Abstract
Human leukocyte antigen (HLA) class I alleles can be grouped into supertypes according to their shared peptide binding properties. We examined alleles of the HLA-B58 supertype (B58s) in treatment-naïve human immunodeficiency virus type 1 (HIV-1)-seropositive Africans (423 Zambians and 202 Rwandans). HLA-B and HLA-C alleles were resolved to four digits by a combination of molecular methods, and their respective associations with outcomes of HIV-1 infection were analyzed by statistical procedures appropriate for continuous or categorical data. The effects of the individual alleles on natural HIV-1 infection were heterogeneous. In HIV-1 subtype C-infected Zambians, the mean viral load (VL) was lower among B*5703 (P = 0.01) or B*5703-Cw*18 (P < 0.001) haplotype carriers and higher among B*5802 (P = 0.02) or B*5802-Cw*0602 (P = 0.03) carriers. The B*5801-Cw*03 haplotype showed an association with low VL (P = 0.05), whereas B*5801 as a whole did not. Rwandans with HIV-1 subtype A infection showed associations of B*5703 and B*5802 with slow (P = 0.06) and rapid (P = 0.003) disease progression, respectively. In neither population were B*1516-B*1517 alleles associated with more favorable responses. Overall, B58s alleles, individually or as part of an HLA-B-HLA-C haplotype, appeared to have a distinctive impact on HIV-1 infection among native Africans. As presently defined, B58s alleles cannot be considered uniformly protective against HIV/AIDS in every population.
The concept of human leukocyte antigen (HLA) supertypes, categories of alleles that share common peptide-binding motifs, offers a simplification of the complex HLA nomenclature by consolidating the huge spectrum of individual alleles into relatively few groups. Individual alleles assigned to each supertype have either proven or predicted ability to present antigenic peptides with similar anchoring residues at the second and C-terminal positions of peptide ligands. Initially proposed in the late 1990s, four HLA-A and five HLA-B supertype categories encompass the majority of known HLA class I alleles (24, 25). There have been subsequent efforts to improve supertype categorization (8, 22). However, independent of the supertype assignment method, the HLA epitopes for just a few of the major supertypes have been estimated to provide substantial population coverage for antigenic peptides. If conserved peptides that cross-react with representative HLA class I supertypes can be identified, a supertype-guided approach could simplify the development of human immunodeficiency virus type 1 (HIV-1) subunit vaccines. Furthermore, designing a multiepitope HIV-1 vaccine candidate to target a few supertypes would be considerably easier than tailoring one to highly population-specific allelic differences. However, the success of such a strategy would depend on numerous factors (13, 15), including the degree of uniformity in the nature and magnitude of the CD8+ cytotoxic T-lymphocyte (CTL) responses to viral peptides.
The associations of CTL responses and control of HIV-1 viremia appear to differ from one HLA class I supertype to another (20, 23, 31). However, shared epitope recognition by alleles within a single supertype would be expected to produce relatively uniform associations of immunologic or clinically relevant CTL responses. On the other hand, if functional differences among alleles within a single HLA supertype are substantial, then the knowledge of those differences would be crucial for predicting responses to vaccines designed on the basis of supertype. Since not all HLA class I supertypes have been uniform in their associations with HIV-1 outcomes (4, 31), the systematic assessment of alleles within the same supertype may identify important differences in their epitope-binding characteristics.
The HLA-B58 supertype (B58s) is associated with a favorable response in Caucasians infected with HIV-1 clade B and in Africans infected with clade C (20, 23, 31). Among B58s alleles (all B*57 alleles, B*5801-*5802, and B*1516-*1517) (25). The HLA-B*57 alleles have consistently been associated with a favorable disease course in populations with different HIV-1 viral subtypes and ethnic backgrounds (1, 6, 11, 28, 30). However, assessment of the other B58s alleles has been relatively sparse (4, 10, 14). We evaluated the degree of control of HIV-1 infection exerted by the different B58s alleles in two cohorts of infected native Africans. Our findings of contrasting associations for certain B58s alleles, along with apparent effect modification by their accompanying HLA-C alleles, highlight the distinctive contributions of individual B58s alleles and their local haplotypes.
