever, while the duck virus replicated in multiple organs and caused symptoms of respiratory illness. Importantly, H5N1 viruses isolated in Mongolia, Russia, Inner Mongolia, and the Liaoning Province of China after August 2005 were genetically closely related to one of the genotypes isolated during the Qinghai outbreak, suggesting the dominant nature of this genotype and underscoring the need for worldwide intensive surveillance to minimize its devastating consequences.
Highly pathogenic H5N1 influenza viruses have repeatedly caused serious outbreaks of disease at poultry farms since 1997 and pose a significant threat to human health due to their ability to infect humans, resulting in high mortality (3, 16, 20). Since late 2003, H5N1 viruses have spread in an unprecedented manner across Asia, resulting in more than 60 human fatalities in Thailand, Vietnam, Cambodia, and Indonesia and in the slaughter or infectious deaths of more than 150 million birds. Despite extensive efforts to contain these outbreaks, H5N1 viruses continue to circulate among poultry in Asia (Office International des Epizooties [http://www.oie.int]) and remain a threat to both veterinary and human public health. In fact, the viruses have now spread to Europe, increasing the likelihood of an H5N1 pandemic.
Wild aquatic birds harbor all 16 hemagglutinin (HA) and all nine neuraminidase (NA) subtypes of influenza A virus and therefore serve as the natural reservoir for this pathogen. Although influenza viruses in wild aquatic birds are occasionally transmitted to avian (e.g., chickens and turkeys) and mammalian (e.g., humans, pigs, horses, minks, whales, and seals) species, where they may produce outbreaks of severe disease, they persist in evolutionary equilibrium (stasis) in their natural reservoir and do not generally cause disease in wild waterfowl (22).
Highly pathogenic H5N1 viruses do not appear to have entered the wild-bird populations to any appreciable extent until late April to June 2005, when a large outbreak of H5N1 infection occurred in Qinghai Lake in western China (2, 12), a major breeding site for migratory birds whose flyways extend to Southeast Asia, India, Siberia, Australia, and New Zealand (4). Initial reports (2, 12) of this outbreak identified a single introduction of an H5N1 virus into four species of waterfowl, including bar-headed geese (Anser indicus), brown-headed gulls (Larus brunnicephalus), great black-headed gulls (Larus ichthyaetus), and great cormorants (Phalacrocorax carbo). The virus was shown to be pathogenic in chickens and mice and was shown to possess a Lys-to-Glu substitution at position 627 of PB2, an alteration previously associated with high virulence in mice and found only in H1N1 and H3N2 human isolates as well as some H5N1 isolates from humans and tigers (7, 9, 10, 19, 23) but not from wild birds.
In this report, we provide information on the magnitude of the Qinghai Lake outbreak of H5N1 viral disease, the evolution of the viruses during the course of the outbreak, subsequent transmission to birds in other remote locations, and the pathological properties of the viruses in animal models, including nonhuman primates.
MATERIALS AND METHODS
Virus isolation and identification.
Organ samples and cloacal swabs were collected within 24 h from birds that had died and were inoculated into 10-day-old embryonated specific-pathogen-free eggs for virus isolation, as described previously (1). The HA and NA subtypes were determined by conventional hemagglutination inhibition and neuraminidase inhibition tests as described previously (15). All experiments with the H5N1 isolates were performed in a biosafety level 3 laboratory, and animal experiments were performed in high-efficiency particulate air-filtered isolators. Abbreviations of the isolates are given in Table 1.
TABLE 1.
H5N1 viruses isolated from wild birds in Qinghai Lake in the present study
| Virus | Isolation datea | Abbreviation | Genotype |
|---|---|---|---|
| A/bar-headed goose/Qinghai/1/05 | 5/5/05 | BHGs/QH/1/05 | A |
| A/bar-headed goose/Qinghai/2/05 | 5/5/05 | BHGs/QH/2/05 | B |
| A/bar-headed goose/Qinghai/3/05 | 5/22/05 | BHGs/QH/3/05 | C |
| A/bar-headed goose/Qinghai/4/05 | 5/22/05 | BHGs/QH/4/05 | C |
| A/bar-headed goose/Qinghai/5/05 | 5/22/05 | BHGs/QH/5/05 | C |
| A/bar-headed goose/Qinghai/8/05 | 5/22/05 | BHGs/QH/8/05 | C |
| A/bar-headed goose/Qinghai/9/05 | 5/22/05 | BHGs/QH/9/05 | C |
| A/bar-headed goose/Qinghai/10/05 | 5/22/05 | BHGs/QH/10/05 | C |
| A/brown-headed gull/Qinghai/1/05 | 5/22/05 | BHGull//QH/1/05 | C |
| A/great black-headed gull/ Qinghai/1/05 | 5/22/05 | GBHGull/QH/1/05 | C |
| A/great black-headed gull/ Qinghai/2/05 | 5/22/05 | GBHGull/QH/2/05 | C |
| A/great black-headed gull/ Qinghai/3/05 | 5/22/05 | GBHGull/QH/3/05 | C |
| A/great cormorant/Qinghai/3/05 | 5/22/05 | GC/QH/3/05 | C |
| A/whooper swan/Qinghai/1/05 | 6/16/05 | WS/QH/1/05 | C |
| A/ruddy shelduck/Qinghai/1/05 | 7/16/05 | RS/QH/1/05 | D |
Isolation date is shown as month/day/year.
