Abstract
In an attempt to experimentally define the roles of viral proteins encoded by the B19 genome in the viral life cycle, we utilized the B19 infectious clone constructed in our previous study to create two groups of B19 mutant genomes: (i) null mutants, in which either a translational initiation codon for each of these viral genes was substituted by a translational termination codon or a termination codon was inserted into the open reading frame by a frameshift; and (ii) a deletion mutant, in which half of the hairpin sequence was deleted at both the 5′ and the 3′ termini. The impact of these mutations on viral infectivity, DNA replication, capsid protein production, and distribution was systematically examined. Null mutants of the NS and VP1 proteins or deletion of the terminal hairpin sequence completely abolished the viral infectivity, whereas blocking expression of the 7.5-kDa protein or the putative protein X had no effect on infectivity in vitro. Blocking expression of the proline-rich 11-kDa protein significantly reduced B19 viral infectivity, and protein studies suggested that the expression of the 11-kDa protein was critical for VP2 capsid production and trafficking in infected cells. These findings suggest a previously unrecognized role for the 11-kDa protein, and together the results enhance our understanding of the key features of the B19 viral genome and proteins.
Parvovirus B19 is the only member of the Parvoviridae confirmed to cause disease in humans and is the type member of the Erythrovirus genus. B19 is highly erythrotropic, with infection of erythroid progenitor cells leading to cytotoxicity and interruption of erythrocyte production (27). The physiological conditions of the host and the extent of the immune antiviral response then contribute to the evolution and clinical manifestation of the infection (39). Infection causes fifth disease in children (1, 2), polyarthropathy syndromes in adults (23, 26), transient aplastic crisis in patients with underlying chronic hemolytic anemia (31, 35), and chronic anemia due to persistent infection in immunocompromised patients (18, 19). Infection during pregnancy can lead to hydrops fetalis with possible fetal loss (16) and/or congenital infection (6).
In common with other parvoviruses, B19 has a small (22 nm), nonenveloped, icosahedral capsid, encapsidating a single-stranded DNA genome of 5,596 nucleotides (nt). The ends of the genome are long inverted terminal repeats (ITRs) of 383 nt, of which the distal 365 nt form an imperfect palindrome (9). Transcription of the B19 viral genome is controlled by the single promoter (p6) located at map unit 6, which regulates the synthesis of all nine viral transcripts (4, 29). The single nonspliced transcript encodes the nonstructural protein (NS) and, by a combination of different splicing events, the other eight transcripts encode the two capsid proteins (VP1 and VP2) and two smaller proteins of unknown function (7, 29, 38). In addition, a short open reading frame (ORF) putatively encoding protein X was found in the VP1 region of the B19 genome. However, the specific roles of these viral proteins in B19 infectivity have not been experimentally defined due to difficulties in in vitro culture of the virus and the lack of an infectious clone. Current knowledge regarding the functions of B19 viral proteins is mainly based on postulation from studies of other parvoviruses. The B19 NS protein is a multifunctional protein: besides transregulation of the p6 promoter (10, 32), sequence analysis has shown that NS contains the motifs for nucleoside triphosphate (NTP) binding and hydrolysis (25) associated with helicase activity, suggesting a role of NS in B19 DNA replication. Accumulating evidence also suggests that the NTP-binding motifs of NS are involved in the induction of apoptosis in erythroid lineage cells during B19 infection (24). The major capsid protein, VP2, which comprises 95% of the capsid, is a 58-kDa protein (30). Earlier studies have shown that VP2 expressed in insect cells self-assembles into virus-like particles (14) and VP2 binds directly to blood group P antigen, the cellular receptor of B19 virus (5). The minor capsid protein, VP1, has the same amino acid sequence as VP2 plus an additional 227 amino acids at the amino terminus, the VP1-unique region (VP1u) (30). Previous studies have shown that the main neutralizing epitopes of B19 are in VP1u (34), which is located on the outside of the capsid (15, 33). Recently, a conserved phospholipase A2 (sPLA2) motif was identified in the VP1u of members of the Parvoviridae, including parvovirus B19 (20, 40). The two small proteins, 7.5 and 11 kDa, are encoded by the small abundant mRNA of B19 (21, 37, 38) and are unique among the parvoviruses characterized to date. The 11-kDa protein contains several proline-rich motifs that are conserved to Src homology 3 (SH3) bind domain of eukaryotic proteins (12), but it is not known what functions this or the 7.5-kDa protein play in B19 replication and/or pathogenesis.
Previously, we reported the construction of an infectious clone of parvovirus B19 (43). Transfection of the clone into permissive cells led to the production of viral transcripts, DNA replication, and the production of infectious virus. In the present study, we constructed B19 mutants of the infectious clone to systematically knock out the expression of all of the B19 viral proteins. The effects of these mutations on infectious virus production, viral DNA replication, RNA processing, and protein production were examined.
MATERIALS AND METHODS
Cell lines, virus, and antiserum.
UT7/Epo-S1 cells, a subclone of UT7/Epo previously reported to have an increased sensitivity for parvovirus B19 (17, 36), were kindly provided by Kazuo Sugamura (Tohoku University Graduate School of Medicine [Japan]). The cells were maintained in Iscove modified Dulbecco medium containing 10% fetal calf serum and 2 U of recombinant human erythropoietin (Epo; AMGEN, Thousand Oaks, CA)/ml and grown at 37°C with 5% CO2.
Parvovirus B19 J35 isolate (GenBank AY386330) was obtained from the serum of a child with sickle cell anemia undergoing aplastic crisis and sent to the National Institutes of Health (Bethesda, MD) for diagnostic purposes. The serum was found by dot blot assay (28) to contain approximately 1012 genome copies of B19 virus per ml.
Polyclonal rabbit antisera against the B19 NS and 11-kDa proteins were generated by immunization of a rabbit with keyhole limpet hemocyanin-conjugated synthetic oligopeptides PepNS (KAILGGQPTRVDQKMRGSVC) and Pep11K (PIRQPNTKDIDNVEFKYLTC), respectively (Alpha Diagnostic, San Antonio, TX). For the detection of the enhanced green fluorescent protein (EGFP), polyclonal rabbit antisera (BD Biosciences/Clontech, Palo Alto, CA) to full-length EGFP were used.
Two mouse monoclonal antibodies (MAb) against VP2 capsid proteins were used: MAb 521-5D specific to a conformational epitope (a gift from Larry Anderson, Centers for Disease Control and Prevention, Atlanta, GA) and MAb 8293 (Chemicon, Temecula, CA) specific to a linear epitope.
Generation of B19 mutant genomes based on the infectious clone.
Plasmid pB19-4244, containing the full-length B19 sequence (isolate J35; GenBank AY386330), and plasmid pB19-N8, containing the entire coding region of B19 genome but lacking the distal 204 nt in both ITRs, have previously been described (43).
Null mutations of the key B19 proteins were produced by site-specific mutagenesis of pB19-4244. Specifically, the expression of VP1, VP2, 7.5-kDa, 11-kDa, or X protein was knocked out by either substituting the start codon (ATG) with a stop codon (TAA or TAG) or inserting a termination codon into the ORF (Table 1).