MATERIALS AND METHODS
Subjects.
We analyzed data from participants in two cohorts whose available biologic materials permitted resolution of HLA B58s: one included HIV-1-infected Zambian sex partners, and the other included sexually active HIV-1-infected Rwandan women. The design and conduct of these studies have been described elsewhere (9, 18, 28). The selection of mostly HIV-1 subtype C-infected Zambians (n = 423) was based on the availability of HIV-1 RNA measurements. The selection of mostly HIV-1 subtype A-infected Rwandan women (n = 202) was based on the availability of clinical and hematological indicators of disease progression.
HIV-1 outcomes.
HIV-1 RNA levels (viral loads [VLs]) were measured by a Roche Amplicor 1.0 assay (Roche Diagnostic Systems Inc., Branchburg, N.J.). Logarithmic (log10) transformation allowed modeling of VL as a continuous, normally distributed variable. Categorical VL analysis involved groups with <10,000, 10,000 to 100,000, and >100,000 copies/ml VL. Rwandan women were monitored from the date when infection was first documented until death or for ≥8 years and categorized as relatively slow (n = 101), intermediate (n = 86), or rapid (n = 15) progressors (27).
HLA class I typing and haplotype assignment.
HLA class I alleles were detected by PCR with sequence-specific primers (Pel-Freez Clinical Systems, Brown Deer, Wis.) for both cohorts. B58s alleles were resolved to their four-digit specificities by automated reference-strand conformation analysis. HLA-B and HLA-C haplotypes in Zambians were manually assigned after linkage disequilibrium analysis (28).
Statistical analysis.
For the Zambian cohort, the Mann-Whitney test was used to compare the median log10 VLs among subjects with various B58s alleles. For Zambian couples with a previously established correlation between VL levels in epidemiologically linked transmitters and seroconverters (29), we used generalized estimating equation (GEE) methodology (32) to assess the association between B58s alleles and VL. Because VLs in subjects with preexisting seropositivity at enrollment or seroconverters during follow-up could also reflect time-sensitive effects of HLA alleles, we stratified the subjects by serostatus to account for such differences in the analysis of the effects of B58s alleles. In separate stratification, we also aimed to dissect the effects of individual B58s alleles from those of their closely linked HLA-C alleles.
For the Rwandan cohort, the Jonckheere-Terpstra test was used to compare the proportions of subjects in the HIV-1 disease progression categories. Proportional odds regression analysis was used to measure the proportional odds ratios (POR) and 95% confidence intervals (CI) for the B58s alleles.
SAS version 9.01 (SAS Institute, Cary, N.C.) and GraphPad Prism version 4.0 were used for all statistical analyses and graphs.
RESULTS
B58s alleles and HIV-1 outcomes in Zambians.