Genetic and phylogenetic analyses.
Viral RNA was extracted with the RNeasy Mini kit (QIAGEN, Valencia, CA) and was reverse transcribed. PCR amplification was performed by using segment-specific primers (primer sequences are available upon request). The PCR products were purified with the QIAquick PCR purification kit (QIAGEN) and sequenced by using the CEQ DTCS-Quick Start kit on a CEQ 8000 DNA sequencer (Beckman Coulter). Sequence data were compiled with the SEQMAN program (DNASTAR, Madison, WI), and the phylogenetic analysis was carried out with the PHYLIP program of the CLUSTALX software package (version 1.81), implementing a neighbor-joining algorithm.
Animal experiments.
The intravenous pathogenicity index (IVPI) of the isolates in chickens was determined according to the recommendations of the Office International des Epizooties (14). Briefly, groups of 10 specific-pathogen-free 6-week-old White Leghorn chickens housed in isolator cages were inoculated intravenously with 0.2 ml of a 1:10 dilution of bacterium-free virus-containing allantoic fluid (see Table 2 for titers of the isolates).
TABLE 2.
Lethality of the H5N1 avian influenza isolates in chickens and mice
| Virus | Titer of virus stock (log10 EID50/ml) | IVPIc | Virus titers in organs of mice (log10 EID50/ml ± SD)a
|
MLD50 (log EID50)b | |||
|---|---|---|---|---|---|---|---|
| Lung | Spleen | Brain | Kidney | ||||
| BHGs/QH/1/05 | 6.7 | 3.0 | 5.7 ± 0.6 | 1.3 ± 0.1 | 2.2 ± 0.3 | 1.8 ± 0.2 | <0.5 |
| BHGs/QH/2/05 | 6.3 | 3.0 | 4.3 ± 0.1 | + | < | < | >6.0 |
| BHGs/QH/3/05 | 8.5 | 3.0 | 6.3 ± 0.9 | 2.0 ± 0.3 | 2.9 ± 0.7 | 2.5 ± 0.3 | <0.5 |
| BHGull/QH/1/05 | 7.3 | 3.0 | 5.6 ± 0.7 | 1.6 ± 0.3 | 1.6 ± 0.1 | 1.6 ± 0.1 | <0.5 |
| GBHGull/QH/1/05 | 7.0 | 3.0 | 6.1 ± 0.2 | 1.6 ± 0.3 | 1.6 ± 0.1 | 1.8 ± 0.2 | <0.5 |
| GC/QH/3/05 | 7.8 | 3.0 | 5.9 ± 0.5 | 1.3 ± 0.0 | 1.6 ± 0.2 | 1.6 ± 0.2 | <0.5 |
| WS/QH/1/05 | 7.5 | 3.0 | 6.5 ± 0.7 | 1.5 ± 0.1 | 1.5 ± 0.1 | 1.8 ± 0.4 | <0.5 |
| RS/QH/1/05 | 7.3 | 3.0 | 6.7 ± 1.0 | 3.3 ± 0.3 | 2.9 ± 0.6 | 3.3 ± 0.3 | <0.5 |
Six-week-old BALB/c female mice were infected intranasally with 106 EID50 of virus in a 50-μl volume. Three mice from each group were euthanized on day 3 postinoculation, and virus in the organs was titrated in eggs. SD, standard deviation; +, virus was detected only in undiluted samples; <, virus was not detected.
The MLD50 dose was determined by inoculating groups of five 6-week-old female mice intranasally with 10-fold serial dilutions of each virus containing doses ranging from 101 to 106 EID50 in 50 μl.
The IVPI was determined according to recommendations of the Office International des Epizooties (14).
Groups of eight 7-week-old female BALB/c mice (Beijing Experimental Animal Center, Beijing, People's Republic of China) were lightly anesthetized with CO2 and inoculated intranasally with 106 50% egg infectious doses (EID50) of the virus in a volume of 50 μl. On day 3, three of the eight mice were euthanized for virus titration. The remaining five mice were monitored daily for weight loss and mortality. To determine the 50% mouse lethal doses (MLD50) of the viruses, we inoculated six groups of mice (n = 5 mice each) intranasally with 10-fold serial dilutions of the virus containing 101 to 106 EID50 in a 50-μl volume (1). The MLD50 was calculated by the method of Reed and Muench (17).
Three-year-old, colony-bred, female rhesus macaques (Macaca mulatta) were individually placed in a negative-pressure isolator. The macaques were infected intranasally with 107 EID50 of the viruses in 2 ml of phosphate-buffered saline (four animals infected with A/bar-headed goose/Qinghai/1/05 [BHGs/QH/1/05], four animals infected with A/great cormorant/Qinghai/3/05 [GC/QH/3/05], and three animals infected with A/duck/Guangxi/35/2001 [H5N1] [DK/GX/35/01]). One animal from each group was euthanized on days 4 and 7 postinfection by exsanguination under ketamine anesthesia. Nasal swabs, bronchoalveolar lavage specimens, and organs from the euthanized animals were collected for virus titration and for histological and immunohistochemical examinations. The remaining animals were observed for 2 weeks. Sera collected from all animals prior to infection and 2 weeks postinfection were used to detect anti-influenza virus antibodies.
Histopathological studies.
