Abstract
The Cucumber necrosis virus (CNV) particle is a T=3 icosahedron composed of 180 identical coat protein (CP) subunits. Each CP subunit includes a 34-amino-acid (aa) arm which connects the RNA binding and shell domains. The arm is comprised of an 18-aa “β” region and a 16-aa “ɛ” region, with the former contributing to a β-annular structure involved in particle stability and the latter contributing to quasiequivalence and virion RNA binding. Previous work has shown that specific regions of the CNV capsid play important roles in transmission by zoospores of the fungal vector Olpidium bornovanus and that particle expansion is essential for this process. To assess the importance of the two arm regions in particle accumulation, stability, and virus transmission, five CP arm deletion mutants were constructed. Our findings indicate that β(−) mutants are capable of producing particles in plants; however, the arm(−) and ɛ(−) mutants are not. In addition, β(−) particles bind zoospores less efficiently than wild-type CNV and are not fungally transmissible. β(−) particles are also less thermally stable and disassemble under swelling conditions. Our finding that β(−) mutants can accumulate in plants suggests that other features of the virion, such as RNA/CP interactions, may also be important for particle stability.
The capsids of many plant and animal viruses are multifunctional (2), having roles in genome protection, cell-to-cell and long-distance movement within plants (4), vector transmission (9, 16, 21, 25), replication (1), and suppression of gene silencing (22, 34). Structural studies of several plant viruses have revealed that the overall architecture of the T=3 capsid can be highly conserved between otherwise divergent virus groups, including those of several animal viruses (14). The structures of many plant virus particles have been obtained, and several in vitro studies have been conducted to assess the roles of the different structural domains in particle integrity and assembly, but few in vivo studies have been conducted. Moreover, fewer studies that relate the various structural domains to other possible functions of viral capsids have been reported.
Cucumber necrosis virus (CNV), a member of the Tombusviridae family, is a 33-nm spherical virus that encapsidates a monopartite, positive-sense RNA genome (28). In nature, transmission of CNV occurs via zoospores of the fungus Olpidium bornovanus, in which zoospore-bound particles are transmitted to cucumber following zoospore entry into root cells (3, 25). Based on structural homology to Tomato bushy stunt virus (12, 15), CNV is a T=3 icosahedron consisting of 180 identical 41-kDa coat protein (CP) subunits. Each subunit consists of three major structural domains: the R domain, which extends interiorly in the capsid; the S domain, which forms the shell of the capsid; and the P domain, which projects outward from the capsid. The P and S domains are joined by a 5-amino-acid (aa) hinge, and the R and S domains are connected by a 34-aa arm (Fig. 1A). The CP subunit exists in three different conformations, designated A, B, and C. In the C subunit, the arm is ordered and the R domain is spatially disordered, whereas in the A and B subunits, both the arm and R domain are disordered. The CP arm can be divided into two structurally distinct regions, “β” and “ɛ” (Fig. 1B). The β regions from three C subunits interdigitate at the particle threefold axis to form a β-annular structure (Fig. 1C) that contributes to particle stability and possibly to assembly as well. Similar structural regions within the CPs of various other T=3 icosahedral viruses are believed to function as scaffolds important for particle stability and virus assembly (5, 32, 33). The ɛ region of the CNV arm extends from the β region and continues along the inner face of the S domain. It has been suggested that the ɛ region may serve to bind RNA within the capsid, contribute to the stability of the swollen form of the particle at the quasi twofold axis, and, due to its ordered or disordered state in C or A/B subunits, respectively, contribute to quasi-equivalence (12, 24, 35).
FIG. 1.
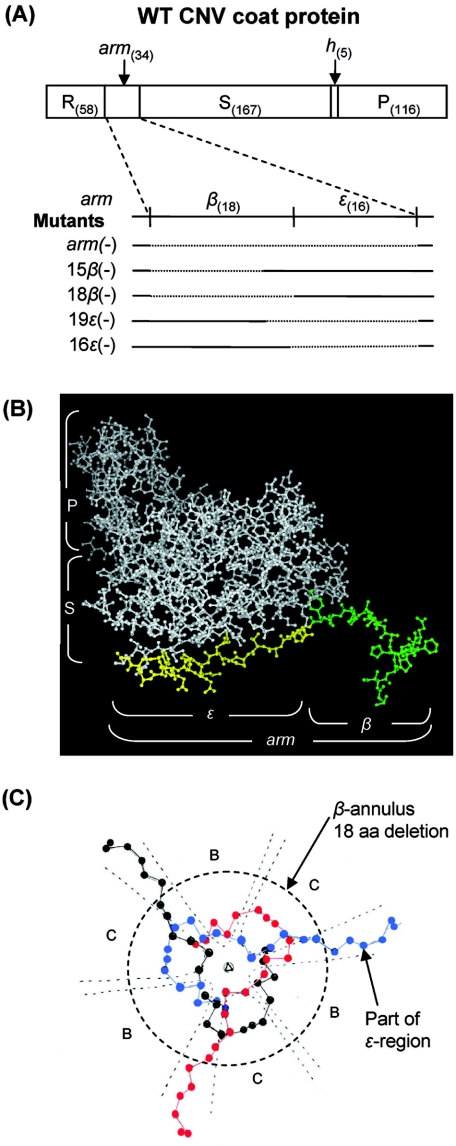
Description of the locations of the CNV CP arm and deletion mutants. (A) Linear order of the CP domains and the β and ɛ regions of the arm as well as the locations of the deleted regions of each arm mutant. The numbers of amino acids comprising each CP domain and the β and ɛ regions are shown in parentheses. R, RNA binding; S, shell; h, hinge; P, protruding domain. The dotted regions indicate the deleted regions in the arm. (B) Ball-and-stick representation of the C subunit of CNV CP showing the locations of the ɛ (yellow) and β (green) regions of the arm. The R domain is not shown. (C) Schematic representation of the β annulus plus six residues of the ɛ region (the partial arms of the three C subunits are in black, blue, and red) viewed down the particle threefold axis. The region within the circle (dashed lines) indicates the deleted residues of the 18β(−) mutant. The balls represent α-carbon positions. (Diagram adapted from reference 12.)