TABLE 1.
Mutagenesis in the B19 genomea
| Plasmid | Mutation target(s) | Change in nt sequence | Intended change(s) in expressed proteins | Coincidental changes in other expressed proteins |
|---|---|---|---|---|
| pB19-N8 | ITRs | Deletion of nt 204 in the ITR at both 5′ (1-204) and 3′ (5392-5596) ends | None | None |
| pB19-M20/NS(−) | NS | AfIII digestion and fill in resulted in the addition of AATT after nt 757 | Frameshift mutation resulted in a TAA codon; truncated 54-aa NS | None |
| pB19-M20/VP1(−) | VP1 | G2633T | GAA to TAA; no VP1 expression | None |
| pB19-M20/VP2/(−) | VP2 | A3305T and T3306A | ATG to TAG; no VP2 expression | Truncated VP1 protein |
| (VP1u expressed) | ||||
| pB19-M20/7.5(−) | 7.5-kDa protein | A2084T and T2085A | ATG to TAG; no 7.5-kDa protein expression | N490I in NS |
| pB19-M20/X(−) | ORF X | A2874 T and T2875A | ATG to TAG; no X protein expression | H84L in VP1 |
| pB19-M20/11(−) | 11-kDa protein | A4917T and T4918A | ATG to TAG; no 11-kDa protein expression | Y765L in VP1 or Y538L in VP2 |
All nucleotide positions are based on the J35 genome (GenBank AY386330). aa, amino acids.
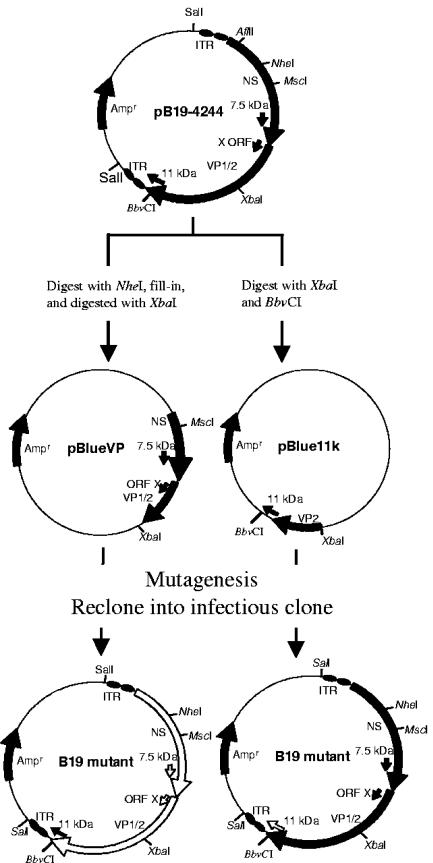
Due to the instability of the ITRs, it was not possible to perform direct mutagenesis on plasmids containing the intact ITR sequences. Therefore, the B19 DNA fragments of interest were first subcloned into a plasmid vector (Fig. 1) and, after successful mutagenesis, were then reinserted into pB19-M20. In preparation of the null mutants of VP1, VP2, 7.5-kDa, or X protein, the full-length plasmid pB19-M20 was cut with NheI and the 5′ overhang filled in using T4 polymerase. The linearized plasmid was redigested with XbaI and the B19 fragment (from nt 1246 to 3425 in the genome of J35 isolate) and ligated into an XbaI-Ecl36II-digested pBluescript II KS(+) cloning vector (Stratagene, La Jolla, CA). The resulting intermediate construct was named pBlueVP (Fig. 1). After site-specific mutagenesis (Table 1), the plasmid with the B19 fragments containing the desired mutations was double digested with MscI and XbaI, and the B19 fragments were ligated into the MscI-XbaI-digested pB19-M20 plasmid.
FIG. 1.
Schematic diagram of the construction of the B19 mutants based on infectious clone pB19-4244. Plasmid pB19-4244 contains 5,596 nt of the full-length B19 genome in pProEx HTb vector. NheI-XbaI or XbaI-BbvCI fragment of pB19-4244 were subcloned into pBluescript II KS(+) cloning vector (Stratagene). After mutagenesis, the DNA fragments containing mutations were recloned into pB91-M20. The solid arrows indicate the wild-type genes in the B19 genome, and the open arrows indicate the mutants.
The ORF encoding the 11-kDa protein is located at the 3′ end of the B19 genome and overlaps with capsid genes and their 3′ untranslated region (3′UTR). In order to construct an 11-kDa protein null mutant, the pB19-M20 was digested with XbaI and BbvCI, and the B19 fragment (from nt 3425 to 4986 in the genome of the J35 isolate) was ligated into an XbaI-BbvCI-digested pBluescript II KS(+) cloning vector (Stratagene). The resulting intermediate construct was named pBlueVP (Fig. 1). After mutagenesis (Table 1), the plasmid was digested with XbaI and BbvCI, and the B19 fragment was ligated into the XbaI-BbvCI-digested pB19-M20 plasmid.
An NS-protein null mutant was constructed by digestion of full-length plasmid pB19-M20 with AfIII (nt 567 in B19 J35 genome), and the 5′ overhang was filled in by using T4 polymerase. The linearized plasmid was self-ligated to generate a frameshift, which resulted in a translation termination codon (TAA) at nt 778 of the B19 genome (Table 1).
Transfection of UT7/Epo-S1 cells.
Transfection and infection of the UT7/Epo-S1 cells were performed as previously The full-length B19 genome was released from the plasmid by restriction enzyme digestion with SalI, the DNA fragments were separated by agarose gel electrophoresis, and the 5.6-kb B19 genome was excised and purified by using QIAEX II Gel Extract Kit (QIAGEN, Santa Clarita, CA). The cells (2 × 106) were transfected with 5 μg of purified B19 DNA by using the AMAXA Cell Line Nucleofector Kit R (AMAXA Biosystems, Inc., Nattermannallee, Germany). The cells were harvested at various times posttransfection and used for infection, DNA, RNA, and immunofluorescence (IF) studies.
Infection of UT7/Epo-S1 cells.
Cells were harvested at 72 h posttransfection and washed in fresh culture medium, and cell lysate was prepared by three cycles of freeze-thawing. After centrifugation at 10,000 × g for 10 min, the clarified supernatant was treated with RNase at a final concentration of 1 U/μl (Roche, Indianapolis, IN), and then 10 μl of treated supernatant was mixed with an equal volume UT7/Epo-S1 cells (2 × 104) in Iscove modified Dulbecco medium for 2 h at 4°C to allow a maximum virus-cell interaction. The cells were diluted to 2 × 105 cells/ml in the culture medium, followed by incubation at 37°C in 5% CO2. Cells were harvested at 3 days postinfection and tested for evidence of infection by detection of spliced viral capsid transcripts and capsid protein. The permissivity of UT7/Epo-S1 cells was confirmed in each experiment by infecting the cells with B19 virus (J35) as described elsewhere (43).
RT-PCR for B19 transcripts.