Samples of the lung, liver, spleen, kidney, pancreas, heart, muscle, trachea, proventriculus, and large intestine from naturally infected bar-headed geese and samples of tonsil, lung, liver, spleen, and heart from nonhuman primates experimentally infected with the H5N1 viruses were fixed in 10% neutral buffered formalin solution at necropsy. They were dehydrated, embedded in paraffin, and cut into 5-μm sections, which were then stained with routine hematoxylin and eosin (HE). For viral antigen detection, sections were processed for immunohistochemistry by a two-step dextran polymer method (DAKO Japan Inc., Kyoto, Japan) using polyclonal rabbit antibody to an H5 virus. Nonhuman primate tissues infected with DK/GX/35/01 were subjected to immunostaining to detect viral antigens; in these studies, polyclonal anti-H5 goat serum was used as a primary antibody, and peroxidase-labeled anti-goat rabbit serum served as a secondary antibody. Immunoreactions were visualized with diaminobenzidine.
Nucleotide sequence accession numbers.
The nucleotide sequences analyzed in this study are available at the Influenza Sequence Database under accession numbers ISDN137990 to ISDN138151.
RESULTS
Course of the avian influenza outbreak at Qinghai Lake.
The islets and wetlands of Qinghai Lake, located in western China, are part of a protected natural reserve for wild birds. More than 100,000 wild birds, representing 189 species, spend the spring and summer at this reserve every year. Since the end of April 2005, bar-headed geese arriving at Qinghai Lake from southern Asia have shown signs of disease, including tremor and torticollis.
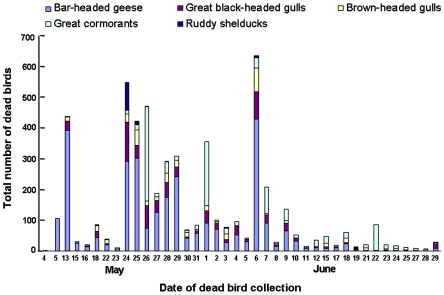
On 4 May 2005, two bar-headed geese (Anser indicus) were found dead in the wetlands of Qinghai Lake, with 105 dead geese reported on the following day (Fig. 1 and 2). On 13 May, a total of 437 dead birds were collected. The species identified extended to great black-headed gulls (Larus ichthyaetus) and brown-headed gulls (Larus brunnicephalus), whose habitats on the lake overlap closely with those of bar-headed geese. Disease signs and deaths were observed among ruddy shelducks (Tadorna ferruginea) beginning on 13 May, with 90 and 12 dead shelducks collected on 24 and 25 May, respectively. A limited number of dead great cormorants (Phalacrocorax carbo), gathered on two islets located 2 miles away from concentrations of bar-headed geese and gulls, were first observed on 16 May, and a large number of these birds were found dead on 24 to 26 May and 1 June (Fig. 1 and 2). Altogether, 6,184 dead gulls, geese, great cormorants, and ruddy shelducks were found from 4 May to 29 June; bar-headed geese accounted for more than half of this total. A limited number of whooper swans (Cygnus cygnus), black-headed cranes (Grus nigricollis), and pochards (Aythya ferina) also died during this outbreak.
FIG. 1.
Course of migratory waterfowl deaths due to H5N1 viruses at Qinghai Lake. A total of 6,184 dead birds were collected from 4 May to 29 June 2005: 3,282 bar-headed geese, 929 great black-headed gulls, 570 brown-headed gulls, 1,302 great cormorants, and 145 ruddy shelducks.
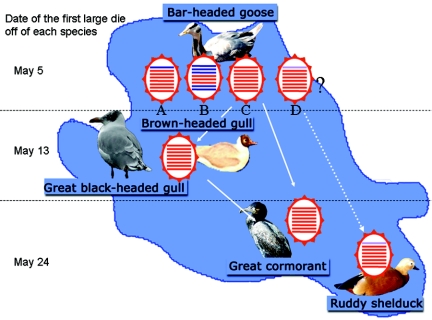
FIG. 2.
Pattern of spread of H5N1 viruses of different genotypes among wild birds at Qinghai Lake. The disease began in bar-headed geese, spread to brown-headed gulls and great black-headed gulls, and then spread to great cormorants and ruddy shelducks. The viruses from these species represent four genotypes, two of which appear to have spread from bar-headed geese to the other three avian species, although it is uncertain whether the genotype D virus originated in the bar-headed goose population.
Pathological examination.
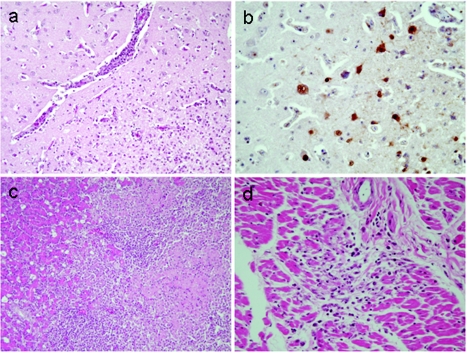
We observed severe hyperemia and edema in the brain, hemorrhage and necrosis in the pancreas, and severe cloudy swelling of the kidneys in the bar-headed geese, great black-headed gulls, brown-headed gulls, and great cormorants that were examined immediately after death. Histologic analysis of organ samples from two moribund bar-headed geese revealed typical nonsuppurative encephalitis characterized by perivascular cuffing of mononuclear cells, microgliosis, degeneration of nerve cells, and edema (Fig. 3a), all indicative of viral infection. Influenza virus infection was confirmed by immunohistochemical studies with antibodies to an H5 virus (Fig. 3b). There was also evidence of coagulative necrosis with nonsuppurative inflammation in the pancreas (Fig. 3c) as well as degenerative foci of cardiomyocytes with nonsuppurative inflammation (Fig. 3d).