The CP of CNV, as well as that of several other tombusviruses, is not required for cell-to-cell movement (29, 31); however, systemic movement of unencapsidated viral RNA is delayed compared with that of encapsidated RNA (31). Previous work has shown that specific regions of the S and P domains of the CNV capsid are involved in fungal transmission (15, 23) and that the defects are due, at least in part, to inefficient attachment of virions to the zoospore surface (15, 23). Kakani et al. (17) have shown that zoospore-bound CNV is conformationally different from native CNV particles and that a CP mutant with a Pro-to-Gly change in the arm near the β and ɛ junction that is incapable of swelling was not being fungally transmitted, suggesting that swelling is required for transmission. These findings have been related to the possible role of particle swelling during virus disassembly as has been suggested by others (14, 17). In this study, we have generated CNV CP arm mutants to examine the role of the “β” and “ɛ” regions of the arm in virus infection, focusing on particle accumulation, stability, and vector transmission.
MATERIALS AND METHODS
Isolation and purification of virus.
CNV (pK2/M5) was maintained by mechanical passage in Nicotiana benthamiana or Nicotiana clevelandii as previously described (23). A “miniprep” procedure was employed to partially purify wild-type (WT) CNV and CNV mutants from single infected leaves for use in the initial screenings and for subsequent assays (23). The concentration of virus was determined by electrophoresis of several dilutions through a 1% agarose gel buffered in 80 mM Tris-80 mM borate, pH 8.3, followed by staining with ethidium bromide (EtBr) in electrophoresis buffer containing 1 mM EDTA. Purification of virus from larger amounts of leaf tissue (100 to 250 g) was carried out by a differential centrifugation method previously described (15). Virus pellets were resuspended in 10 mM sodium acetate buffer (pH 5.0), and virus concentration was determined spectrophotometrically (absorbance at 260 nm of a 1-mg/ml suspension of CNV is 4.5.)
In vitro transcription.
T7 polymerase runoff transcripts from full-length cDNA clones of CNV were generated by a method previously described (26). Transcripts were used immediately to inoculate leaves of 3- to 4-week-old N. benthamiana plants. Aliquots of transcript reaction mixtures were routinely examined by agarose gel electrophoresis to assess the quality and quantity of transcripts produced.
Leaf RNA extraction.
Infected leaves were ground to a powder in liquid nitrogen, and RNA was extracted with phenol-chloroform in buffer containing 100 mM Tris, 100 mM NaCl, and 10 mM EDTA, pH 7.5, along with 0.5% sodium dodecyl sulfate (SDS) and 5% 2-mercaptoethanol.
Site-directed mutagenesis.
In vitro mutagenesis was used to produce the following CNV CP arm mutants: arm(−), 15β(−), 18β(−), 16ɛ(−), and 19ɛ(−). An EcoRI/NcoI fragment, encompassing the CNV CP and flanking regions in a full-length infectious cDNA clone of CNV (pK2/M5) (26), was excised and subcloned into EcoRI/NcoI-digested pT7 blue (Novagen) and used as a template for oligonucleotide-directed in vitro mutagenesis (8). Primers used to create the deletion mutants are described in Table 1. Subclones bearing mutations were digested with EcoRI and NcoI and cloned into pK2/M5, which was then used as a template to form full-length infectious clones. The sequences between the EcoRI and NcoI sites were confirmed by sequence analysis.
TABLE 1.
Primers used for PCR mutagenesis of CNV CP arm mutants
| Construct name | Primer sequences | Amino acids deleteda |
|---|---|---|
| Arm(−) | Negative sense, 5′ AGCACCGTTTCCATTCTTACC 3′; positive sense, 5′ TCTGTGCGAATAACCCATAGAG 3′ | 59 to 92 |
| 15β(−) | Negative sense, 5′ AGCACCGTTTCCATTCTTACC 3′; positive sense, 5′ ATCTCTTATGCCTATGCGG 3′ | 59 to 73 |
| 18β(−) | Negative sense, 5′ AGCACCGTTTCCATTCTTACC 3′; positive sense, 5′ GCCTATGCGGTTAAAGGAAGG 3′ | 59 to 76 |
| 16ɛ(−) | Negative sense, 5′ ATAAGAGATTGGCGCGGC 3′; positive sense, 5′ TCTGTGCGAATAACCCATAGAG 3′ | 77 to 92 |
| 19ɛ(−) | Negative sense, 5′ TGGAGCAGCAATAGCCCCAGG 3′; positive sense, 5′ TCTGTGCGAATAACCCATAGAG 3′ | 74 to 92 |
Refers to CP amino acids beginning at the amino terminus.
Analysis of the CP subunit.
Electrophoresis of protein from purified mutant particles was performed using SDS containing 12% polyacrylamide gels according to the method of Laemmli (18) to verify correct protein size.
Virion RNA extraction.