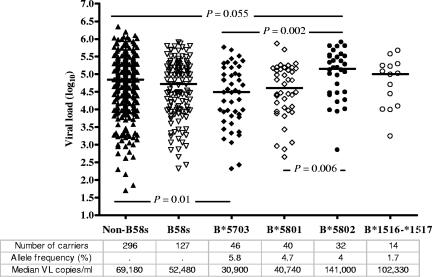
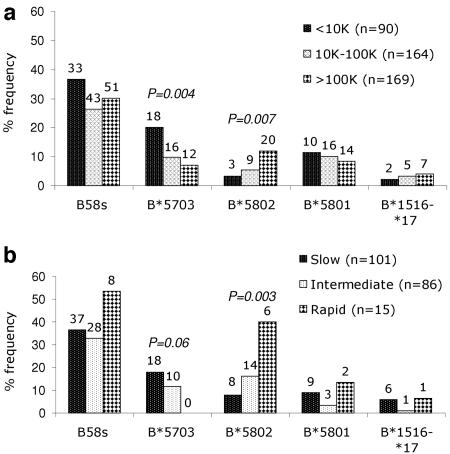
Of those Zambians whose specimens were typed at the HLA-B locus, 127 (30%) carried at least one B58s allele. Compared with Caucasians, Zambians were more likely to carry B58s alleles, including B*5802. Frequencies of individual B58s alleles in this population (2N = 846) ranged from 1.7% for B*1516-*1517 to 5.8% for B*5703 (Fig. 1). The mean log10 VL for all seropositive individuals was 4.67 (∼47,000 copies/ml). Overall, subjects with B58s alleles showed a slightly lower log10 VL than those without it (Δlog10 VL = −0.12, P = 0.18). However, VLs differed markedly across groups carrying certain individual B58s alleles. HLA-B*5703 was associated with low VLs in both linear (mean VL = 4.36, Δlog10 VL = −0.34, P = 0.01) and categorical (P = 0.004) analyses (Fig. 2a). Conversely, B*5802 was associated with higher VLs in both linear (mean VL = 4.97, Δlog10 VL = +0.33, P = 0.02) and categorical (P = 0.007) analyses. For subjects with B*5801, the median log10 VL (4.61) was not appreciably different from that found for subjects with B*5703 (4.49) (P = 0.54) and not significantly lower than the median for subjects with non-B58s alleles (P = 0.23). The small number of individuals with B*1516-*1517 had a nonsignificantly higher mean VL (mean VL = 4.77, Δlog10 VL = +0.12, P = 0.53) than those without any B58s allele.
FIG. 1.
Viral load distribution of HLA-B58 supertype alleles in Zambians. Horizontal bars correspond to median viral loads. The P values were derived from the Mann-Whitney test, and only P values of <0.1 are shown.
FIG. 2.
(a) Distribution of HLA-B58 supertype alleles within three VL categories in Zambians. The vertical bars represent the proportions of subjects with a particular B58s allele within each category. The number of subjects with a B58s allele is shown at the top of the bar for each category. P values were calculated with the Jonckheere-Terpstra test. (b) Distribution of HLA-B58 supertype alleles within three HIV-1 disease progression groups of Rwandans. The vertical bars represent the proportions of subjects with a B58s allele within each group. The number of subjects with a B58s allele is shown at the top of the bar for each category. P values were derived from the Jonckheere-Terpstra test.
To assess the effect of duration of HIV-1 infection on our findings, we compared transmitting partners who were HIV-1 positive at study entry (i.e., seroprevalent subjects) with partners who seroconverted after entry (i.e., seroconverters). The contrasting effects of B*5703 and B*5802 on VL were consistently apparent following stratification (Table 1), although the magnitude and significance of the B*5802 disadvantage were diminished among seroconverters. The protective effect for B*5801 was suggested among seroprevalent subjects (mean VL = 4.5, Δlog10 VL = −0.28, P = 0.06) and particularly among those with the B*5801-Cw*03 haplotype (mean VL = 4.19, Δlog10 VL = −0.65, P = 0.02) but absent in seroconverters. However, low numbers of seroconverters with B58s alleles may have accounted for the observed differences in the magnitude of the effects. The association with VL could not be meaningfully assessed in B*1516-*1517 seroconverters (n = 4).
TABLE 1.
Effects of HLA-B58s alleles on HIV-1 VL, stratified according to heterosexual HIV-1 transmission categories in Zambiansa
| HLA-B58s allele | Seroprevalent subjectsb (n = 276)
|
Seroconvertersc (n = 147)
|
Combined groups (n = 423)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Mean | ΔVLd | P | No. | Mean | ΔVL | P | No. | Mean | ΔVL | P | |
| B58s | 85 | 4.64 | −0.16 | 0.14 | 42 | 4.47 | −0.08 | 0.59 | 127 | 4.58 | −0.12 | 0.18 |
| B*5801 | 27 | 4.49 | −0.28 | 0.06 | 13 | 4.56 | +0.04 | 0.86 | 40 | 4.52 | −0.16 | 0.23 |
| B*5802 | 18 | 5.17 | +0.46 | 0.001 | 14 | 4.74 | +0.24 | 0.28 | 32 | 4.97 | +0.33 | 0.02 |
| B*5703 | 35 | 4.5 | −0.28 | 0.07 | 11 | 3.98 | −0.59 | 0.01 | 46 | 4.36 | −0.34 | 0.01 |
| B*1516-*1517 | 10 | 4.66 | −0.09 | 0.69 | 4 | —e | — | — | 14 | 4.77 | +0.12 | 0.53 |
All estimates are from the univariate GEE models.