FIG. 3.
Histopathologic analysis of a moribund bar-headed goose from the H5N1 virus outbreak at Qinghai Lake. (a) Brain showing scattered nonsuppurative inflammatory foci characterized by perivascular cuffing of mononuclear cells, microgliosis, degeneration of nerve cells, and edema (HE stain). (b) Brain with numerous nerve and glial cells positive for viral antigen (brown pigments) by immunohistological staining with an anti-H5 polyclonal antibody. (c) Pancreas showing scattered coagulative necrotic foci in parenchyma with nonsuppurative inflammation (HE stain). (d) Heart showing small nonsuppurative inflammatory foci with degenerating cardiomyocytes (HE stain).
Virus isolation and identification.
To isolate the virus, we inoculated cloacal swab samples and homogenates of organs from sick and dead birds into 10-day-old embryonated specific-pathogen-free chicken eggs. Fifteen H5N1 viruses were isolated from multiple organs (brain, lung, spleen, kidney, and intestine) and cloacal swabs of bar-headed geese, a brown-headed gull, a great cormorant, great black-headed gulls, and a whooper swan and from the feces of a ruddy shelduck (Table 1).
Sequence analysis.
To understand the genetic relationship between the Qinghai Lake isolates and other H5N1 viruses, we sequenced the entire genomes of the 15 isolates. Previously published sequences of H5N1 viruses isolated from the same outbreak by Chen et al. (2) and Liu et al. (12) were included in the analysis for comparison. The HA, NA, and nucleoprotein genes of all viruses isolated during the Qinghai Lake outbreak were similar to each other and closely resembled those of the A/chicken/Jiangxi/25/04 virus isolated during the 2004 outbreak in China (Fig. 4; also see Fig. S1 and S2a and b in the supplemental material). As reported previously (2, 12), all of these viruses had a series of basic amino acids at the HA cleavage site (RRRKKR) characteristic of other influenza viruses that are highly pathogenic in chickens; they also had a 20-amino-acid deletion in the NA stalk (residues 49 to 68) compared with the NA of the Goose/Guangdong/1/96 virus.
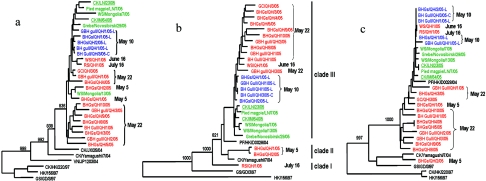
FIG. 4.
Phylogenetic analyses of the H5N1 viruses isolated during the Qinghai Lake outbreak. The phylogenetic trees were generated with the PHYLIP program of the CLUSTALX software package (version 1.81) by using the neighbor-joining algorithm and bootstrap values of 1,000. (a) HA (nucleotides 105 to 1659); (b) PB2 (nucleotides 82 to 2264); (c) PA (nucleotides 67 to 2151). The phylogenetic tree of HA was rooted to A/mallard/Denmark/64650/03 (H5N7), and the PB2 and PA phylogenetic trees were rooted to A/Memphis/1/90 (H3N2). The sequences of the wild-bird viruses obtained in this study are shown in red, and the viruses isolated after the Qinghai Lake outbreak are shown in green, while those of the wild-bird viruses reported previously by Liu et al. and Chen et al. are shown in blue. Dates of virus isolation during Qinghai Lake outbreak are also shown.
Unlike previous reports (2, 12), the PB2 genes of viruses isolated during the Qinghai Lake outbreak differed from each other (Fig. 4b). The PB2 genes of BHGs/QH/1/05 and A/bar-headed goose/Qinghai/2/05 (BHGs/QH/2/05) viruses shared 99.8% identity but were less than 97% identical to those of the other isolates. Phylogenetically, the PB2 genes of the Qinghai isolates were divided into three clades. The A/ruddy shelduck/Qinghai/1/05 (RS/QH/1/05) virus formed a clade by itself; BHGs/QH/1/05 and BHGs/QH/2/05, both isolated early in the Qinghai Lake outbreak, formed the second clade with A/peregrine falcon/Hong Kong/04 and A/chicken/Yamaguchi/7/04 (CK/Yamaguchi/7/04) viruses; and the remaining isolates, including those reported by previously by Chen et al. (2) and Liu et al. (12), formed the third clade (Fig. 4b). The PB2 protein of all the Qinghai Lake viruses in the last clade had a lysine at position 627, which is conserved in authentic human viruses and is associated with the high virulence of H5N1 viruses in mice (8). Interestingly, however, two of the index isolates in the second clade, together with other viruses in this clade and in clade 1, contain glutamic acid at position 627.