RNA was extracted from purified β(−) mutant particles using 10 mM EDTA and phenol-chloroform-1% SDS in 50 mM Tris buffer (pH 8.8) as previously described (27).
In vitro swelling.
WT CNV, 15β(−), and 18β(−) virions (500 ng virus/20 μl) were swollen in 50 mM sodium phosphate buffer (pH 7.6) containing 25 mM EDTA at room temperature for 30 min.
Electron microscopy.
Purified virus was negatively stained with 2% uranyl acetate. Each sample was applied to a collodion-covered copper grid and allowed to absorb for 1 min. Samples were viewed at 27,000× to 80,000× magnification in a JEOL 100CX transmission electron microscope (TEM) operated at 80 kV.
Fungus transmission assay.
O. bornovanus isolate SS196 was maintained on cucumber roots (Cucumis sativus cv. Poinsette 76) as previously described (23). Purified 15β(−) and 18β(−) virions were tested for transmission by O. bornovanus zoospores as previously described (15). Virus (1 μg) was incubated with 10 ml of zoospores (105/ml in 50 mM glycine, pH 7.6). After a 15-min acquisition period, the mixture was poured into pots containing 14- to 16-day-old cucumber seedlings. Five days later, roots of cucumber seedlings were tested for the presence of CNV by double-antibody-sandwich enzyme-linked immunosorbent assay (ELISA) using polyclonal antisera raised to CNV particles. Each transmission experiment included a WT CNV control, to determine any background level of CNV transmission in the absence of zoospores, and an aviruliferous zoospore control to confirm the absence of contaminating virus in zoospore preparations. Each assay was replicated three times.
In vitro binding assay.
An in vitro binding assay was performed to determine the ability of mutant virus to bind to zoospores of O. bornovanus (15). One hundred micrograms of purified virus was incubated with 5 × 105 zoospores in 1 ml of 50 mM sodium phosphate binding buffer (pH 7.6) for 1 h at room temperature. Following incubation, zoospores were pelleted by centrifugation at ∼2,700 × g for 7 min. The pellet was washed with 1.5 ml of binding buffer and then resuspended in sterile, deionized water. The zoospore pellet was assayed for the presence of virus by Western blot analysis using CNV monoclonal antibody 57.2 (specific to the CNV R domain) (23) and an enhanced chemiluminescence detection system (Amersham Biosciences). The quantity of virus in the pellet was assessed by densitometric analysis of exposed film using the ImageQuant TL program (Amersham Biosciences).
Thermal stability and RNase sensitivity assays.
Purified WT CNV and 18β(−) virions were assayed for thermostability by incubating 1 μg of virus (in 10 mM sodium acetate, pH 7.2) for 30 min at 4°C, 26°C, 37°C, 42°C, 55°C, 60°C, 70°C, and 100°C. Particles were immediately examined by electrophoresis using a 0.7% agarose gel at 90 V for 1.5 h. Virion RNA was visualized by staining with ethidium bromide and virion protein by subsequent staining with Coomassie brilliant blue. RNase assays were conducted by incubating 1 μg of 18β(−) virions with 10 ng of pancreatic RNase A after incubation at 70°C for 30 min or after incubation at 26°C for 30 min, followed by incubation at 26°C for 30 min. A negative control assay in which particles were incubated without RNase A at 70°C for 30 min, followed by incubation at 26°C for 30 min, was performed. Virions were electrophoresed and stained as described above.
RESULTS
Role of the β and ɛ regions of the CNV CP arm in particle accumulation.
Five CP arm deletion mutants were produced to assess the role of the β and ɛ regions of the arm in particle accumulation (Fig. 1A). Transcripts of each arm mutant were used to inoculate leaves of N. benthamiana plants, and plants were monitored for symptom induction. Both the 15β(−) and 18β(−) mutants produced symptoms typical of WT CNV, resulting in necrotic lesions on inoculated leaves and subsequent systemic necrosis and death of the plants within 5 to 6 days postinoculation (dpi) (data not shown). The arm(−), 16ɛ(−), and 19ɛ(−) mutants produced necrosis on the inoculated leaves, but systemic necrosis was delayed in comparison to that of WT CNV, appearing 10 to 14 dpi (data not shown). Mutant-infected plants were then tested for the presence of viral genomic RNA and production of virions. Figure 2A shows an electropherogram of the total leaf RNA extracts of each of the mutants and indicates that viral genomic RNA is readily observed in each mutant-infected plant. In addition, truncated genomic RNA (which lacks the CP-coding regions [data not shown]) was occasionally observed in ɛ(−) mutants (Fig. 2A, lane 8). Agarose gel electrophoresis of partially purified virus particles showed that particles accumulated in leaves infected with 15β(−) and 18β(−) mutants (Fig. 2B, lanes 4 and 5), whereas particles were not observed in leaves infected with arm(−), 16ɛ(−), and 19ɛ(−) mutants (Fig. 2B, lanes 3, 6, and 7). The yields of 15β(−) and 18β(−) virions from infected plants at 5 dpi were determined, and the yields of both were found to be approximately 18% of that of WT CNV. SDS-polyacrylamide gel electrophoresis analysis was conducted to confirm the presence of the deletions in the CP subunits of purified 15β(−) and 18β(−) particles. As seen in Fig. 2C, the CPs of both mutants show the expected increase in mobility relative to that of WT CNV CP. Virions of β(−) deletion mutants purified from infected plants were analyzed for the presence of viral genomic RNA. Agarose gel electrophoresis demonstrated that genomic RNA was present in both 15β(−) and 18β(−) virions (data not shown).
FIG. 2.