Total RNA was extracted from either transfected or infected UT7/Epo-S1 cells (2 × 105) using RNA STAT60 (Tel-Test, Inc., Friendswood, TX). Residual DNA was removed by DNase I treatment (1 U per μg of total RNA) for 15 min at room temperature. RNA was converted to cDNA with random hexamers and SuperScript II (Invitrogen/Life Technologies, San Diego, CA), and reverse transcription-PCR (RT-PCR) for the spliced capsid transcripts was performed with primers the B19-9 (from nt 384 to 402 in the J35 genome) and B19-1 (from nt 2139 to 2120 in the J35 genome) as previously described (28).
Southern blot analysis of B19 DNA.
B19 DNA was extracted from transfected or infected UT7/Epo-S1 cells (5 × 105) as previously described (36). Briefly, 5 × 105 cells were treated with 100 mM NaCl, 5 mM EDTA, 10 mM Tris-HCl (pH 7.5), 0.5% sodium dodecyl sulfate, and 200 μg of proteinase K/ml overnight at 37°C, followed by phenol-chloroform extraction. For some experiments, high- and low-molecular weight DNA were separated by the Hirt method (13). Purified DNA (400 ng) was digested with 20 U of BamHI (single cut in B19) or EcoRI (no cut in B19) at 37°C for 4 h, and the fragments were separated by agarose-electrophoresis, transferred to a nylon membrane (Amersham Biosciences, Piscataway, NJ), and hybridized with a 32P-random-primed probe of the B19 coding region as previously described (36). The densities of detected bands were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
RPA.
The cells were harvested at 72 h posttransfection, and total RNA was extracted from the UT7/Epo-S1 cells (5 × 105) using RNA STAT60. A probe (PNSVP) to simultaneously detect transcripts of both NS and capsids was constructed by designing two primer pairs: p949 (5′-GGAGCTTGCCATGCCAAGAA-3′)-p1169 (5′-CATACCGCGGAGTAAAGTGTACTGGTTAAAG-3′) and p3199 (5′-CATACCGCGGAGTAAAAGACTACTTTACTTTA-3′)-p3389 (5′-CCCCCTCACTCCACATGCTT-3′), targeting NS and capsid genes, respectively, with SacII restriction sites (underlined). PCR amplification with these two primer pairs was performed using pB19-M20 as a template (43). After SacII digestion of the amplicons, they were ligated together, and the ligated products were reamplified using p949 and p3389. The PCR products were cloned into a pCRII vector (Invitrogen/Life Technologies). The probe was generated from linearized template by in vitro transcription with SP6 polymerase with MAXIscript In Vitro Transcription kit (Ambion, Austin, TX). RNA hybridizations for RNase protection assays (RPAs) were done by using the RPA III kit according to the manufacturer's instruction. The densities of protected bands were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Immunoblot analysis.
The procedure for immunoblot analysis was essentially the same as described elsewhere (42). Briefly, the cells were harvested at 72 h posttransfection, and the proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were transferred to nitrocellulose membrane and then blocked by immersion in TBS buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.4]) containing 5% milk and 0.05% Tween 20 at room temperature for 2 h to saturate protein-binding sites. Antigens were detected by incubation of the membrane with MAb 8293 or rabbit polyclonal antibodies (anti-NS, 11-kDa protein, or GFP) (1:1,000 dilution), followed by incubation with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibody (1:10,000 dilutions) (BD Biosciences Clontech, Palo Alto, CA). Bands were visualized with enhanced chemiluminescence by incubating the membrane with SuperSignal chemiluminescent reagent (Pierce, Rockford, IL) and exposing it to X-ray film. The densities of detected bands were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
IF for B19 capsid proteins.
Transfected cells were harvested and cytocentrifuged at 1,500 rpm for 8 min in a cytospin funnel (Shandon). The cells were fixed in mixture of acetone and methanol (1:1) at −20°C for 5 min, washed twice in phosphate-buffered saline, and then incubated with MAb 521-5D, MAb 8293 (1:500 dilution), or a rabbit polyclonal antibody to 11-kDa protein (1:200 dilution) in phosphate-buffered saline with 10% fetal calf serum for 1 h at 37°C. For double IF staining, a lissamine rhodamine-labeled goat anti-mouse immunoglobulin G (IgG) and fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit IgG (BD Biosciences/Clontech) were used as secondary antibodies, The nucleus was counterstained with propidium iodide or dihydrochloride hydrate (DAPI [4′,6′-diamidino-2-phenylindole]; Vector Laboratories, Inc., Burlingame, CA), and the slides were examined by confocal microscopy (LSM 510; Leica).
Generation of GFP-11-kDa expression plasmid and cell lines expressing GFP-11-kDa fusion protein.
The 11-kDa protein gene was PCR amplified using primers p11f (5′-GACTCTCGAGATGCAAAACAACACCA-3′) and p11r (reverse 5′-GACTGGATCCTTTTTCTAAATTTTGAT-3′). XhoI and BamHI restriction sites (underlined) were engineered into the p11f and p11r primers, respectively. The PCR products were digested with XhoI and BamHI and cloned “in frame” with the GFP gene in a pEGFP-N3 expression vector (BD Biosciences/Clontech) to produce plasmid p11EGFP.
Cell lines stably expressing GFP-tagged 11-kDa protein or GFP alone were produced by transfecting linearized p11EGFP or pEGPP-N3 (as a control) into UT7/Epo-S1 cells as described above, and 24 h after transfection the GFP-expressing cells were sorted by flow cytometry and cultured for 9 days in medium containing 600 μg of G418 (Invitrogen/Life Technologies)/ml. The production of GFP-tagged 11-kDa protein was tested by immunoblot analysis with the polyclonal rabbit antiserum specific for 11-kDa protein or GFP as described above. Both cells stably expressing either GFP-tagged 11-kDa protein or GFP alone were transfected with pB19-M20 and pB19-M20/11(−). The production of capsid protein was tested by immunoblot analysis as described above.
RESULTS
Generation of B19 mutant genome.
The purpose of this study was to generate B19 mutant genomes and compare the phenotypes of the different viruses produced. The first mutant, pB19-M20/NS(−), contained a TAA after the 54th amino acid codon of the NS reading frame. This mutation blocked expression of the C-terminal 616 amino acid residuals, including the region encoding the ATPase motif (Table 1). Blocking of full-length NS protein production was confirmed by immunoblot analysis: a 74-kDa band of NS protein was detected in the UT7/Epo-S1 cells transfected with pB19-M20 but was absent in the cells transfected with pB19-M20/NS(−) (data not shown). The second mutant, pB19-M20/VP1(−), contained a TAG after the second amino acid codon of VP1; this was done to avoid mutation of the NS protein expressed from the overlapping ORF of NS. As the ORF encoding VP2 protein is completely overlapped with that of VP1 in the same frame, the third mutant, pB19-M20/VP2(−), in which a TAG was substituted for the ATG of VP2, also resulted in a termination of translation of VP1 at the 227th amino acid codon. Blocking of VP1 and/or VP2 production, respectively, was confirmed by immunoblot analysis with MAb 8293 that recognizes a linear epitope in the VP1/VP2 protein. After the transfection of UT7/Epo-S1 cells with pB19-M20, two proteins of 85 and 58 kDa that corresponded to the molecular masses of VP1 and VP2, respectively, were detected. In contrast, the 85-kDa VP1 protein was absent in the cells transfected with pB19-M20/VP1(−) and both the 85- and the 58-kDa proteins were undetectable in the cells transfected with pB19-M20/VP2(−) (data not shown).