The phylogenetic trees of the PA (Fig. 4c), PB1, M, and NS genes (see Fig. S2c to e in the supplemental material) of these H5N1 viruses were similar to each other. BHGs/QH/2/05 and CK/Yamaguchi/7/04 were closely related to each other and formed a single clade, while the remaining viruses formed another clade. The PA gene of the BHGs/QH/2/05 virus shared less than 93% identity with other wild-bird viruses isolated at Qinghai Lake but was 98.2% identical to the corresponding gene of the CK/Yamaguchi/7/04 virus. The PB1 gene of the BHGs/QH/2/05 virus shared less than 93% identity with the other wild-bird viruses but was 98.7% identical to that of the CK/Yamaguchi/7/04 virus. Thus, the Qinghai Lake isolates represented four genotypes, genotypes A to D (Fig. 2). The BHGs/QH/2/05 (genotype B) isolate shares the PB2 gene with genotype A viruses, but four of its other genes appear to be unique among the wild-bird viruses detected in this study.
Experimental infection of chickens, mice, and nonhuman primates.
Using chickens, we next tested the pathogenicity of eight H5N1 viruses, including at least one virus of each genotype, according to recommendations of the Office International des Epizooties (14). All viruses killed chickens within 24 h and had an intravenous pathogenicity index of 3.0, the highest value possible (Table 2). Similarly, in mice, all isolates, with the exception of BHGs/QH/2/05, were highly lethal when administered intranasally with an MLD50 of less than 0.5 log EID50 (Table 2). This group of lethal viruses was readily recovered from each of the organs tested, indicating its ability to cause systemic infection.
To test the pathogenic potential of these isolates in primates, we intranasally infected rhesus macaques with 107 EID50s of BHGs/QH/1/05 or GC/QH/3/05. Half of the animals infected with either virus showed an increased body temperature for 1 to 3 days (see Fig. S3a and b in the supplemental material) and anorexia on day 1 or 2 postinfection. Increased respiratory rates were observed on days 5 to 6 postinfection in all animals, but none died or showed severe symptoms during the 2-week observation period. Surprisingly, virus was not recovered by nasal swabs, lung lavages, or organ samples collected on day 4 or 7 postinfection from animals infected with either virus (Table 3), even though sera collected at 2 weeks postinfection had antibody titers of more than 1:2,000 as determined by an enzyme-linked immunosorbent assay.
TABLE 3.
Titers of viruses isolated from rhesus macaques infected with H5N1 avian influenza virusesa
| Virus | Days postinfection | Virus titers in organs of nonhuman primates (log10 EID50/ml)a
|
||||||
|---|---|---|---|---|---|---|---|---|
| Nasal swab | Lung lavage | Tonsil | Lung | Spleen | Liver | Heart | ||
| BHGs/QH/1/05 | 4 | < | < | < | < | < | < | < |
| 7 | < | < | < | < | < | < | < | |
| GC/QH/3/05 | 4 | < | < | < | < | < | < | < |
| 7 | < | < | < | < | < | < | < | |
| DK/GX/35/01 | 4 | 2.5 | 3.5 | ND | 1.5 | 4.5 | + | 1.3 |
| 7 | < | 3.8 | ND | 1.5 | < | < | < | |
Three-year-old, colony-bred, female rhesus macaques (Macaca mulatta) were infected intranasally with 107 EID50 of the viruses in 2 ml of phosphate-buffered saline. One animal from each group was euthanized on days 4 and 7 postinfection by exsanguination under ketamine anesthesia. Nasal swabs, bronchoalveolar lavage specimens, and organs from the euthanized animals were collected for virus titration. <, virus was not detected; ND, not determined; +, virus was detected only in the original samples.
By contrast, when infected with the DK/GX/35/01 H5N1 virus isolated from a healthy duck in Guangxi Province in 2001 during routine surveillance (1, 11), all three macaques developed fever within the first 2 days postinfection (see Fig. S3c in the supplemental material) and became anorexic on day 1. Two of the animals recovered on day 3, while the third animal remained ill until day 6. The DK/GX/35/01 virus was isolated from nasal swabs, lung lavages, and samples of lung, spleen, liver, and heart from animals euthanized on day 4 and from the lungs of animals euthanized on day 7, but it was not recovered from other organs tested (cerebellum, cerebrum, kidney, pancreas, intestines, inguinal lymph nodes, mesenteric lymph gland, and blood). These findings confirm the susceptibility of rhesus macaques to avian H5N1 virus infection and indicate that the DK/CX/35/01 virus, but not BHGs/QH/1/05 and GC/QH/3/05, can cause systemic disease in nonhuman primates.