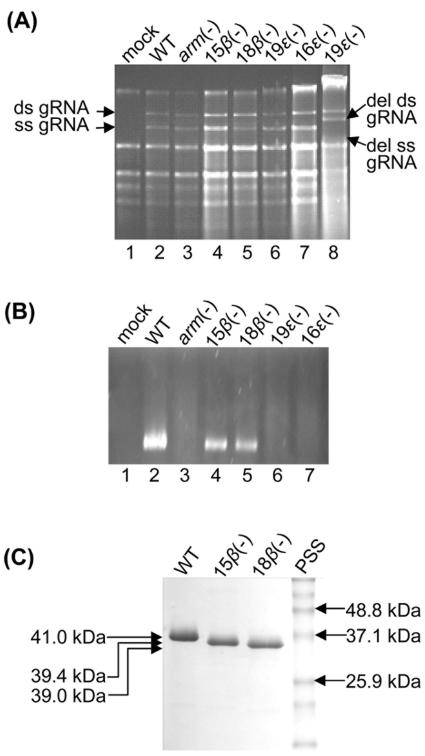
Gel electrophoresis of leaf RNA, virions, and CP subunits of arm mutants. (A) Agarose gel electrophoresis of total RNA extracted from inoculated leaves at 4 dpi and visualized with EtBr. Lane 8 corresponds to an independent extract of 19ɛ(−) in which genomic RNA was found to contain a deletion in the CP open reading frame. The positions of the double-stranded (ds) and single-stranded (ss) genomic RNAs (gRNA) are indicated on the left, and the deleted (del) ds and ss gRNA are indicated on the right. (B) Agarose gel electrophoresis of partially purified virions. Virions in 1% gel buffered in Tris-borate buffer (pH 8.3) were visualized by EtBr staining in the presence of 1 mM EDTA. (C) SDS-polyacrylamide gel electrophoresis gel of 5 μg of purified WT CNV, 15β(−), and 18β(−) virions. Gels were stained with Coomassie brilliant blue. The numbers on the right indicate the molecular masses (in kDa) of the protein size standards (PSS). The numbers on the left indicate the predicted sizes of WT, 15β(−), and 18β(−) CP subunits.
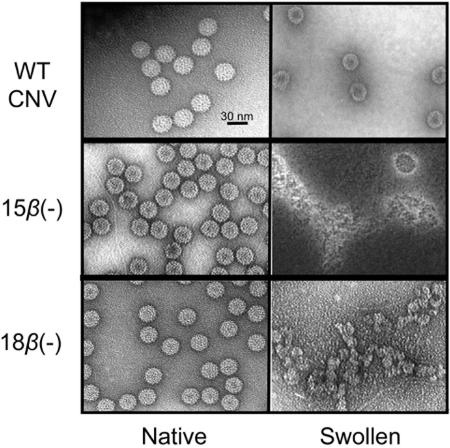
β(−) mutant particles were also visualized using TEM. Negatively stained particles were found to be grossly similar to those of WT CNV particles; however, β(−) mutant particles appeared more hexagonal in shape than CNV particles and also exhibited more densely stained centers (Fig. 3).
FIG. 3.
Electron micrographs of negatively stained native and swollen β(−) mutant particles. Virions were swollen in 50 mM Tris (pH 8.0)-25 mM EDTA for 30 min at room temperature and immediately examined by TEM. Native particles are in the left column, and swollen particles are in the right column. The virus particles used are shown on the left of each pair of treatments.
Particles of CNV β(−) mutants are not transmissible by O. bornovanus.
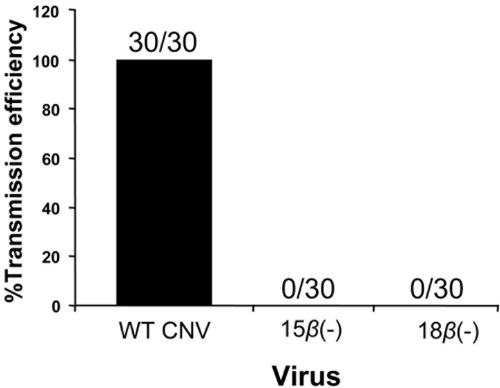
Particles purified from plants infected with 15β(−) and 18β(−) mutants were analyzed for their ability to be transmitted by O. bornovanus to roots of cucumber seedlings. Transmission efficiency was scored by assessing the number of pots infected with virus (as determined by ELISA of cucumber root extracts) versus the number of pots inoculated. Results of the transmission assays indicated that 30 of 30 pots (100% efficiency) inoculated with CNV-zoospore mixtures became infected with WT CNV, whereas 0 of 30 pots (0% efficiency) became infected when either 15β(−) or 18β(−) zoospore mixtures were used (Fig. 4). These results suggest that the β annulus may have an important role in virus transmission, an effect that may be confounded by the relatively low level of particle accumulation as described above.
FIG. 4.
Summary of fungus transmission assays of β(−) mutant particles. One microgram of either WT CNV, 15β(−), or 18β(−) particles was incubated with 1 × 106 zoospores. After 15 min, the mixture was poured into pots containing cucumber seedlings. Five days later, the roots of the seedlings were tested for the presence of CNV by double-antibody-sandwich ELISA using polyclonal antisera raised to CNV particles. The percentage of pots showing transmission for each virus is indicated on the y axis. The numbers on the columns indicate the numbers of pots showing transmission versus the numbers of pots inoculated with virus-zoospore mixtures. The data represent a compilation of three separate experiments, with two replicates per experiment.