Seroprevalent subjects, seropositive index partners who were HIV-1 positive at the time of study entry.
Seroconverters, partners of discordant couples who seroconverted during follow-up.
ΔVL values correspond to beta coefficients from the GEE models.
—, not reported due to instability of estimates.
There were 143 haplotype combinations consisting of a B58s allele and its imputed linked HLA-C allele identified among all Zambians studied, each with two presumed haplotypes (2N = 844). Notably, (i) the association of B*5703 with lower VLs was strong in the presence of Cw*18 but entirely lost in its absence; (ii) B*5801 commonly formed haplotypes with both Cw*03 and Cw*07, but only the B*5801-Cw*03 haplotype was associated with appreciably lower VLs (Table 2) ; and (iii) the association of the B*5802-Cw*06 haplotype and that of B*5802 overall with higher VLs were strong and indistinguishable due to the very strong disequilibrium between these B and Cw alleles. Because the carriage of Cw*18 appeared statistically to modify the effect of B*5703 in the Zambian population, we explored the role of Cw*18 further. Its carriage was associated with lower VLs among all subjects (mean VL = 4.3, Δlog10 VL = −0.43, P < 0.0001) as well as in the subsets of those with and without B*5703 (mean VL = 4.25, Δlog10 VL = −0.72, P < 0.01, and mean VL = 4.39, Δlog10 VL = −0.34, P = 0.03, respectively). However, because Cw*18 also shows linkage disequilibrium with B*8101, another allele associated with lower VLs in Zulu/Xhosa populations (14) and in our Zambian population (mean VL = 4.34, Δlog10 VL = −0.35, P = 0.03), the independent Cw*18 effect could be assessed only in the absence of both B*5703 and B*8101. Even in that small subgroup (n = 6), Cw*18 showed an effect (Δlog10 VL = −0.33), albeit statistically insignificant (P = 0.3), of a magnitude similar to that seen with B*5703 and B*8101 carriers.
TABLE 2.
Linear associationa of HLA-B58s and HLA-C alleles with HIV-1 viral load in Zambians
| B58s and Cw alleles carried by group | Meanb | ΔVLc | P |
|---|---|---|---|
| B*5703-Cw*18 haplotype (n = 38)d | 4.24 | −0.47 | <0.001 |
| Cw*18+ B*5703+ (n = 38) vs. Cw*18+ B*5703− (n = 28) | 4.25 | −0.14 | 0.52 |
| Cw*18− B*5703+ (n = 8) vs. Cw*18− B*5703− (n = 348) | 5.0 | +0.27 | 0.31 |
| B*5703+ Cw*18+ (n = 38) vs. B*5703+ Cw*18− (n = 8) | 4.25 | −0.72 | <0.01 |
| B*5703− Cw*18+ (n = 28)f vs. B*5703− Cw*18− (n = 349) | 4.39 | −0.34 | 0.03 |
| B*5802-Cw*06 haplotype (n = 30)d | 4.95 | +0.31 | 0.03 |
| Cw*06+ B*5802+ (n = 30) vs. Cw*06+ B*5802− (n = 59) | 4.95 | +0.47 | <0.01 |
| Cw*06− B*5802+ (n = 2) vs. Cw*06− B*5802− (n = 332) | —e | — | — |
| B*5802− Cw*06+ (n = 59) vs. B*5802− Cw*06− (n = 332) | 4.48 | −0.19 | 0.14 |
| B*5801-Cw*03 haplotype (n = 13)d | 4.18 | −0.5 | 0.05 |
| Cw*03+ B*5801+ (n = 13) vs. Cw*03+ B*5801− (n = 39) | 4.23 | −0.52 | 0.07 |
| Cw*03− B*5801+ (n = 27) vs. Cw*03− B*5801− (n = 344) | 4.68 | +0.01 | 0.92 |
| B*5801+ Cw*03+ (n = 13) vs. B*5801+ Cw*03− (n = 27) | 4.23 | −0.43 | 0.12 |
| B*5801− Cw*03+ (n = 39) vs. B*5801−Cw*03− (n = 344) | 4.75 | +0.08 | 0.61 |
Assessed by GEE.