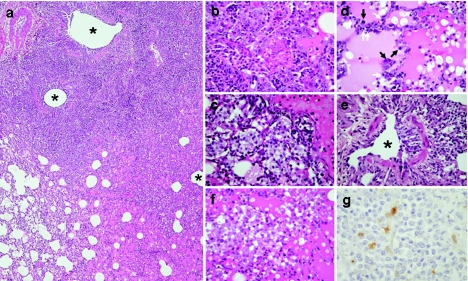
Necropsy of macaques on day 7 postinfection revealed a spectrum of macroscopic lesions in the lungs. A GC/QH/3/05 virus-infected animal had focal discoloration in the medial lobe, and large foci of consolidation on the accessory and lower lobes in a BHGs/QH/1/05 virus-infected animal were seen, together with prominent swelling of the lymph nodes, while extensive pulmonary consolidation was apparent on the medial and lower lobes in DK/GX/35/01 virus-infected animals. Histologically, the consolidated area seen in the BHGs/QH/1/05-infected macaque was consistent with prominent features of bronchointerstitial pneumonia with massive recruitment of lymphocytes (Fig. 5a). Prominent features within and on the periphery of the lesions included proliferative and reactive hyperplasia of alveolar cells (Fig. 5b), an accumulation of foamy macrophages in alveolar spaces (Fig. 5c), severe alveolar edema (Fig. 5d), and thickening of alveolar walls with lymphocyte recruitment (Fig. 5d, arrows). In one of the animals infected with the DK/GX/35/01 virus, foci of peribronchiolitis detected at 4 days postinfection (Fig. 5e) had progressed to a massive accumulation of foamy macrophages within alveolar spaces and severe alveolar edema by 7 days postinfection (Fig. 5f), but lymphocyte recruitment into the alveolar wall and regenerative changes of alveolar cells were rare compared with findings in the BHGs/QH/1/05-infected macaque. In sharp contrast to these observations, the lungs of animals infected with the GC/QH/3/05 virus had only small lesions consisting of focal alveolitis accompanied by hyperplasia of alveolar cells. In extrapulmonary organs, suppurative tonsillitis and systemic activation of lymph follicles were prominent by 4 days postinfection in the animal infected with BHGs/QH/1/05 virus. Viral antigens could be detected only in tonsilar epithelium on day 4 postinfection in animals infected with BHGs/QH/1/05 or GC/QH/3/05 (Fig. 5g); however, animals infected with DK/GX/35/01 showed positive reactions to anti-H5 serum in several tissues, including tonsil, lung, and spleen (data not shown). These results demonstrate the varied pathogenic potential in primates of the H5N1 viruses isolated from waterfowl.
FIG. 5.
Histological findings from rhesus macaques infected with H5N1 viruses. (a) Section from a consolidated area from lungs shows bronchointerstitial pneumonia with severe infiltration of inflammatory cells (BHGs/QH/1/05 virus, day 7 postinfection) (HE stain). The lung lesions were distributed around the bronchioli. Asterisks indicate lumen of bronchioli. (b) Severe alveolar damage was observed within and along the periphery of the consolidated area (BHGs/QH/1/05 virus, day 7 postinfection) (HE stain). Severe proliferative and reactive hyperplasia of alveolar cells with massive recruitment of lymphocytes, fibrin exudates, and alveolar edema are shown. (c) A strong reaction with macrophages was one of the prominent findings in the lungs (BHGs/QH/1/05 virus, day 7 postinfection) (HE stain). (d) Severe alveolar edema, thickening of alveolar wall with lymphocyte recruitment (white arrow), and regeneration of alveolar cells (black arrow) were also observed (BHGs/QH/1/05 virus, day 7 postinfection) (HE stain). (e) The lung lesions were detected as peribronchiolitis in a macaque infected with DK/GX/35/01 virus at 4 days postinfection (DK/GX/35/01 virus, day 4 postinfection) (HE stain). The asterisk indicates lumen of bronchioles. (f) Prominent alveolar edema and strong reaction with foamy macrophages but scant regenerative change and scant lymphocytic recruitment in a macaque infected with DK/GX/35/01 virus (DK/GX/35/01 virus, day 7 postinfection) (HE stain). (g) Viral antigens in tonsilar epithelium on day 4 postinfection (brown) (BHGs/QH/1/05 virus, day 4 postinfection) (immunohistochemistry).
Subsequent spread of the Qinghai Lake-like viruses.
Subsequent to the outbreak in Qinghai Lake from April to June 2005, H5N1 viruses have continued to cause outbreaks in Asia and have now even caused outbreaks in Europe (WHO report, http://www.who.int). We sequenced the entire genome of some H5N1 viruses isolated from wild birds in Mongolia in August 2005 and chickens during major outbreaks in the Liaoning Province and Inner Mongolia (see Fig. S1 in the supplemental material) in October and November 2005, respectively. Phylogenetic analysis of these viruses and a virus isolated from a wild bird in Russia in August 2005 (GenBank access numbers DQ230521, DQ230523, DQ234073, DQ230575, DQ230577, DQ232605, DQ232607, and DQ232609) showed that these viruses belong to genotype C (Fig. 2; see Fig. S2 in the supplemental material); moreover, all of these viruses possessed Lys at position 627 in PB2. Taken together, these findings suggest that H5N1 viruses have spread to wild-bird populations and have been disseminated.
DISCUSSION
Here, we report a detailed analysis of a large outbreak of H5N1 avian influenza virus occurring in migratory waterfowl from late April through June 2005 in the Qinghai Lake region of western China. In contrast to previous reports of viruses isolated during this outbreak (2, 12), our studies reveal a marked heterogeneity among the causative viruses. For example, sequence analyses of 15 viruses isolated from six different avian species showed that at least four genotypes were responsible for the outbreak. All of the viruses were highly lethal to chickens, and seven of the eight test viruses replicated systemically and were highly lethal in mice. We also found that the viruses isolated early in the outbreak possessed a typical avian virus signature amino acid at position 627 of PB2, Glu, unlike later isolates, which had Lys at this position (2, 12). Moreover, these index viruses possessed a phylogenetically distinct PB2 gene compared with those of other Qinghai isolates. This suggests that the virus introduced into the Qinghai Lake waterfowl population may have possessed the index virus-like PB2 gene but that during the outbreak, it acquired a PB2 gene with Lys at position 627. Alternatively, the newly introduced virus may have already possessed PB2 with Lys at this position, but viruses with the mutant type of PB2 were not detected until 10 May.