CNV 18β(−) mutants show decreased binding to zoospores in vitro.
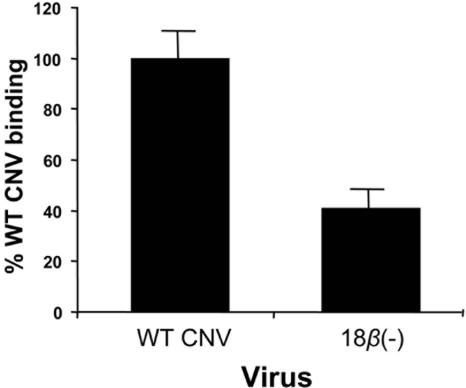
The 18β(−) mutants were assessed to determine whether the lack of transmissibility may be at least partly due to an inefficient ability of mutant particles to bind zoospores. To do this, a previously developed in vitro virus/zoospore binding assay was utilized (15, 23). One hundred micrograms of either WT CNV or 18β(−) particles was incubated with 5 × 105 zoospores for 1 h, followed by low-speed centrifugation to pellet zoospores and washing to remove unbound or nonspecifically bound virus (15). The amount of bound virus in the pellet was then determined by Western blot analysis, followed by densitometry. Figure 5 shows that a lower level of 18β(−) particles (approximately 41% of that of WT CNV) was found in the zoospore pellet, suggesting that the lack of transmission of CNV 18β(−) particles may be at least partly due to a reduced ability to stably attach to zoospores during the transmission process.
FIG. 5.
Summary of in vitro virus/zoospore binding assays. One hundred micrograms of either WT CNV or 18β(−) particles was added to 4 × 105 O. bornovanus zoospores and incubated for 1 h. Following incubation, zoospores were pelleted and washed. The amount of zoospore-bound virus was determined by Western blot analysis using a previously described CNV monoclonal antibody (57.2) corresponding to the R domain (23; unpublished data). The quantity of virus in the pellet was assessed by densitometric analysis of the exposed film. Percent 18β(−) binding values were normalized against WT CNV. The results are averages of samples from three separate experiments, using triplicate samples of each virus per experiment.
β(−) particles disassemble at alkaline pH in the presence of EDTA.
Many icosahedral viruses undergo structural expansion in vitro when incubated at alkaline pH in the presence of EDTA (24). To assess the role of the β annulus in particle stability under swelling conditions, β(−) mutant particles were incubated with EDTA at pH 8.0 and analyzed by TEM. WT CNV particles were slightly enlarged with electron-dense centers, as expected. However, few intact particles were observed in similarly treated β(−) particles; instead, discrete areas of stained material with no specific structural features were observed (Fig. 3). These results suggest that particles lacking the CP β annulus disassemble under swelling conditions.
18β(−) particles are less thermally stable than WT CNV.
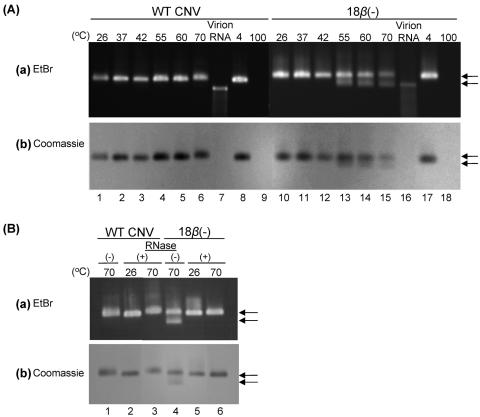
The stability of 18β(−) mutant particles was further investigated by analyzing virions subjected to heat treatment. Mutant particles were incubated at increasing temperatures, followed by assessment of particle integrity by agarose gel electrophoresis. Virion RNA was visualized by ethidium bromide staining and capsids by subsequent staining with Coomassie brilliant blue (Fig. 6). Stained gels of WT CNV revealed a single band at all temperatures examined (except at 100°C, at which no band was observed). However, stained gels of 18β(−) mutant particles incubated at 55°C, 60°C, and 70°C revealed two bands, one that comigrated with WT CNV particles and a second that migrated slightly faster (Fig. 6A, panels a and b, lanes 13 to 15). The faster-moving band stained with both EtBr and Coomassie blue, indicating that it is a ribonucleoprotein (Fig. 6A, panels a and b, lanes 13 to 15). In addition, the second band did not comigrate with virion RNA of 18β(−) mutant particles (Fig. 6A, panel a, compare lanes 13 to 15 with lane 16), indicating that it is not simply viral RNA that was released from the heated particles. To further evaluate the nature of the second band, 18β(−) mutant particles were subjected to RNase treatment after incubation at 70°C. Agarose gel electrophoresis showed that the addition of RNase to 18β(−) mutant particles immediately after incubation at 70°C resulted in the disappearance of the faster-moving ribonucleoprotein species in both the EtBr- and the Coomassie blue-stained gels (Fig. 6B, panels a and b, compare lane 4 with lane 6); however, particles incubated at 26°C were not RNase sensitive (Fig. 6B, panel a, lane 5). Also, WT CNV particles were not RNase sensitive at either temperature (Fig. 6B, lanes 2 and 3). Together, these results suggest that the β region of the CP arm contributes to the thermal stability of CNV virions and that virion RNA of 18β(−) virions becomes exposed in the thermally destabilized state. In addition, the complete loss of 18β(−) particle integrity following RNase treatment indicates that virion RNA contributes, at least in part, to structural stability.
FIG. 6.