The ORF encoding the 7.5-kDa protein and X protein are completely overlapped by those of NS and VP1, respectively. Substitution of the ATG with a TAG in the ORF of the 7.5-kDa and X proteins resulted in point mutations in the NS and VP1, respectively (Table 1). However, due to the unavailability of antibody specific for the 7.5-kDa protein or the X protein, we were unable to confirm that the expression of these proteins had been successfully blocked.
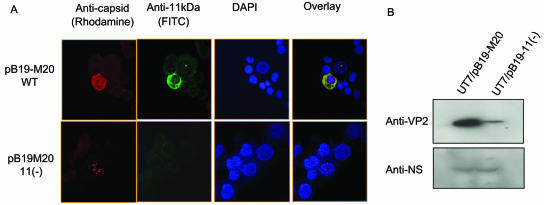
The 5′ end of the ORF encoding 11-kDa protein contains three closely spaced ATG codons, and all of them appeared to be functional (37). Therefore, in order to completely knock out the expression of the 11-kDa protein, we introduced two substitutions at A4917T and T4918A to replace the third ATG with a TAG codon. Since the ORF encoding the 11-kDa protein is partially overlapped with that of VP1 and VP2, these substitutions resulted in point mutations in VP1 (Y765L) and VP2 (Y538L). Blockage of 11-kDa protein expression was confirmed by double IF staining of the UT7/Epo-S1 cells transfected with either pB19-M20 or pB19-M20/11(−) using MAb 521-5D (for capsid protein) and the rabbit anti-11-kDa protein serum. The 11-kDa protein was predominantly detected in the cytoplasm and appeared to be colocalized with the capsid protein in the cells transfected with pB19-M20 but was completely absent in cells transfected with the 11-kDa protein null mutant (see Fig. 6A), confirming that the production of 11-kDa proteins was blocked in pB19-M20/11(−). Production of VP1 and/or VP2 in the 11-kDa protein null mutant was examined by immunoblot analysis with MAb 8293. After the transfection of UT7/Epo-S1 cells with pB19-M20/11(−), two proteins of 85 and 58 kDa that corresponded to the molecular masses of VP1 and VP2, respectively, were detected. (data not shown).
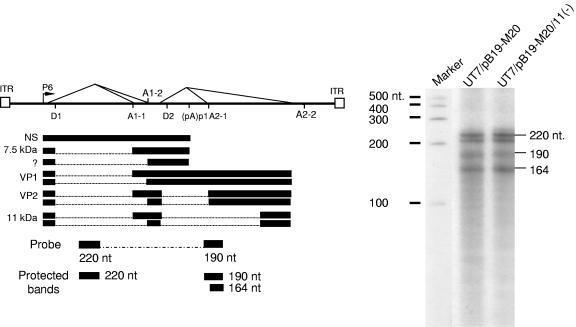
FIG. 6.
Blocking expression of the 11-kDa protein dose not influence relative abundance of major viral transcripts. (A) Schematic diagram of the B19 genome and probe used. The promoter (p6), the intron donors (D1 and D2), and acceptors (A1-1, A1-2, and A2) are shown. The locations of the PNSVP probe and the expected protected bands are depicted. (B) RPA analysis of B19 transcripts. Twenty micrograms of total RNA isolated at 72 h posttransfection of UT7/Epo-S1 cells was hybridized with probe PNSVP, and the sizes of the protected fragments are indicated by arrows on the right. The molecular marker is a 32P-labeled RNA ladder (Ambion) with the respective sizes indicated in nucleotides on the left.
Null mutations of NS, VP1, or 11-kDa protein or deletions in ITR block production of infectious virus.
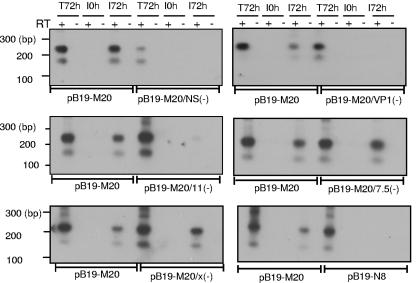
In order to determine which gene products were essential for the production of infectious virus, UT7/Epo-S1 cells were transfected with the different B19 mutant plasmids, and clarified supernatants prepared from the cell lysates of transfected cells were used to infect fresh UT7/Epo-S1 cells (Table 2). The detection of spliced capsid transcripts by RT-PCR and capsid protein by IF were used as markers for successful infectious viral production. After the initial transfection, spliced transcripts were detected in all samples (T72h, Fig. 2). No transcripts were detected immediately after inoculation of UT7/Epo-S1 cells with the RNase-treated clarified supernatant, confirming that there was no carryover of the RNA from the transfected cells. At 72 h postinoculation, spliced capsid transcripts were detected in the samples derived from the cells transfected with pB19-M20, pB19-M20/7.5(−), and pB19-M20/X(−) but not from the cells transfected with pB19-M20/NS(−), pB19-M20/VP1(−), and pB19-N8 (Fig. 2). This indicated that the NS and VP1 proteins, as well as intact ITR sequences, were required for the production of infectious virus. In the samples derived from cells transfected with pB19-M20/11(−), a very weak band was detected at 72 h postinoculation (Fig. 2), suggesting that the 11-kDa protein was required for efficient production of infectious virus.
TABLE 2.
Summary of phenotype changes in B19 mutantsa
| Protein mutation/ plasmid | Infectious virus production | B19 DNA replication | Capsid protein production and distribution |
|---|---|---|---|
| ITR(−)/ | None | None | No change |
| pB19-M20/N8 | |||
| NS(−)/ | Marked decrease | ||
| pB19-M20/NS(−) | None | None | Nuclear only |
| VP1(−)/ | None | No significant | Nuclear only |
| pB19-M20/VP1(−) | change | ||
| VP2(−)/ pB19-M20/VP2(−) | Not determined | No significant change | Not determined |
| 11-kDa protein(−)/ pB19-M20/11(−) | Significant decrease | No significant change | Decrease Nuclear only |
| 7.5-kDa protein(−)/ pB19-M20/7.5(−) | No significant change | No significant change | No change |
| ORF X(−)/ pB19-M20/x(−) | No significant change | No significant change | No change |
All results are relative to infection with the wild-type infectious clone pB19-M20.
FIG. 2.
Determining the B19 genes critical for infectious virus production in permissive cells by RT-PCR analysis of the B19 transcripts. At 72 h posttransfection, the cell lysate was prepared by freezing and thawing, and the clarified supernatant from wild-type infectious clone (pB19-M20) or B19 mutant transfected cells was used to infect fresh UT7/Epo-S1 cells. Total RNA was extracted from the cells at 72 h posttransfection (T72h) and at 0 and 72 h postinfection (I0h and I72h, respectively). RT-PCR was performed with the primer pair B19-1 and B19-9. The products were analyzed by agarose gel electrophoresis. “+” and “−” indicate the presence or absence, respectively, of reverse transcriptase in the reaction. The numbers on the left indicate molecular sizes in base pairs.