A distinct temporal pattern of infection of different avian species by the H5N1 viruses was also apparent (Fig. 1). Bar-headed geese were the first species to be affected, followed by brown headed gulls and great black-headed gulls about 10 days later (13 May) and then by ruddy shelducks and great cormorants after another 10 days (24 and 25 May). The time between the detection of small numbers of deaths (13 and 16 May) and the detection of considerably larger numbers of deaths (24 and 26 May) of ruddy shelducks and great cormorants was also about 10 days. These findings could be interpreted to indicate stepwise introduction of the virus into different avian species in the lake. Our sequence analyses revealed that at least three genotypes of H5N1 viruses were circulating among bar-headed geese, while the viruses isolated from great black-headed gulls, brown-headed gulls, great cormorants, and whooper swans were similar to each other and belonged to only one of the genotypes found in bar-headed geese (Fig. 4). Viruses representing genotypes A and B were isolated from the bar-headed geese that died early during the outbreak and were likely not spread to other species. We speculate that viruses of genotype D may also have been present in bar-headed geese at the beginning of the outbreak but were not identified because of the limited number of dead birds analyzed.
The origin of the virus responsible for the Qinghai Lake outbreak remains unclear. The disease was first recognized in bar-headed geese (Fig. 1 and 2), suggesting at least two possible mechanisms for the introduction of the H5N1 virus into wild-bird populations by this species using the lake as a habitat. One possibility is that the virus was carried to the lake by other wild birds not susceptible to H5N1 infection and was then transmitted to bar-headed geese. Another possibility is that bar-headed geese infected elsewhere were the species that brought the virus to Qinghai Lake, presumably via the East Asian-Australian flyway or the Central Asian-Indian flyway. If the first scenario is correct, the virus should have been transmitted to all susceptible species at the same time, including brown-headed gulls and great black headed gulls, which congregate with bar-headed geese in the islet where H5N1 virus-infected bar-headed geese were found. The fact that the disease was identified in these two species of gulls approximately 10 days after the discovery of fatal cases of H5N1 infection among bar-headed geese supports the second scenario.
Importantly, a genotype C virus, which was found in multiple species in Qinghai Lake, was responsible for the wild-bird outbreak of H5N1 infection in Mongolia and Russia in August 2005 and also caused major outbreaks in chickens in the Liaoning Province and Inner Mongolia in October and November 2005 (Fig. 4; see Fig. S2 in the supplemental material), suggesting that viruses of this genotype may be more pernicious than those of other genotypes. These findings call for intensive surveillance of wild migrating birds as biologic vectors that possibly spread H5N1 viruses over a wide range of territories.
It is important that viruses of genotype C possess Lys at position 627 in PB2, unlike any other avian viruses, and that this residue is found in some human H5N1 (7, 9, 10) and H7N7 (5) isolates as well as in the virus responsible for the Spanish influenza pandemic (21). Thus, it is worrisome that H5N1 viruses with a trait associated with human adaptation have entered into migrating waterfowl populations.
In the present study, seven of eight test viruses replicated systemically and killed mice. Among these seven lethal viruses, five have a lysine at position 627 of the PB2 protein and one has asparagine at position 701 of this molecule. Although both of these changes are associated with high virulence in mice (6, 8, 11), our findings indicate the existence of additional mutations that contribute to the virulence of avian H5N1 viruses in mammals.
Several different animal models have been used to evaluate the virulence of avian H5N1 influenza viruses in mammals. Maines et al. demonstrated the general equivalence of mice and ferrets for assessing the pathogenic potential of H5N1 viruses isolated from chickens and humans in Thailand and Vietnam (13). Experimental infection of cynomolgous macaques with the index H5N1 virus from the 1997 outbreak in Hong Kong resulted in acute respiratory distress syndrome and multiple-organ dysfunction, which was similar to findings in humans (18, 24). By contrast, the clinical signs produced in rhesus macaques by infection with two of the Qinghai Lake isolates were quite mild. Although the A/duck/Guangxi/35/01 virus did cause systemic infection and symptoms of influenza-like illness, the extent of the disease as judged by both its clinical symptoms and histopathology was milder than that reported previously by Rimmelzwaan et al. (18). This discrepancy may reflect the experimental procedures used by Rimmelzwaan et al. and our group. While we intranasally infected macaques with 2 ml of virus in fluid, Rimmelzwaan and colleagues used 5 ml of viral fluid, applying 4 ml intratracheally, 0.5 ml to the tonsils, and 0.25 ml to each of the conjunctiva, which would be expected to induce more severe disease than that seen in our study.
In conclusion, the H5N1 viruses that caused a massive outbreak of lethal disease among wild birds at Qinghai Lake in western China represent a phylogenetically and biologically heterogeneous group reiterating the features of H5N1 viruses now circulating in nature. The fact that viruses with a PB2 mutation associated with human adaptation of avian viruses are circulating in migratory waterfowl and that an avian H5N1 virus was capable of causing systemic infection in primates is worrisome. Moreover, migratory waterfowl may possibly spread these viruses over a wide range of territories. If viruses with the ability to replicate systemically in primates establish in migratory waterfowl, there would be an even more critical need for increased surveillance of poultry and the development of control measures.