Agarose gel electrophoreses illustrating the thermostabilities and RNase sensitivities of WT CNV and 18β(−) particles. (A) One microgram of either WT CNV or 18β(−) virions was incubated at a range of temperatures for 30 min in 10 mM sodium acetate buffer, pH 7.2. Immediately thereafter, virions were electrophoresed through a 0.7% agarose gel buffered in Tris-borate, pH 8.3, and visualized with EtBr (a). The gel was then stained with Coomassie brilliant blue (b). (B) RNase sensitivity assays in which 1 μg of either WT CNV or 18β(−) particles was incubated with 10 ng of pancreatic RNase A after incubation at 70°C for 30 min or after incubation at 26°C for 30 min, followed by an additional incubation at 26°C for 30 min, were conducted. Virions were electrophoresed and stained with EtBr (a) followed by Coomassie blue (b), as shown in panel A. (−) and (+) denote the absence and presence of RNase, respectively. Arrows point to the two bands that are discernible in the EtBr- and Coomassie blue-stained gels of 18β(−) mutants at 55°C, 60°C, and 70°C.
Heating WT CNV particles to 70°C resulted in slightly slower mobility on agarose gels (Fig. 6A, panels a and b, lane 6, and Fig. 6B, panels a and b, lanes 1 and 3). The slower mobility might be due to expansion similar to that observed when WT particles are subjected to alkaline pH and EDTA (Fig. 3).
TEM analysis was performed to directly visualize the effects of 55°C and 70°C treatment on 18β(−) mutant particles (data not shown). Alterations to the particles were not apparent. However, due to our observation that the majority of the particles remain unchanged, as determined by agarose gel electrophoresis (Fig. 6A and B, lanes 13 to 15 and lane 4, respectively), and the possibility that the heat treatment could make the particles morphologically unidentifiable, it would be difficult to assess whether any change in structure could actually be observed.
DISCUSSION
There are few in vivo studies that delineate the role of the individual CP domains of icosahedral plant viruses in particle assembly and transmission. Based on structural studies, the tombusvirus particle has been postulated to be stabilized, in part, by the β annulus located at the threefold axis as well as by calcium-mediated subunit-subunit interactions and RNA (10, 12). The remaining residues of the arm (the ɛ region) are likely important for virion RNA binding (35) and for the conformational switching required for T=3 particle formation (10). In this study, we examined the significance of the β and ɛ regions of the CNV arm in particle accumulation in plants as well as in the ability of the particles to be fungally transmitted. Our studies indicate that the β region is not essential for CNV particle accumulation in plants, whereas the ɛ region is. However, the presence of the β region does contribute to both particle stability and fungal transmissibility.
CNV β(−) particles accumulate to 18% of those in WT virus in N. benthamiana, suggesting that the β region of the CP arm plays an important role in particle assembly and/or stability but is not absolutely required. It has been suggested that Turnip crinkle virus assembly initiates with the formation of a complex consisting of a trimer of C/C dimers that is stabilized by the β annulus and possibly virion RNA attachment as well (13, 32, 36). The formation of CNV T=3 particles that lack the CP β region in plants demonstrates that the β annulus is not absolutely required for CNV particle stability or assembly initiation, and therefore, additional factors, such as interactions with viral RNA, may also play an important role. The CP β region is highly hydrophobic and therefore is unlikely to interact directly with viral RNA. However, the upstream R domain sequences and the downstream ɛ region that immediately flank the β region likely do bind RNA, as they contain clusters of basic amino acids (32, 35); thus, RNA-protein interactions that are expected to occur in each of the three C-C subunits at the threefold axis could substantially increase the stability of this complex during assembly initiation and/or within assembled particles. However, it is still possible that CNV assembly may be initiated by a pentamer of A/B subunits, possibly as an alternative mode of assembly. Comparable to our findings, mutational analyses of Cowpea chlorotic mosaic virus mutants lacking the β hexamer (which is functionally similar to the CNV β annulus) revealed that the β hexamer is not required for virion formation, suggesting that assembly initiation can occur via pentamers of A/B dimers (33, 37). Similar results obtained by in vitro studies of the assembly of a sobemovirus capsid (Sesbania mosaic virus), where it was shown that the β annulus is also not required for formation of T=3 capsids, have recently been reported, suggesting that assembly could initiate via pentamers as suggested for Cowpea chlorotic mosaic virus (30).
Our observation that β(−) mutant particles accumulate at a relatively low level could be due to reduced particle stability following assembly. This notion is supported by our findings that β(−) mutant particles disassemble under in vitro swelling conditions (Fig. 3) and that they are more heat labile than WT CNV particles (Fig. 6A) (see below). The lack of particle accumulation in plants infected with either arm(−) or ɛ(−) mutants is consistent with the role of the ɛ region of the arm in virion RNA binding and quasi-equivalence (10, 11). This result is also consistent with our previous finding (17) that a proline-to-glycine mutation at CNV CP aa 85, which lies in the ɛ region close to the arm/shell junction and which may control the flexibility of the arm, which is required for quasiequivalent interactions, also results in little or no observable particle accumulation. The 18-amino-acid sequence of the ɛ region is highly basic, containing five Lys or Arg residues, and is therefore likely to bind RNA along the inner lining of the shell (35). Thus, an additional crucial role for this region may be in the assembly initiation and/or stabilization of subunit interactions during or following assembly, as has been suggested in the case of TCV (32, 37). Our in vivo experiments would not be able to distinguish between an alternate or dual role for the ɛ region.