Null mutation of NS blocks viral DNA replication.
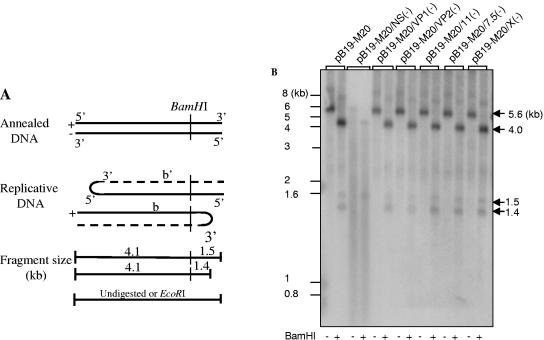
During the replication of parvovirus B19, the viral single-stranded DNA is converted to a double-stranded replicative form, which has either an “extended” or a “turnaround” form at the terminal regions. These intermediate structures provide evidence for viral DNA replication and can be distinguished by BamHI restriction enzyme digestion (Fig. 3A) (8). Detection of this replicative form was therefore used to determine whether the block in production of infectious virus was due to the inability of the viral DNA to replicate. After transfection, the distinct doublets of 1.5 and 1.4 kb characteristic of B19 replication were detected in UT7/Epo-S1 cells transfected with pB19-M20, pB19-M20/7.5(−), and pB19-M20/X(−), all of which had produced infectious virus (Fig. 3B). This doublet was also detected in the cells transfected with pB19-M20/VP1(−), pB19-M20/VP2(−), and pB19-M20/11(−), which were unable to produce infectious virus. However, in the sample transfected with NS null mutant [pB19-M20/NS(−)], the 1.4-kb fragment representing the turnaround replication form of viral DNA was not detected (Fig. 3B). In addition, although the same amount of B19 DNA was used in all transfections, the density of the 5.6-kb band corresponding to the full-length B19 genome detected in the sample of NS null mutant was 60-fold lower than that of pB19-M20 or the other mutants. Together, these results confirm the critical role of B19 NS protein in viral DNA replication. Since there was no block in viral DNA replication in the B19 virus with VP1, VP2, or 11-kDa protein null mutants, this suggested that the inability to produce infectious virus from the cell transfected with these mutants was due to deficiencies in other stages of the viral life cycle.
FIG. 3.
Determination of B19 gene(s) essential for viral DNA replication. (A) Schematic representation of the replication of B19 viral genome. (B) Southern blot analysis of B19 genome replication. DNA was purified from UT7/Epo-S1 cells transfected with purified SalI-digested fragments of plasmids containing either wild-type B19 genome or mutants. Purified DNA was digested with BamHI or EcoRI, and the fragments were separated by agarose-electrophoresis, transferred to a nylon membrane, and hybridized with a 32P-random-primed probe of the complete B19 coding region. The numbers on the left indicate molecular sizes in kilobases.
Null mutations of NS, VP1, and 11-kDa protein alter expression and distribution of viral capsid protein.
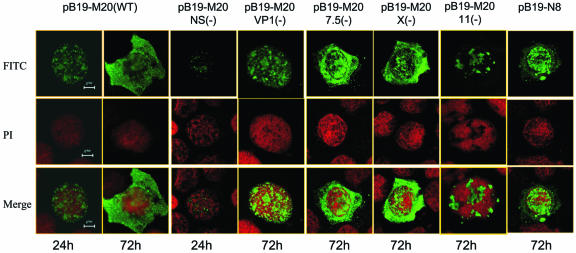
UT7/Epo-S1cells transfected with pB19-M20 were examined by IF staining with MAb 521-5D, which recognizes a conformational epitope in the VP2. Capsid proteins were detected first in the cell nucleus (24 h posttransfection), where they appeared either to be evenly distributed or formed small clusters (Fig. 4). At later stages of infection (72 h posttransfection) capsid proteins were found mainly in the cytoplasm. A similar time course and localization was observed in the cells transfected with pB19-M20/7.5(−) and pB19-M20/X(−). In the cells transfected with pB19-M20/NS(−), only weak expression of capsid proteins was detected at 24 h posttransfection and then became completely undetectable by 72 h, suggesting a role of NS proteins in the regulation of B19 capsid gene expression. In cells transfected with pB19-M20/VP1(−), the capsid proteins accumulated in the nucleus and were not transported into the cytoplasm. In the cells transfected with pB19-N8, the majority of the capsid protein appeared to accumulate in the nucleus. In cells transfected with the 11-kDa protein null mutant [pB19-M20/11(−)], the production of viral capsid protein was significantly decreased. In addition, the viral capsid proteins formed rough clusters in the nucleus and were not transported into the cytoplasm at later stages of infection (Fig. 4 and Fig. 5A). When the cells transfected with wild-type B19 infectious clone or these mutants were examined by IF staining with MAb 8293, which recognizes a linear epitope in VP2, the pattern of IF staining for capsid proteins was similar to that seen with MAb 521-5D (data not shown). Taken together, our results suggested that expression of the VP1 specific region and the 11-kDa protein, as well as intact ITRs, appeared to be crucial for the process of nuclear exit of viral capsids.
FIG. 4.
Changes in viral capsid protein expression and distribution in cells transfected with B19 mutants. At 24 or 72 h posttransfection, the viral capsid proteins were detected in cells transfected with wild-type infectious clone (pB19-M20) or with B19 mutants by IF with MAb 521-5D. An FITC-labeled goat anti-rabbit IgG (BD Biosciences) was used as a secondary antibody, the nucleus was counterstained with propidium iodide, and the slides were examined by confocal microscopy.
FIG. 5.
Changes in viral capsid protein production and distribution in UT7/Epo-S1 cells transfected with 11-kDa protein null mutant. (A) IF staining. UT7/Epo-S1 cells were transfected with pB19-M20 or pB19-M20/11(−) plasmids, and at 72 h posttransfection capsid proteins were detected by IF with MAb 521-5D/anti-mouse rhodamine conjugate and 11-kDa protein/anti-rabbit-FITC conjugate antibodies. The nuclei were counterstained with DAPI, and the slides were examined by confocal microscopy. (B) Immunoblot analysis. The cells were transfected with pB19-M20 or pB19-M20/11(−), collected 72 h posttransfection, and tested by immunoblot analysis with a MAb to viral capsid protein (MAb 8293) or rabbit antibody to the NS protein.
Null mutation of the 11-kDa protein alters the production of viral capsid protein but has no effect on viral RNA transcription and splicing.