Supplementary Material
Acknowledgments
We thank Peirong Jiao, Gongxun Zhong, Krisna Wells, and Martha McGregor for excellent technical assistance; John Gilbert for editing the manuscript; and Kangzhen Yu for critical discussion. We appreciate R. Sodnomdarjaa for providing the viruses isolated from wild birds in Mongolia for comparison.
This work was supported by the Animal Infectious Disease Control Program of the Ministry of Agriculture of China; by Chinese National Natural Science Foundation 30440008; by the Chinese National Key Basic Research Program (973) 2005CB523005 and 2005CB523200; by the Chinese National S&T Plan; by Public Health Service research grants from the National Institute of Allergy and Infectious Diseases; by grants-in-aid for Scientific Research on Priority Areas from the Ministries of Education, Culture, Sports, Science, and Technology, Japan; and by CREST (Japan Science and Technology Agency).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Chen, H., G. Deng, Z. Li, G. Tian, Y. Li, P. Jiao, L. Zhang, Z. Liu, R. G. Webster, and K. Yu. 2004. The evolution of H5N1 influenza viruses in ducks in southern China. Proc. Natl. Acad. Sci. USA 101:10452-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, H., G. J. D. Smith, S. Y. Zhang, K. Qin, J. Wang, K. S. Li, R. G. Webster, J. S. M. Peiris, and Y. Guan. 2005. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436:191-192. [DOI] [PubMed] [Google Scholar]
- 3.Claas, E. C., A. D. M. E. Osterhaus, R. van Beek, J. C. De Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472-477. [DOI] [PubMed] [Google Scholar]
- 4.Clements, J. F. 2000. Birds of the world: a checklist. Ibis, Vista, Calif.
- 5.Fouchier, R., P. Schneeberger, F. Rozendaal, J. Broekman, S. Kemink, V. Munster, T. Kuiken, G. Rimmelzwaan, M. Schutten, G. van Doornum, K. Guus, B. Arnold, K. Marion, and A. D. M. E. Osterhaus. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. USA 101:1356-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabriel, G., B. Dauber, T. Wolff, O. Planz, H.-D. Klenk, and J. Stech. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. USA 102:18590-18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, P., S. Watanabe, T. Ito, H. Goto, K. Wells, M. McGregor, A. J. Cooley, and Y. Kawaoka. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J. Virol. 73:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of H5N1 Hong Kong influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 9.Katz, J. M., X. Lu, T. M. Tumpey, C. B. Smith, M. W. Shaw, and K. Subbarao. 2000. Molecular correlates of influenza A H5N1 virus pathogenesis in mice. J. Virol. 74:10807-17810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keawcharoen, J., K. Oraveerakul, T. Kuiken, R. Fouchier, A. Amonsin, S. Payungporn, S. Noppornpanth, S. Wattanodorn, A. Theamboonlers, R. Tantilertcharoen, R. Pattanarangsan, N. Arya, P. Ratanakorn, A. D. M. E. Osterhaus, and Y. Poovorawan. 2004. Avian influenza H5N1 in tigers and leopards. Emerg. Infect. Dis. 10:2189-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Z., H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li, E. Hoffmann, R. G. Webster, Y. Matsuoka, and K. Yu. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79:12058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, J., H. Xiao, F. Lei, Q. Zhu, K. Qin, X. Zhang, X. Zhang, D. Zhao, G. Wang, Y. Feng, J. Ma, W. Liu, J Wang, and G. Gao. 2005. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309:1206. [DOI] [PubMed] [Google Scholar]
- 13.Maines, T., X. Lu, S. Erb, L. Edwards, J. Guarner, P. W. Greer, D. Nguyen, K. Szretter, L. Chen, P. Thawatsupha, M. Chittaganpitch, S. Waicharoen, D. T. Nguyen, T. Nguyen, H. H. T. Nguyen, J. Kim, L. T. Hoang, C. Kang, L. S. Phuong, W. Lim, S. Zaki, R. O. Donis, N. J. Cox, J. M. Katz, and T. M. Tumpey. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79:11788-11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Office International des Epizooties. 2004. OIE manual of diagnostic tests and vaccines for terrestrial animals. Office International des Epizooties, Paris, France.
- 15.Palmer, D. F., M. T. Coleman, W. R. Dowdle, and G. C. Schild. 1975. Advanced laboratory techniques for influenza diagnosis, p. 51-52. Immunology series no. 6. U.S. Department of Health, Education, and Welfare, Washington, D.C.
- 16.Peiris, J. S., W. C. Yu, C. W. Leung, C. Y. Cheung, W. F. Ng, J. M. Nicholls, T. K. Ng, K. H. Chan, S. T. Lai, W. L. Lim, K. Y. Yuen, and Y. Guan. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed, L., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 18.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2001. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 75:6687-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 21.Tanbenberger, J., A. Reid, R. Lourens, R. Wang, G. Jin, and T. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889-893. [DOI] [PubMed] [Google Scholar]
- 22.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization Global Influenza Program Surveillance Network. 2005. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 11:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen, K. Y., P. K. Chan, M. Peiris, D. N. Tsang, T. L. Que, K. F. Shortridge, C. P. T. Heung, W. K. To, E. T. Ho, R. Sung, and A. F. Cheng. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467-471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.