Corresponds to mean log10 VL of haplotype or the first group of comparison strata.
Corresponds to beta coefficients from the GEE models.
Compared to the absence of haplotype.
Not reported due to instability of estimates.
Six Cw*18-positive subjects had lower VLs (mean = 4.4, Δlog10 VL = −0.33, P = 0.3) after exclusion of 22 B*8101-positive subjects
B58s alleles and HIV disease progression in Rwandans.
Of 202 seropositive women whose specimens were typed at the HLA-B locus, 73 (36.1%) carried at least one B58s allele. Frequencies of individual B58s alleles in this population (2N = 404) ranged from 2% for B*1516-*1517 to 3.7% for B*5801, 7.4% for B*5703, and 8.2% for B*5802. The proportions of each HIV-1 disease progression group who carried B58s alleles did not differ (P = 0.81). B*5703 was associated with a relatively favorable disease course (POR = 0.47; 95% CI = 0.21 to 1.07; P = 0.06), whereas B*5802 was strongly associated with accelerated HIV-1 progression (POR = 3.46; 95% CI = 1.6 to 7.7; P = 0.003) (Fig. 2b). No trends were apparent for B*5801 (P = 0.46) or B*1516-*1517 (P = 0.25).
DISCUSSION
Our observations for two populations of Africans infected with different HIV-1 subtypes demonstrate functional heterogeneity for individual alleles within the B58 supertype. We found no appreciable advantage of the B58 supertype as a whole on HIV/AIDS, in contrast to several previous studies (3, 16, 23, 31). Different HIV-1 subtypes or HLA class I supertype frequencies could have accounted for the observed population-specific effects of supertypes on viral control and immune escape (31). The more likely reason, however, is that contributions of the individual component alleles of the B58s were not examined (3, 16) or that their analysis was limited by the rarity of certain B58s alleles (23, 31). In particular, the very low frequency of B*5802 among Caucasians (5) precluded assessment of its contribution to the protection by B58s seen in the Multicenter AIDS Cohort Study.
Our results confirm the favorable effect of B*5703 and unfavorable effect of B*5802 on VL previously reported for HIV-1 subtype C in South Africa (14). Additionally, our findings extend the evidence to include the acceleration by B*5802 of the disease progression among Rwandans with the HIV-1 clade A. An earlier report on the failure of B*5802 to prsent immunodominant HIV-1 subtype C Gag peptides in subjects from Botswana (19) accords with poor control of viremia in Zambians and disease progression in Rwandans with B*5802. The available experimental and epidemiologic data point to structural and functional features of B*5802 that set it apart from the other members of the B58 supertype with regard to its capacity to respond to HIV-1 subtype C peptides. Specifically, it has been suggested that changes in amino acid side chains (94I→T, 95I→L, and 97R→W) in the α-2 helix of HLA class I molecules affect the key structures of the antigen binding groove such as the tyrosine bed and the F-pocket (2, 12, 21, 26), thereby impairing the presentation of immunodominant HIV-1 peptides.
While B*5802 has structurally been predicted to function inadequately, B*5801 would be expected to resemble B*57 alleles in controlling HIV-1 infection. However, we did not detect a uniform advantage for all Zambian B*5801 carriers. The protective effect of B*5801 was particularly apparent among seroprevalent subjects in conjunction with a closely linked Cw*03 allele. The absence of a B*5801 benefit among seroconverters may imply a later effect of B*5801 as distinct from the early protection well established for B*57 in a study of recent seroconverters (1) and in our own. Thus, despite their similarity in both predicted and reported binding motifs, B*5801 differed somewhat from B*5703 in its associations with HIV-1 outcomes in our study. Together, our data from Africans suggest important functional differences between B*58 and B*57 in the context of HIV/AIDS.