The production of viral proteins in cells transfected with wild-type B19 genome or the 11-kDa protein null mutant was compared by immunoblot analysis with antibodies specific to NS or capsid protein. Compared to pB19-M20, the band density of the capsid protein was ∼30-fold lower in the sample derived from the cells transfected with pB19-M20/11(−). No difference was found in NS protein expression between these two samples (Fig. 5B). These results suggested that nonexpression of the 11-kDa protein had a significant effect on the production of viral capsid protein. Therefore, to determine whether the reduction of capsid protein production was due to an effect of the 11-kDa protein on transcriptional processing, an RPA was performed using a hybrid probe (PNSVP), which simultaneously detected the NS and VP transcripts (Fig. 6A). The probe PNSVP protected bands of 220, 190, and 164 nt, which represented unspliced mRNA encoding NS and spliced mRNA encoding VP1 and VP2, respectively (Fig. 6B). The ratios of unspliced NS to spliced VP1 and of unspliced NS to spliced VP2 were 1:2.5 and 1:3, respectively, in the cells transfected with pB19-M20/11(−) (Fig. 6B). There was no difference in the relative abundance of unspliced NS versus spliced VP between pB19-M20 and with pB19-M20/11(−), suggesting that the expression of the 11-kDa protein had no role in viral RNA processing.
Restoration of B19 capsid production by in trans complementation of the 11-kDa protein.
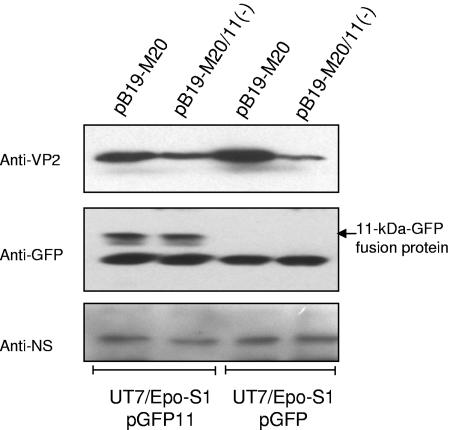
Cell lines which stably expressed either a GFP-tagged form of the 11-kDa protein or GFP protein alone were generated. The expression of GFP-tagged 11-kDa- protein in the cells was tested by immunoblot analysis with the anti-GFP antibody. A protein of 37 kDa corresponding to the GFP-tagged 11-kDa protein was detected in the cells with plasmid pGFP11 but absent in the cells with pGFP (vector control) (Fig. 7), indicating a stable expression of GFP-tagged 11-kDa protein in these cells. The 37-kDa protein was also recognized by the anti-11-kDa protein antibody (data not shown), confirming expression of desired fusion protein in these cells. After transfection of these cell lines with pB19-M20 or pB19-M20/11(−), the relative amounts of capsid and NS protein were analyzed by immunoblot analysis. In the cells expressing GFP only (vector control), the difference of VP2 between the cells transfected with pB19-M20 and pB19-M20/11(−) remained similar to that seen in native UT7/Epo-S1 cells (∼30-fold). However, after transfection of pB19-M20/11(−) in the GFP-tagged 11-kDa protein-expressing cells, the expression of VP2 was substantially increased and the band density of VP2 was only approximately twofold lower than that in pB19-M20-transfected cells (Fig. 7), suggesting that expression of the GFP-tagged 11-kDa protein had complemented VP2 protein production and confirming the specific role of 11-kDa protein in production of viral capsid protein.
FIG. 7.
Expression of 11-kDa protein complements production of viral capsid protein in trans. Cell lines stably expressing GFP-tagged 11-kDa protein or GFP alone were transfected with pB19-M20 or pB19-M20/11(−), and samples were collected 72 h posttransfection and tested by immunoblot analysis with an MAb to the viral capsid protein (MAb 8293), a rabbit antibody to the viral NS protein, or a rabbit antibody to the GFP. The numbers on the left indicate the molecular masses in kilodaltons based on the broad-range prestained standards (Bio-Rad). Bands were visualized by using SuperSignal chemiluminescent reagent (Pierce) and exposure to X-ray film.
DISCUSSION
Due to the difficulty in culturing B19 in vitro, little has been experimentally determined of the role of the known and putative B19 viral proteins in infectivity. The production of an infectious clone has now permitted a systematic study of the function of the different B19 viral proteins, and we describe here our construction of B19 mutant genomes and the application of these B19 mutants to investigate the roles of viral proteins in the B19 life cycle and set the framework for a better understanding of the functions of viral proteins encoded by B19 (Table 2).
Based on studies of AAV and the minute virus of mice (MVM) (3), it has been postulated that B19 DNA replication is initiated from short double-stranded regions provided by self-annealed, terminal hairpin structure. DNA synthesis proceeds from these palindromes to produce high-molecular-weight intermediates through a rolling hairpin model. This mechanism suggests that NS may play important roles in two critical steps of this process. First, the NS protein is involved in the assembly of a cellular replication complex on the origin for replication. Second, the binding of the NS protein to the origin will introduce a site- and strand-specific nick at the terminal resolution site, providing a new 3′ end used to replicate back through the ITR. Although the B19 NS sequence does contain the conserved helicase motif and putative NTP binding site (25), due to the lack of a full-length infectious clone and difficulties in producing recombinant B19 NS protein this model had not been experimentally tested in B19 infection. In the present study we showed that no infectious viruses were produced from the cells transfected with B19 mutants carrying the NS knockout mutation or ITR deletion. Furthermore, Southern blot analysis demonstrated that no replication form or newly synthesized viral DNA was detected in the cells transfected with NS null mutant, confirming the critical role of the NS protein in B19 viral replication.
Although it did not influence viral DNA replication, “knocking out” the expression of VP1 completely abolished the infectivity of B19 virus. The minor capsid protein, VP1, has the same amino acid sequence as VP2 plus an additional 227 amino acids at the amino terminus, the VP1-unique region (VP1u) (7). Previous studies have shown that the main neutralizing epitopes of B19 are in VP1u (34), which is located on the outside of the capsid virion (15, 33). Recently, a conserved secreted phospholipase A2 (sPLA2) motif was identified in the VP1u of members of the Parvoviridae, including B19 (40). In porcine parvovirus (PPV), the sPLA2 motif appeared to be critical for efficient transfer of the viral genome from late endosomes or lysosomes to the nucleus to initiate replication (11, 40), and the motif was required for PPV infectivity (40). Our data also confirmed the importance of the VP1 protein in the B19 life cycle, since the VP1u null mutant completely lost its infectivity. However, B19 mutants containing point mutations in the sPLA2 motif (P133R in the Ca2+ binding loop or D175A in the enzyme catalytic site) are still infectious, albeit the infectivity is reduced (N. Zhi et al., unpublished data), and further mutagenesis studies are required to map out the functional domains in VP1u that are required for B19 infectivity.