In vitro transmission assays showed that β(−) mutant particles are not transmitted by zoospores of O. bornovanus (Fig. 4). In addition, in vitro binding assays indicate that fewer β(−) particles bound zoospores (41% of those found using WT CNV virions) (Fig. 5). Loss of transmissibility could be partly a result of reduced accumulation of virus following transmission, although the 85% reduction in accumulation observed in N. benthamiana plants may not be sufficient on its own to completely account for the loss of transmission. Figure 3 shows that β(−) mutant particles partially disassemble under swelling conditions. This observation, in conjunction with the observation that WT CNV particles undergo swelling upon zoospore binding (17), suggests that at least part of the basis for reduced binding to zoospores is due to the instability of β(−) mutant particles following zoospore binding. We have recently postulated that the β region of the CNV A or B subunit may contribute to the stability of virus/zoospore binding, which is similar to that suggested for the analogous region of the poliovirus CP upon virion binding to host cells (17). Thus, the approximately twofold reduction of zoospore binding may be a result of the absence of the postulated stabilizing effect of the β region.
Our observation that 18β(−) particles heated to high temperatures have altered electrophoretic mobilities and are RNase sensitive suggests that the β region contributes to particle stability and that virion RNA is exposed in the heat-destabilized form. It is likely that the absence of the β annulus is largely responsible for the decreased stability, which is similar to that observed under swelling conditions. The absence of the β regions in the A and B subunits might also contribute to some loss of stability by influencing other aspects of virus particle structure. Attempts were made to observe the morphology of the heat-destabilized form using TEM. Particles with discernibly different structures were not observed; however, as Fig. 6 shows, these may simply correspond to the unaltered particles present in 50°C- and 70°C-treated preparations. It is noted that thermal particle stability in vitro was assessed at temperatures not typical for normal virus replication, and therefore, the significance of these results for virion assembly or stability under natural conditions is not known.
Interestingly, WT CNV virions heated to 70°C result in a slower-migrating form of the particles that are not RNase sensitive (Fig. 6A and B, lanes 6 and 3, respectively). WT CNV particles that have been swollen in vitro also migrate more slowly on agarose gels (data not shown), raising the possibility that the heat-labile form of WT CNV is similar in some respects to the expanded form.
It has been found that T=1 particles can form from the modified CPs of several T=3 viruses by the removal of the portion of the capsid that enables the quasi-equivalent interactions between subunits required for T=3 particle formation (6, 19, 20, 32). Flexibility of the ɛ region of the CNV CP arm is required for quasi-equivalent subunit interactions in T=3 particle formation, whereas such flexibility is not required for T=1 formation. The presence of T=1 particles in arm(−) and ɛ(−) mutant infections was evaluated using a variety of methods, including TEM and sucrose gradient fractionation; however, unequivocal evidence for the presence of T=1 particles was not obtained. T=1 particle formation also was not observed following the deletion of a Flock house virus CP region believed responsible for ordered subunit contacts (5).
It has been estimated that a T=1 capsid can accommodate a maximum of approximately 1 kb of RNA (5). Therefore, it is possible that the inability to form T=1 particles in arm(−) and ɛ(−) infections is due to the lack of small viral RNA species in infected leaves. To test this possibility, plants were coinoculated with the arm(−) or ɛ(−) mutants and either of two CNV defective interfering RNAs (425 and 622 nucleotides) (7). Although both defective interfering RNAs were found to accumulate to high levels in infections with either the arm(−) or the ɛ(−) mutant, T=1 particles were not observed (unpublished observations). Since the ɛ region is highly basic and is likely to play a role in binding RNA (35), it is unlikely that T=1 particles can be formed during infection. It should be noted that many of the studies that have demonstrated the production of T=1 particles by various virus capsids were conducted in vitro, and thus, the conditions leading to the assembly of these particles may not be as stringent as those encountered in vivo.
Our study is unique in that it provides important in vivo information and refines our understanding of the role of the β and ɛ regions of the CP arm in virus particle accumulation and stability. Our data are also consistent with the notion that viral RNA plays an important role in particle assembly and/or stability. In addition, our findings further suggest that the β annulus and/or the β region itself may be important for the stability of the expanded form of the CNV particle during fungal transmission.
Acknowledgments
This work was partially supported by a Canada study grant for female doctorates, the UBC John M. Yorston Memorial Prize, a UBC graduate fellowship (to E.H.), and NSERC Operating Grant 43840-2001.
We also thank Kishore Kakani, Ron Reade, Steve Orban, and Jane Theilmann for technical assistance.
REFERENCES
- 1.Bol, J. F. 1999. Alfalfa mosaic virus and ilarviruses: involvement of coat protein in multiple steps of the replication cycle. J. Gen. Virol. 80:1089-1102. [DOI] [PubMed] [Google Scholar]
- 2.Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39:419-460. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, R. N. 1996. Fungal transmission of plant viruses. Annu. Rev. Phytopathol. 34:87-108. [DOI] [PubMed] [Google Scholar]
- 4.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 72:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson, J. W., and M. J. Rossmann. 1982. Assembly and crystallization of a T = 1 icosahedral particle from trypsinized southern bean mosaic virus coat protein. Virology 116:128-136. [DOI] [PubMed] [Google Scholar]
- 7.Finnen, R. L., and D. Rochon. 1995. Characterization and biological activity of DI RNA dimers formed during cucumber necrosis virus coinfections. Virology 207:282-286. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, C. L., and G. K. Pei. 1997. Modification of a PCR-based site-directed mutagenesis method. BioTechniques 23:570-574. [DOI] [PubMed] [Google Scholar]
- 9.Gray, S. M., and N. Banerjee. 1999. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison, S. C. 1983. Virus structure: high-resolution perspectives. Adv. Virus Res. 28:175-240. [DOI] [PubMed] [Google Scholar]
- 11.Harrison, S. C. 1984. Multiple modes of subunit association in the structures of simple spherical viruses. Trends Biochem. Sci. 9:345-351. [Google Scholar]
- 12.Harrison, S. C., A. J. Olson, C. E. Schutt, F. K. Winkler, and G. Brigogne. 1978. Tomato bushy stunt virus at 2.9 Å resolution. Nature 276:368-373. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, S. C., P. K. Sorger, P. G. Stockley, J. Hogle, R. Altman, and R. K. Strong. 1987. Mechanism of RNA virus assembly and disassembly, p. 379-395. In M. A. Brinton and R. Rueckert (ed.), Positive strand RNA viruses. Alan R. Liss, Inc., New York, N.Y.