Although the preeminence of HLA-B alleles in HIV/AIDS has been demonstrated experimentally and epidemiologically (7, 14), in our Zambian cohort certain HLA-C alleles in linkage disequilibrium with their corresponding HLA-B alleles appeared to be contributing to their effect on VL. In particular, Cw*18 showed significant associations with low VL both in the presence and in the absence of B*5703, with which it is in linkage disequilibrium; conversely, VL was higher in subjects carrying B*5703 who lacked Cw*18. Compared with the effect of B*5703 overall, the magnitude of the VL association with the B*5703-Cw*18 haplotype was greater. In Rwandan subjects, neither the number of patients with the B*5703-Cw*18 haplotype nor the strength of linkage disequilibrium between the two alleles of the haplotype was sufficient to assess the effects of the two alleles with appropriate stratification.
The advantage of B*5801 likewise appeared dependent on the presence of the Cw*03 allele in Zambians. For B*5802 and Cw*0602, the linkage disequilibrium was so uniform that an independent deleterious effect of B*5802 could not be established. For reasons that are unclear, in Zambians these HLA-B-HLA-C haplotype combinations may have exerted particularly strong joint effects on the usual class I-mediated CTL pathway. It is also possible that HLA-C-restricted CTL responses may influence the epitopes targeted by relevant HLA-B alleles. A third possibility is that HLA-C alleles are involved through their additional role as ligands for killer immunoglobulin-like receptors (KIRs). However, HLA-KIR interactions are complex; they can drive both activating and inhibiting KIR effects (16, 17). Similar contributions of HLA-C alleles were not observed in Rwandans, probably due to differences in HLA-B-HLA-C haplotype frequencies and the prevalent HIV-1 subtype.
Our study had several strengths and limitations. We were able to examine the effects of B58s alleles in two African populations with distinct circulating viral subtypes. The capability of assessing the effects of B58s alleles among both seroprevalent and recently seroconverted individuals was also advantageous. Our longitudinal study design enabled us to detect the protective effect of B*5801 among the seroprevalent but not the recently seroconverted Zambians. This effect had been predicted in seroprevalent South Africans (14) but may not be generalizable to subjects in earlier stages of infection. The relatively high prevalence of several B*58s alleles among Africans provided sufficient power to evaluate the associations of HIV-1 outcomes with individual B58s alleles, except for B*1516-*1517, which has also been shown to be protective (10). The assessment of this pair of alleles was limited by small numbers in both cohorts. An absence of virologic data for Rwandans further limited our assessment, which was confined to categorical analysis of clinical outcomes.
In summary, B58s alleles or their haplotypes exert effects distinct enough from each other that the properties of all alleles of the B58 supertype should not be considered the same. Because HLA alleles interact with products of other genes inside and outside of the major histocompatibility complex as well as products of the virus itself, it is rather unlikely that their supertype classification based solely on CTL function can entirely capture their pluripotential effects. Further systematic investigation of individual alleles within other known HLA supertypes could prove equally informative for studies of infection and immunity.
Acknowledgments
This work was supported by several grants (AI40591, AI42454, AI41530, and AI41951) from the National Institute of Allergy and Infectious Diseases (NIAID), with additional funding from the Center for AIDS Research at the University of Alabama at Birmingham.
We are grateful to investigators, staff, and participants of the Zambia-Emory HIV-1 Research Project (ZEHRP) and Project San Francisco for their valuable contributions to this work. We also thank I. Brill, G. Cloud, and A. Moore for their help in data management.