The signals and mechanism for the trafficking of nonenveloped icosahedral viruses across the nuclear membrane are not completely understood, particularly whether the mature viral particles actively traffic through the nuclear pore. Studies of MVM infection suggest that, at the initiation of the infection, the amino-terminal domain of VP2 (2Nt) is cleaved during cytoplasmic uptake (22). This serves to externalize the NLS contained in the VP1 amino terminus and contributes to the docking of the incoming viral particles to the nuclear pole complex. For nuclear export, encapsidation of the MVM viral genome triggers the externalization of 2Nt of some VP2 subunits outside the capsid shell, which then drive the active export of mature virus (22). Recently, a nonconventional nuclear import motif has been shown to be necessary for nuclear import of the B19 VP2, but the nuclear export signals in B19 are completely unknown. In the present study, trafficking of B19 viral particles from the nucleus was appeared to be blocked in the VP1 and 11-kDa protein null mutants. Moreover, the infectivity of these B19 mutants was significantly reduced compared to wild-type virus. A similar phenotype change was also observed in the cells transfected with pB19-N8, which lacks half of the ITR sequences at both the 5′ and the 3′ ends. Previous studies have shown that the unique region of VP1 is exposed on the surface of B19 empty virion (15, 33), which may provide a docking site for the interaction between viral particles and host cell exporting machinery.
Our data suggest that intact ITR sequences are necessary for B19 replication and perhaps encapsidation and that encapsidation of the B19 viral genome and exposure of VP1u on the surface of the virion are essential for triggering the export of B19 virus from the nucleus. The role of the 11-kDa protein in this process may be different from that of VP1 and intact ITRs, but the significant reduction of capsid protein in the 11-kDa protein null mutant would directly influence the efficiency of encapsidation of the B19 viral genome.
The B19 genome has two minor ORFs that overlap the 3′ ends of the NS and VP ORFs, and abundant amounts of polyadenylated small mRNAs with these two ORFs are found in infected cells and are translated into the 7.5- and 11-kDa proteins, respectively (21, 37, 38). Analysis of the amino acid sequence indicates that the 11-kDa protein contains three proline-rich regions with the SH3-binding motif often present in signaling molecules within the cell (12). In the present study there was a significantly reduced infectivity of the 11-kDa protein null mutant. Moreover, blocking 11-kDa protein expression not only reduced the amount but also altered the distribution pattern of viral capsid protein in the cells: the capsid proteins formed large clusters in the nucleus and were not exported to the cytoplasm. The mutation introduced to block the 11-kDa protein expression also generated a point mutation in the capsid protein, which might itself be responsible for the observed phenotype. However, although we cannot completely exclude this possibility, the ability to significantly complement this defect by coexpression of a GFP-tagged 11-kDa fusion protein suggests that the nonexpression of 11-kDa protein is responsible. In addition, since the RPA showed no difference in transcription levels between the wild-type infectious clone and the 11-kDa null mutant, the changes in viral capsid proteins expression induced by the 11-kDa protein null mutation likely occurred due to changes at the posttranscriptional level. Other studies have suggested a role of specific protein-protein interaction between the SH3-binding motifs of the 11-kDa protein and the host cell factors, such as receptor-binding protein 2 (Grb2) (12) and a variety of cellular kinases (Zhi et al. unpublished), and perhaps a role of the 11-kDa protein in altering the cellular environment to favor viral replication and maturation.
In addition to the 7.5- and 11-kDa protein-encoding ORFs, a short ORF (encoding a putative “X protein”) was observed in the VP1 coding region of B19. Although there is no evidence showing that this small ORF is expressed in B19, it is structurally similar to the SAT protein that was recently characterized in PPV (41). Our results showed no significant differences between the wild type and knockout mutants of either X protein or the 7.5-kDa protein with respect to infectivity and viral DNA replication. However, with a small genome of 5.6 kb and limited coding capacity, it is reasonable to postulate that the 7.5-kDa protein and the X protein, if it is expressed, may play some role in the B19 life cycle and/or pathogenesis.
The specific tropism of B19 for erythroid progenitor cells suggests that erythroid-specific gene transduction may be feasible if recombinant B19 vectors could be developed, with packaging of “genes of interest” within a B19 capsid. However, we and other researchers have been encountering difficulties expressing B19 viral capsid protein in either permissive or nonpermissive mammalian cells. The results presented here suggest that this may in part be due to the lack of the B19 11-kDa protein in the system to enable efficient production and normal distribution of capsid proteins. In addition, in order to produce recombinant B19 viruses which are infectious, both VP1 and VP2 capsid proteins are necessary and would need to be included in the package system. Taken together, we suggest that a successful packaging system for the production of functional recombinant B19 viral particles would need to include full-length ITR(s), VP1, VP2, and 11-kDa proteins with the NS protein.
Acknowledgments
This study was supported by the Intramural Research Program of the NHLBI, NIH.
REFERENCES
- 1.Anderson, M. J., P. G. Higgins, L. R. Davis, J. S. Willman, S. E. Jones, I. M. Kidd, J. R. Pattison, and D. A. Tyrrell. 1985. Experimental parvoviral infection in humans. J. Infect. Dis. 152:257-265. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. J., S. E. Jones, S. P. Fisher-Hoch, E. Lewis, S. M. Hall, C. L. R. Bartlett, B. J. Cohen, P. P. Mortimer, and M. S. Pereira. 1983. Human parvovirus, the cause of erythema infectiosum (fifth disease)? Letter. Lancet i:1378. [DOI] [PubMed] [Google Scholar]
- 3.Berns, K. I. 1990. Parvovirus replication. Microbiol. Rev. 54:316-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blundell, M. C., C. Beard, and C. R. Astell. 1987. In vitro identification of a B19 parvovirus promoter. Virology 157:534-538. [DOI] [PubMed] [Google Scholar]
- 5.Brown, K. E., S. M. Anderson, and N. S. Young. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114-117. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. E., S. W. Green, J. Antunez de Mayolo, J. A. Bellanti, S. D. Smith, T. J. Smith, and N. S. Young. 1994. Congenital anaemia after transplacental B19 parvovirus infection. Lancet 343:895-896. [DOI] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., V. C. McKie, L. J. Anderson, C. R. Astell, and P. Tattersall. 1986. Identification of the major structural and nonstructural proteins encoded by human parvovirus B19 and mapping of their genes by procaryotic expression of isolated genomic fragments. J. Virol. 60:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1984. Characterization and molecular cloning of a human parvovirus genome. Science 226:1161-1165. [DOI] [PubMed] [Google Scholar]