- 14.Hogle, J. 2002. Poliovirus cell entry: common structural themes in viral entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kakani, K., J.-Y. Sgro, and D. Rochon. 2001. Identification of specific cucumber necrosis virus coat protein amino acids affecting fungus transmission and zoospore attachment. J. Virol. 75:5576-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakani, K., M. Robbins, and D. Rochon. 2003. Evidence that binding of cucumber necrosis virus to vector zoospores involves recognition of oligosaccharides. J. Virol. 77:3922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakani, K., R. Reade, and D. Rochon. 2004. Evidence that vector transmission of a plant virus requires conformational change in virus particles. J. Mol. Biol. 338:507-517. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Leberman, R., and J. T. Finch. 1970. The structures of turnip crinkle virus and tomato bushy stunt viruses. I. A small protein particle derived from turnip crinkle virus. J. Mol. Biol. 50:209-213. [DOI] [PubMed] [Google Scholar]
- 20.Lokesh, G. L., T. D. S. Gowri, P. S. Satheshkumar, M. R. N. Murthy, and H. S. Savithri. 2002. A molecular switch in the capsid protein controls the particle polymorphism in an icosahedral virus. Virology 292:211-223. [DOI] [PubMed] [Google Scholar]
- 21.Lovisolo, O., R. Hull, and O. Rosler. 2003. Coevolution of viruses with hosts and vectors and possible paleontology. Adv. Virus Res. 62:325-379. [DOI] [PubMed] [Google Scholar]
- 22.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbins, M. A., R. D. Reade, and D. M. Rochon. 1997. A cucumber necrosis virus variant deficient in fungal transmissibility contains an altered coat protein shell domain. Virology 234:138-146. [DOI] [PubMed] [Google Scholar]
- 24.Robinson, I. K., and S. C. Harrison. 1982. Structure of the expanded state of tomato bushy stunt virus. Nature 297:563-568. [Google Scholar]
- 25.Rochon, D., K. Kakani, M. Robbins, and R. Reade. 2004. Molecular aspects of plant virus transmission by Olpidium and Plasmodiophorid vectors. Annu. Rev. Phytopathol. 42:211-241. [DOI] [PubMed] [Google Scholar]
- 26.Rochon, D. M., and J. C. Johnston. 1991. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology 181:656-665. [DOI] [PubMed] [Google Scholar]
- 27.Rochon, D. M., and J. H. Tremaine. 1988. Cucumber necrosis virus is a member of the Tombusvirus group. J. Gen. Virol. 69:395-400. [Google Scholar]
- 28.Rochon, D. M., and J. H. Tremaine. 1989. Complete nucleotide sequence of the cucumber necrosis virus genome. Virology 71:251-259. [DOI] [PubMed] [Google Scholar]
- 29.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of the Tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 30.Satheshkumar, P. S., G. L. Lokesh, M. R. N. Murthy, and H. S. Savithri. 2005. The role of arginine-rich motif and β-annulus in the assembly and stability of Sesbania mosaic virus capsids. J. Mol. Biol. 353:447-458. [DOI] [PubMed] [Google Scholar]
- 31.Sit, T. L., J. C. Johnston, M. G. ter Borg, E. Frison, M. A. McLean, and D. Rochon. 1995. Mutational analysis of the cucumber necrosis virus coat protein gene. Virology 206:38-48. [DOI] [PubMed] [Google Scholar]
- 32.Sorger, P. K., P. G. Stockley, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. II. Mechanism of reassembly in vitro. J. Mol. Biol. 191:639-658. [DOI] [PubMed] [Google Scholar]
- 33.Speir, J. A., S. Munshi, G. Wang, T. S. Baker, and J. E. Johnson. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3:63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, C. L., V. Leh, C. Lederer, and A. J. Maule. 2003. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306:33-41. [DOI] [PubMed] [Google Scholar]
- 35.Timmins, P. A., D. Wild, and J. Witz. 1994. The three-dimensional distribution of RNA and protein in the interior of tomato bushy stunt virus: a neutron low-resolution single-crystal diffraction study. Structure 2:1191-1201. [DOI] [PubMed] [Google Scholar]
- 36.Wei, N., L. A. Heaton, and T. J. Morris. 1990. Structure and assembly of Turnip crinkle virus. VI. Identification of coat protein binding sites on the RNA. J. Mol. Biol. 214:85-95. [DOI] [PubMed] [Google Scholar]
- 37.Willits, D., X. Zhao, N. Olson, T. S. Baker, A. Zlotnick, J. E. Johnson, T. Douglas, and M. J. Young. 2003. Effects of the cowpea chlorotic mottle bromovirus β-hexamer structure on virion assembly. Virology 306:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]