REFERENCES
- 1.Altfeld, M., M. M. Addo, E. S. Rosenberg, F. M. Hecht, P. K. Lee, M. Vogel, X. G. Yu, R. Draenert, M. N. Johnston, D. Strick, T. M. Allen, M. E. Feeney, J. O. Kahn, R. P. Sekaly, J. A. Levy, J. K. Rockstroh, P. J. Goulder, and B. D. Walker. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581-2591. [DOI] [PubMed] [Google Scholar]
- 2.Barber, L. D., L. Percival, K. L. Arnett, J. E. Gumperz, L. Chen, and P. Parham. 1997. Polymorphism in the alpha 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J. Immunol. 158:1660-1669. [PubMed] [Google Scholar]
- 3.Bello, G., C. Casado, V. Sandonis, M. Alonso-Nieto, J. L. Vicario, S. Garcia, V. Hernando, C. Rodriguez, J. del Romero, and C. Lopez-Galindez. 2005. A subset of human immunodeficiency virus type 1 long-term non-progressors is characterized by the unique presence of ancestral sequences in the viral population. J. Gen. Virol. 86:355-364. [DOI] [PubMed] [Google Scholar]
- 4.Brander, C. 2004. HLA and HIV: implications for HIV vaccine design. ASHI Q. 28:58-59. [Google Scholar]
- 5.Cao, K., J. Hollenbach, X. Shi, W. Shi, M. Chopek, and M. A. Fernandez-Vina. 2001. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62:1009-1030. [DOI] [PubMed] [Google Scholar]
- 6.Costello, C., J. Tang, C. Rivers, E. Karita, J. Meizen-Derr, S. Allen, and R. A. Kaslow. 1999. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS 13:1990-1991. [DOI] [PubMed] [Google Scholar]
- 7.Dorak, M. T., J. Tang, A. Penman-Aguilar, A. O. Westfall, I. Zulu, E. S. Lobashevsky, N. G. Kancheya, M. M. Schaen, S. A. Allen, and R. A. Kaslow. 2004. Transmission of HIV-1 and HLA-B allele-sharing within serodiscordant heterosexual Zambian couples. Lancet 363:2137-2139. [DOI] [PubMed] [Google Scholar]
- 8.Doytchinova, I. A., P. Guan, and D. R. Flower. 2004. Identifiying human MHC supertypes using bioinformatic methods. J. Immunol. 172:4314-4323. [DOI] [PubMed] [Google Scholar]
- 9.Fideli, U. S., S. A. Allen, R. Musonda, S. Trask, B. H. Hahn, H. Weiss, J. Mulenga, F. Kasolo, S. H. Vermund, and G. M. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res. Hum. Retrovir. 17:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frahm, N., S. Adams, P. Kiepiela, C. H. Linde, H. S. Hewitt, M. Lichterfeld, K. Sango, N. V. Brown, E. Pae, A. G. Wurcel, M. Altfeld, M. E. Feeney, T. M. Allen, T. Roach, M. A. St. John, E. S. Daar, E. Rosenberg, B. Korber, F. Marincola, B. D. Walker, P. J. Goulder, and C. Brander. 2005. HLA-B63 presents HLA-B57/B58-restricted cytotoxic T-Lymphocyte epitopes and is associated with low human immunodeficiency virus load. J. Virol. 79:10218-10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, X., A. Bashirova, A. K. Iversen, J. Phair, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, M. Altfeld, S. J. O'Brien, and M. Carrington. 2005. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 11:1290-1292. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J. 2000. Rapid characterization of HIV clade C-specific cytotoxic T lymphocyte responses in infected African children and adults. Ann. N. Y. Acad. Sci. 918:330-345. [DOI] [PubMed] [Google Scholar]
- 13.Haynes, B. F. 1996. HIV vaccines: where we are and where we are going. Lancet 348:933-937. [DOI] [PubMed] [Google Scholar]
- 14.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 15.Longmate, J., J. York, C. La Rosa, R. Krishnan, M. Zhang, D. Senitzer, and D. J. Diamond. 2001. Population coverage by HLA class-I restricted cytotoxic T-lymphocyte epitopes. Immunogenetics 52:165-173. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Vazquez, A., A. Mina-Blanco, J. Martinez-Borra, P. D. Njobvu, B. Suarez-Alvarez, M. A. Blanco-Gelaz, S. Gonzalez, L. Rodrigo, and C. Lopez-Larrea. 2005. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum. Immunol. 66:285-289. [DOI] [PubMed] [Google Scholar]