- 9.Deiss, V., J. D. Tratschin, M. Weitz, and G. Siegl. 1990. Cloning of the human parvovirus B19 genome and structural analysis of its palindromic termini. Virology 175:247-254. [DOI] [PubMed] [Google Scholar]
- 10.Doerig, C., B. Hirt, J. P. Antonietti, and P. Beard. 1990. Nonstructural protein of parvoviruses B19 and minute virus of mice controls transcription. J. Virol. 64:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorsch, S., G. Liebisch, B. Kaufmann, P. von Landenberg, J. H. Hoffmann, W. Drobnik, and S. Modrow. 2002. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 76:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, M. M., L. Tamburic, C. Shippam-Brett, D. B. Zagrodney, and C. R. Astell. 2001. The small 11-kDa protein from B19 parvovirus binds growth factor receptor-binding protein 2 in vitro in a Src homology 3 domain/ligand-dependent manner. Virology 291:285-291. [DOI] [PubMed] [Google Scholar]
- 13.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cells. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 14.Kajigaya, S., H. Fujii, A. Field, S. Anderson, S. Rosenfeld, L. J. Anderson, T. Shimada, and N. S. Young. 1991. Self-assembled B19 parvovirus capsids, produced in a baculovirus system, are antigenically and immunogenically similar to native virions. Proc. Natl. Acad. Sci. USA 88:4646-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawase, M., M. Momoeda, N. S. Young, and S. Kajigaya. 1995. Most of the VP1 unique region of B19 parvovirus is on the capsid surface. Virology 211:359-366. [DOI] [PubMed] [Google Scholar]
- 16.Kinney, J. S., L. J. Anderson, J. Farrar, R. A. Strikas, M. L. Kumar, R. M. Kliegman, J. L. Sever, E. S. Hurwitz, and R. K. Sikes. 1988. Risk of adverse outcomes of pregnancy after human parvovirus B19 infection. J. Infect. Dis. 157:663-667. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu, N., M. Yamamoto, H. Fujita, A. Miwa, K. Hatake, T. Endo, H. Okano, T. Katsube, Y. Fukumaki, S. Sassa, and Y. Miura. 1993. Establishment and characterization of an erythropoietin-dependent subline, UT-7/Epo, derived from human leukemia cell line, UT-7. Blood 82:456-464. [PubMed] [Google Scholar]
- 18.Kurtzman, G., N. Frickhofen, J. Kimball, D. W. Jenkins, A. W. Nienhuis, and N. S. Young. 1989. Pure red-cell aplasia of 10 years' duration due to persistent parvovirus B19 infection and its cure with immunoglobulin therapy. N. Engl. J. Med. 321:519-523. [DOI] [PubMed] [Google Scholar]
- 19.Kurtzman, G. J., B. Cohen, P. Meyers, A. Amunullah, and N. S. Young. 1988. Persistent B19 parvovirus infection as a cause of severe chronic anaemia in children with acute lymphocytic leukaemia. Lancet ii:1159-1162. [DOI] [PubMed] [Google Scholar]
- 20.Lu, J., N. Zhi, S. Wong, and K. E. Brown. 2006. Activation of synoviocytes by the secreted phospholipase A2 motif in the VP1-unique region of parvovirus B19 minor capsid protein. J. Infect. Dis. 193:582-590. [DOI] [PubMed] [Google Scholar]
- 21.Luo, W., and C. R. Astell. 1993. A novel protein encoded by small RNAs of parvovirus B19. Virology 195:448-455. [DOI] [PubMed] [Google Scholar]
- 22.Maroto, B., N. Valle, R. Saffrich, and J. M. Almendral. 2004. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J. Virol. 78:10685-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumura, M. 2001. Parvovirus-associated arthritis. Am. J. Med. 111:241. [DOI] [PubMed] [Google Scholar]
- 24.Moffatt, S., N. Yaegashi, K. Tada, N. Tanaka, and K. Sugamura. 1998. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J. Virol. 72:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momoeda, M., S. Wong, M. Kawase, N. S. Young, and S. Kajigaya. 1994. A putative nucleoside triphosphate-binding domain in the nonstructural protein of B19 parvovirus is required for cytotoxicity. J. Virol. 68:8443-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, T. L. 2000. Parvovirus-associated arthritis. Curr. Opin. Rheumatol. 12:289-294. [DOI] [PubMed] [Google Scholar]
- 27.Mortimer, P. P., R. K. Humphries, J. G. Moore, R. H. Purcell, and N. S. Young. 1983. A human parvovirus-like virus inhibits hematopoietic colony formation in vitro. Nature 302:426-429. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, Q. T., S. Wong, E. D. Heegaard, and K. E. Brown. 2002. Identification and characterization of a second novel human erythrovirus variant, A6. Virology 301:374-380. [DOI] [PubMed] [Google Scholar]
- 29.Ozawa, K., J. Ayub, Y. S. Hao, G. Kurtzman, T. Shimada, and N. Young. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 61:2395-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozawa, K., and N. Young. 1987. Characterization of capsid and noncapsid proteins of B19 parvovirus propagated in human erythroid bone marrow cell cultures. J. Virol. 61:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattison, J. R., S. E. Jones, J. Hodgson, L. R. Davis, J. M. White, C. E. Stroud, and L. Murtaza. 1981. Parvovirus infections and hypoplastic crisis in sickle-cell anaemia. Lancet i:664-665. [DOI] [PubMed] [Google Scholar]
- 32.Raab, U., K. Beckenlehner, T. Lowin, H. H. Niller, S. Doyle, and S. Modrow. 2002. NS1 protein of parvovirus B19 interacts directly with DNA sequences of the p6 promoter and with the cellular transcription factors Sp1/Sp3. Virology 293:86-93. [DOI] [PubMed] [Google Scholar]
- 33.Rosenfeld, S. J., K. Yoshimoto, S. Kajigaya, S. Anderson, N. S. Young, A. Field, P. Warrener, G. Bansal, and M. S. Collett. 1992. Unique region of the minor capsid protein of human parvovirus B19 is exposed on the virion surface. J. Clin. Investig. 89:2023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saikawa, T., S. Anderson, M. Momoeda, S. Kajigaya, and N. S. Young. 1993. Neutralizing linear epitopes of B19 parvovirus cluster in the VP1 unique and VP1-VP2 junction regions. J. Virol. 67:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serjeant, G. R., J. M. Topley, K. Mason, B. E. Serjeant, J. R. Pattison, S. E. Jones, and R. Mohamed. 1981. Outbreak of aplastic crisis in sickle cell anaemia associated with parvovirus-like agent. Lancet ii:595-597. [DOI] [PubMed] [Google Scholar]
- 36.Shimomura, S., N. Komatsu, N. Frickhofen, S. Anderson, S. Kajigaya, and N. S. Young. 1992. First continuous propagation of B19 parvovirus in a cell line. Blood 79:18-24. [PubMed] [Google Scholar]
- 37.St Amand, J., and C. R. Astell. 1993. Identification and characterization of a family of 11-kDa proteins encoded by the human parvovirus B19. Virology 192:121-131. [DOI] [PubMed] [Google Scholar]
- 38.St Amand, J., C. Beard, K. Humphries, and C. R. Astell. 1991. Analysis of splice junctions and in vitro and in vivo translation potential of the small, abundant B19 parvovirus RNAs. Virology 183:133-142. [DOI] [PubMed] [Google Scholar]
- 39.Young, N. S., and K. E. Brown. 2004. Parvovirus B19. N. Engl. J. Med. 350:586-597. [DOI] [PubMed] [Google Scholar]
- 40.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
- 41.Zadori, Z., J. Szelei, and P. Tijssen. 2005. SAT: a late NS protein of porcine parvovirus. J. Virol. 79:13129-13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhi, N., Y. Rikihisa, H. Y. Kim, G. P. Wormser, and H. W. Horowitz. 1997. Comparison of major antigenic proteins of six strains of the human granulocytic ehrlichiosis agent by Western immunoblot analysis. J. Clin. Microbiol. 35:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhi, N., Z. Zadori, K. E. Brown, and P. Tijssen. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318:142-152. [DOI] [PubMed] [Google Scholar]