- 17.Martin, M. P., X. Gao, J. H. Lee, G. W. Nelson, R. Detels, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, M. Wilson, S. J. O'Brien, and M. Carrington. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429-434. [DOI] [PubMed] [Google Scholar]
- 18.McKenna, S. L., G. K. Muyinda, D. Roth, M. Mwali, N. Ng'andu, A. Myrick, C. Luo, F. H. Priddy, V. M. Hall, A. A. von Lieven, J. R. Sabatino, K. Mark, and S. A. Allen. 1997. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS 11(Suppl. 1):S103-S110. [PubMed] [Google Scholar]
- 19.Novitsky, V., P. O. Flores-Villanueva, P. Chigwedere, S. Gaolekwe, H. Bussman, G. Sebetso, R. Marlink, E. J. Yunis, and M. Essex. 2001. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum. Immunol. 62:146-156. [DOI] [PubMed] [Google Scholar]
- 20.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novitsky, V., N. Rybak, M. F. McLane, P. Gilbert, P. Chigwedere, I. Klein, S. Gaolekwe, S. Y. Chang, T. Peter, I. Thior, T. Ndung'u, F. Vannberg, B. T. Foley, R. Marlink, T. H. Lee, and M. Essex. 2001. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific Elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J. Virol. 75:9210-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reche, P. A., and E. L. Reinherz. 2004. Definition of MHC supertypes through clustering of MHC peptide binding repertoires. Artificial Immune Systems, ICARIS. Lect. Notes Comput. Sci. 3239:189-196. [DOI] [PubMed] [Google Scholar]
- 23.Scherer, A., J. Frater, A. Oxenius, J. Agudelo, D. A. Price, H. F. Gunthard, M. Barnardo, L. Perrin, B. Hirschel, R. E. Phillips, and A. R. McLean. 2004. Quantifiable cytotoxic T lymphocyte responses and HLA-related risk of progression to AIDS. Proc. Natl. Acad. Sci. USA 101:12266-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sette, A., and J. Sidney. 1998. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr. Opin. Immunol. 10:478-482. [DOI] [PubMed] [Google Scholar]
- 25.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201-212. [DOI] [PubMed] [Google Scholar]
- 26.Stewart-Jones, G. B., G. Gillespie, I. M. Overton, R. Kaul, P. Roche, A. J. McMichael, S. Rowland-Jones, and E. Y. Jones. 2005. Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression. J. Immunol. 175:2459-2468. [DOI] [PubMed] [Google Scholar]
- 27.Tang, J., C. Costello, I. P. Keet, C. Rivers, S. Leblanc, E. Karita, S. Allen, and R. A. Kaslow. 1999. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 15:317-324. [DOI] [PubMed] [Google Scholar]
- 28.Tang, J., S. Tang, E. Lobashevsky, A. D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R. A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, J., S. Tang, E. Lobashevsky, I. Zulu, G. Aldrovandi, S. Allen, and R. A. Kaslow. 2004. HLA allele sharing and HIV type 1 viremia in seroconverting Zambians with known transmitting partners. AIDS Res. Hum. Retrovir. 20:19-25. [DOI] [PubMed] [Google Scholar]
- 30.Tang, J., C. M. Wilson, S. Meleth, A. Myracle, E. Lobashevsky, M. J. Mulligan, S. D. Douglas, B. Korber, S. H. Vermund, and R. A. Kaslow. 2002. Host genetic profiles predict virological and immunological control of HIV-1 infection in adolescents. AIDS 16:2275-2284. [DOI] [PubMed] [Google Scholar]
- 31.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
- 32.Zeger, S. L., and K. Y. Liang. 1986. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42:121-130. [PubMed] [Google Scholar]